-
Liver cancer is a frequently occurring malignancy with a poor prognosis and a high mortality rate and is the second leading cause of cancer-related deaths[1]. Many chemotherapeutic drugs are extensively used to treat liver cancer, such as doxorubicin and cisplatin. However, these drugs have shown poor treatment effects in clinical trials because of acquired resistance and the high heterogeneity of liver cancer[2]. Therefore, developing novel and effective drugs to treat liver cancer is important. Natural products have gained considerable attention because they share chemopreventive and chemotherapeutic properties, including antitumor effects. Polyphyllin D (PD) is a steroidal saponin extracted from the rhizomes of Paris polyphylla that has a significant inhibitory effect on tumor cells[3, 4].
In most cancers, the progression of the cell cycle is deregulated by abnormal proteins, including cell cycle-related proteins or their regulators. Liver cancer is largely caused by uncontrolled cell growth and a sequence of disorders in normal cell cycle checkpoints; particularly, regulation of the cell cycle is directly deregulated in the advanced stage of liver cancer[5]. Cyclin/CDK complexes, consisting of the cyclin regulatory subunit and the CDK catalytic subunit, control the cell cycle[6]. Cyclins and CDK inhibitors have been extensively used to treat cancers[7]. The cell cycle offers a new therapeutic direction as a drug target for liver cancer. Increasing evidence supports the close relationship between disordered cholesterol metabolism and the cancer cell cycle[8]. In this study, PD had a strong anticancer effect on liver cancer. The results of a series of experiments showed that PD induced G2/M phase arrest and suppressed the growth of liver cells via dysfunction of cholesterol biosynthesis.
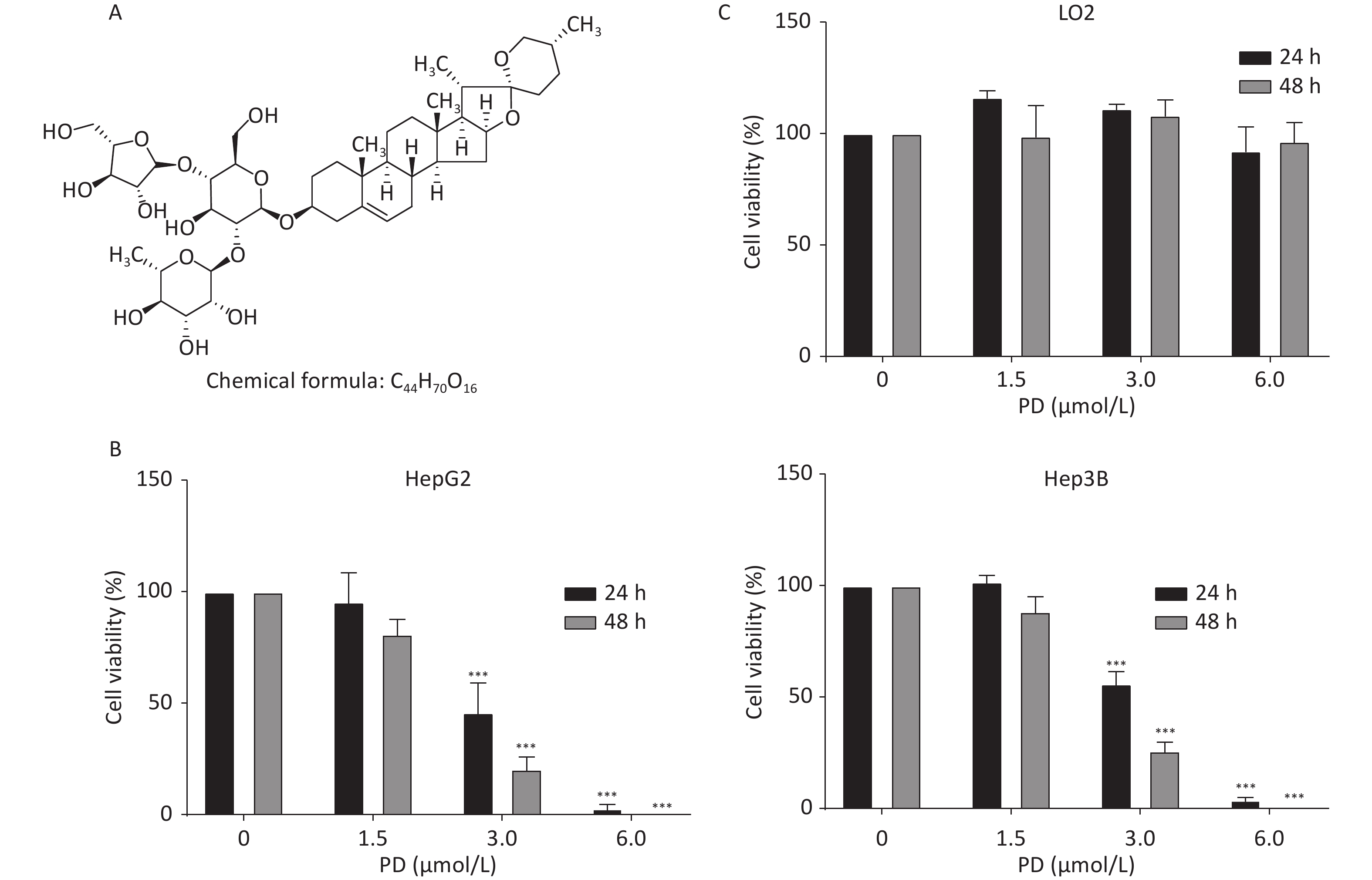
HepG2, Hep3B, and LO2 cells were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (Gibco) at 37 °C in 5% CO2. Supplementary Figure S1A (available in www.besjournal.com) shows the chemical structure of PD. To estimate the inhibitory effect of PD on the growth of liver cancer cells, increasing concentrations (0, 1.5, 3, and 6 μmol/L) of PD were added to HepG2 and Hep3B cells for 24 or 48 h, and the MTT assay was carried out to test cell viability at 570 nm. The data showed that PD markedly inhibited cell growth in a dose-dependent manner (Supplementary Figure S1B). Notably, PD was not toxic to the human normal hepatocyte cell line LO2 (Supplementary Figure S1C). Collectively, these data illustrate that PD may exert an anticancer effect in liver cancer cells but was nontoxic to normal cells.
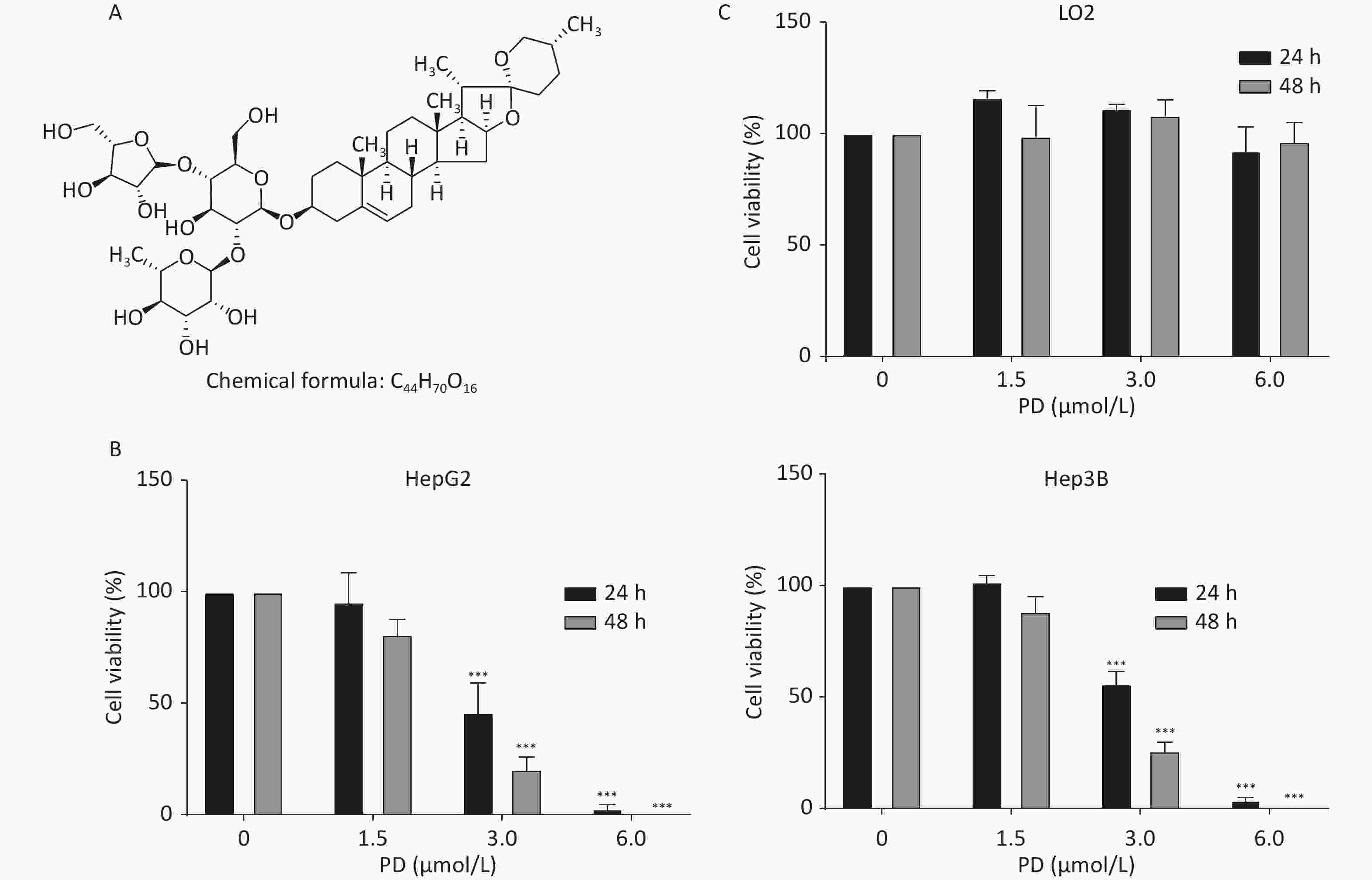
Figure S1. PD inhibits liver cancer cell proliferation. (A) Chemical structure of PD. (B) HepG2 and Hep3B cells were incubated with various concentrations (up to 6 μmol/L) of PD for 24 and 48 h, respectively, and cell viability was determined by the MTT assay. (C) PD did not exert any toxic effects on the normal LO2 hepatocyte cell line. Data (n = 3) are presented as mean ± SEM. ***, P < 0.001 compared with control cells.
The function of PD in the cell cycle was investigated in liver cancer cells. HepG2 and Hep3B cells were respectively exposed to various concentrations of PD (0–6 μmol/L) for 48 h. Cell cycle analysis was performed by C6 flow cytometry (BD Biosciences, San Diego, CA, USA) after incubating with propidium iodide staining buffer in the dark for 30 min. As shown in Figure 1A and 1B, the percentages of the G2/M phase increased significantly in the treated HepG2 and Hep3B cells (P < 0.05).
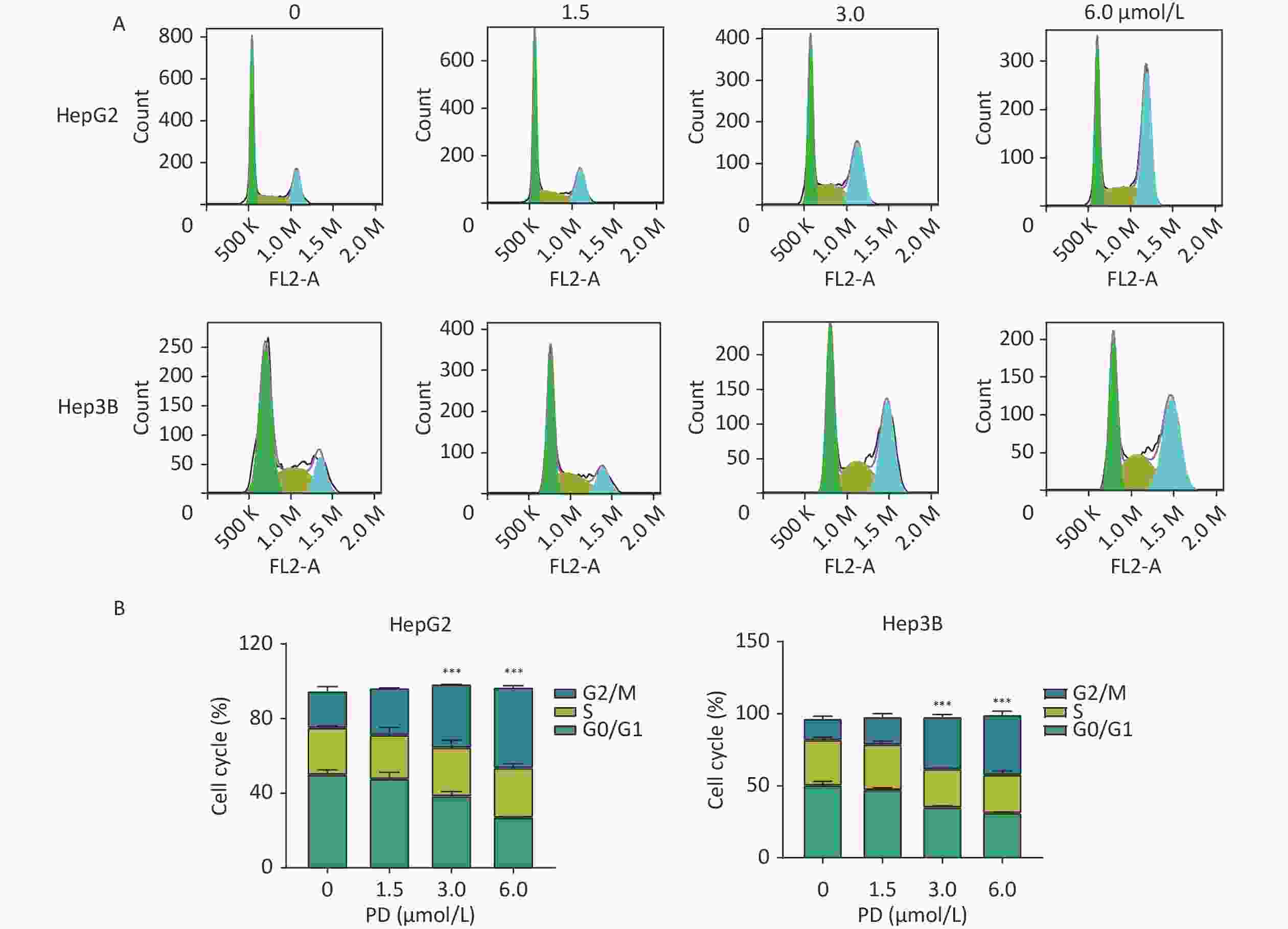
Figure 1. PD causes G2/M arrest in liver cancer cells. (A) HepG2 and Hep3B cells were treated with PD (0, 1.5, 3, and 6 μmol/L) for 48 h, and the cell cycle distribution was determined by flow cytometry. (B) The percentages of cells in the G0/G1, S, and G2/M phases are presented. Data (n = 3) are presented as mean ± SEM. PD, Polyphyllin D.***, P < 0.001 compared with control cells. K, Kilocalorie; M, Million.
The effect of PD on cell cycle arrest was confirmed by the decrease in the Cyclin/CDK markers (Cyclin D3, Cyclin E1, Cyclin A2, Cyclin B1, CDK2, and CDK4) in PD-treated liver cancer cells. In addition, the potent cyclin-dependent kinase inhibitor p21 increased gradually in response to the PD treatment (Figure 2A–C). Taken together, these data reveal that PD generated G2/M phase arrest by contributing to inhibiting the growth of liver cancer cells.
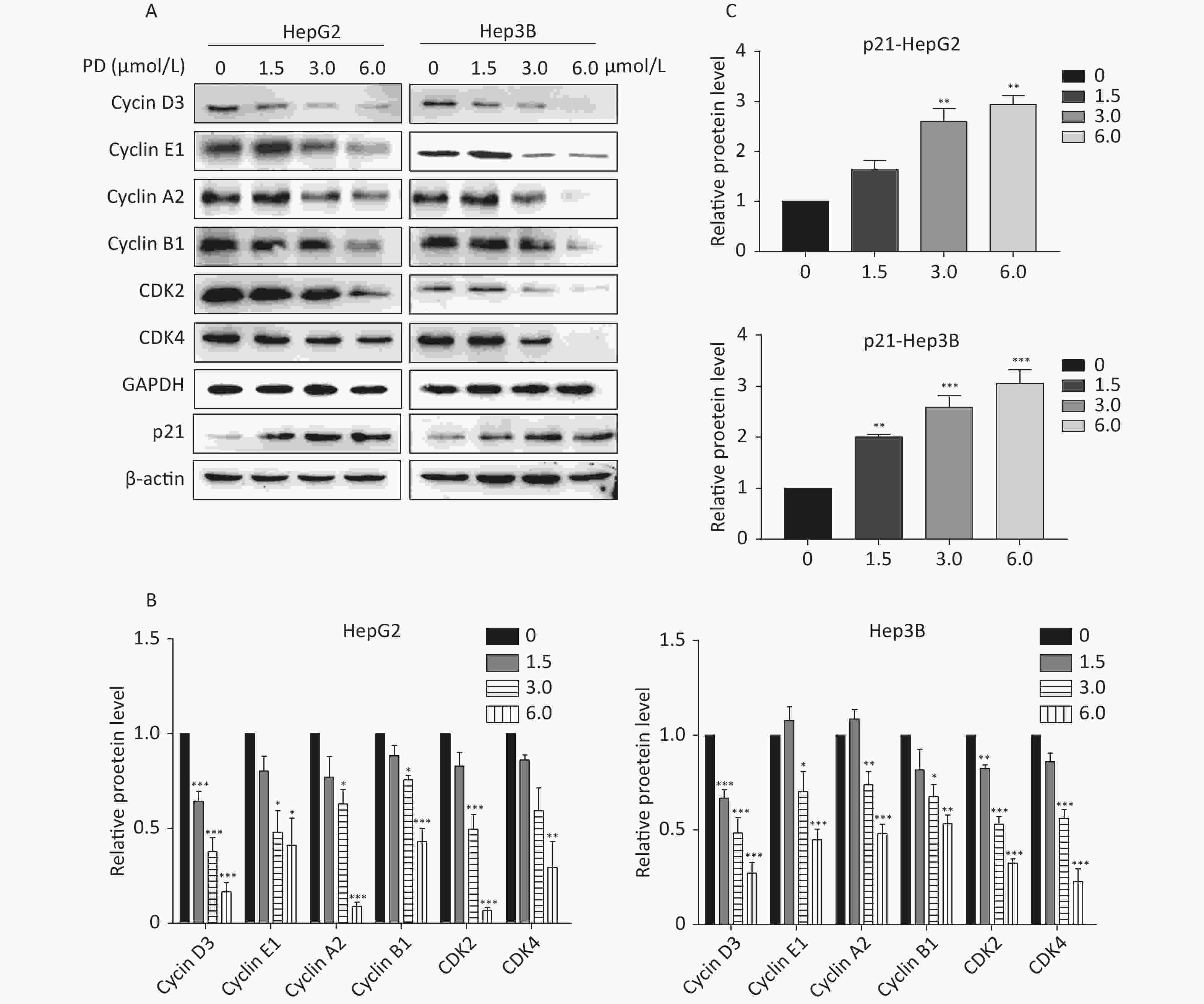
Figure 2. (A–C) Expression levels of Cyclin D3, Cyclin E1, Cyclin A2, Cyclin B1, CDK2, CDK4, and p21 were detected and are presented by western blot. Data (n = 3) are presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with control cells.
To identify the action mechanisms involved in the PD-blocked cell cycle of liver cancer cells, we used data-independent acquisition mass spectrometry to identify the PD-regulated proteins in triplicate samples from the Hep3B cell G2/M fraction (Supplementary Figure S2A, available in www.besjournal.com). The G2/M fraction was resuspended in 1% SDS lysis buffer. The sample was digested with trypsin using the filter-aided sample preparation method. After vacuum-freeze-drying, the peptide samples were desalinated using the MonoTip C18 (GL Sciences, Tokyo, Japan) and further analyzed by mass spectrometry (Thermo Fisher Scientific, Waltham, MA, USA). According to the power law global error model (PLGEM), 3,147 and 3,061 of overlapped proteins were identified and quantified in triplicate samples of the DMSO and PD-treated from the G2/M fraction, respectively (Supplementary Figure S2B). Then, the slope of protein abundance was 0.877, and the adjusted r-squared was 0.996 (Pearson’s r = 0.835, Supplementary Figure S2C). These results suggest that the statistical analysis of the proteomics data was reliable.

Figure S2. DIA-based quantitative proteomics analysis. (A) Experimental flow chart for identifying the PD-regulated proteins (n = 3). (B) Venn diagram showing the number of overlapped proteins in three biological replicates. (C) PLGEM fitting of the abundance of PD-regulated proteins.
A total of 186 differentially expressed proteins (149 upregulated and 37 downregulated) were identified according to fold change (FC) ≥ 1.5, P-value ≤ 0.05 (Figure 3A). To explore the molecular mechanism of PD-induced G2/M arrest, the top canonical pathways and signal channels of the differentially expressed proteins were analyzed by Ingenuity Pathway Analysis and Cytoscape, respectively, suggesting that PD may regulate cholesterol biosynthesis with a P-value of 2.52 x 10-4 (Figure 3B–C). In addition, the up-regulated proteins on cholesterol biosynthesis showed in the Table 1. Most cholesterol biosynthesis-related enzymes, such as HMGCR, CYB5R, TM7SF7, CYP27A1, and the low-density lipoprotein receptor (LDLR), are directly regulated by sterol regulatory element binding transcription factors (SREBPs)[8]. The SREBP-associated proteins involved in fatty acid and cholesterol metabolism, such as LDLR, CYB5R, RUFY1, HMGCR, HACD2, FDFT, TM7SF7, CYP27A1, and MBOA5, increased in response to the treatment with PD and the G2/M-enriched fraction (Figure 3D). Our results show that PD markedly increased free cholesterol and triacylglyceride (TG) levels in liver cancer cells (Figure 3E–F). These data suggest dysfunction of cholesterol biosynthesis is very important in regulating the cell cycle during PD treatment.
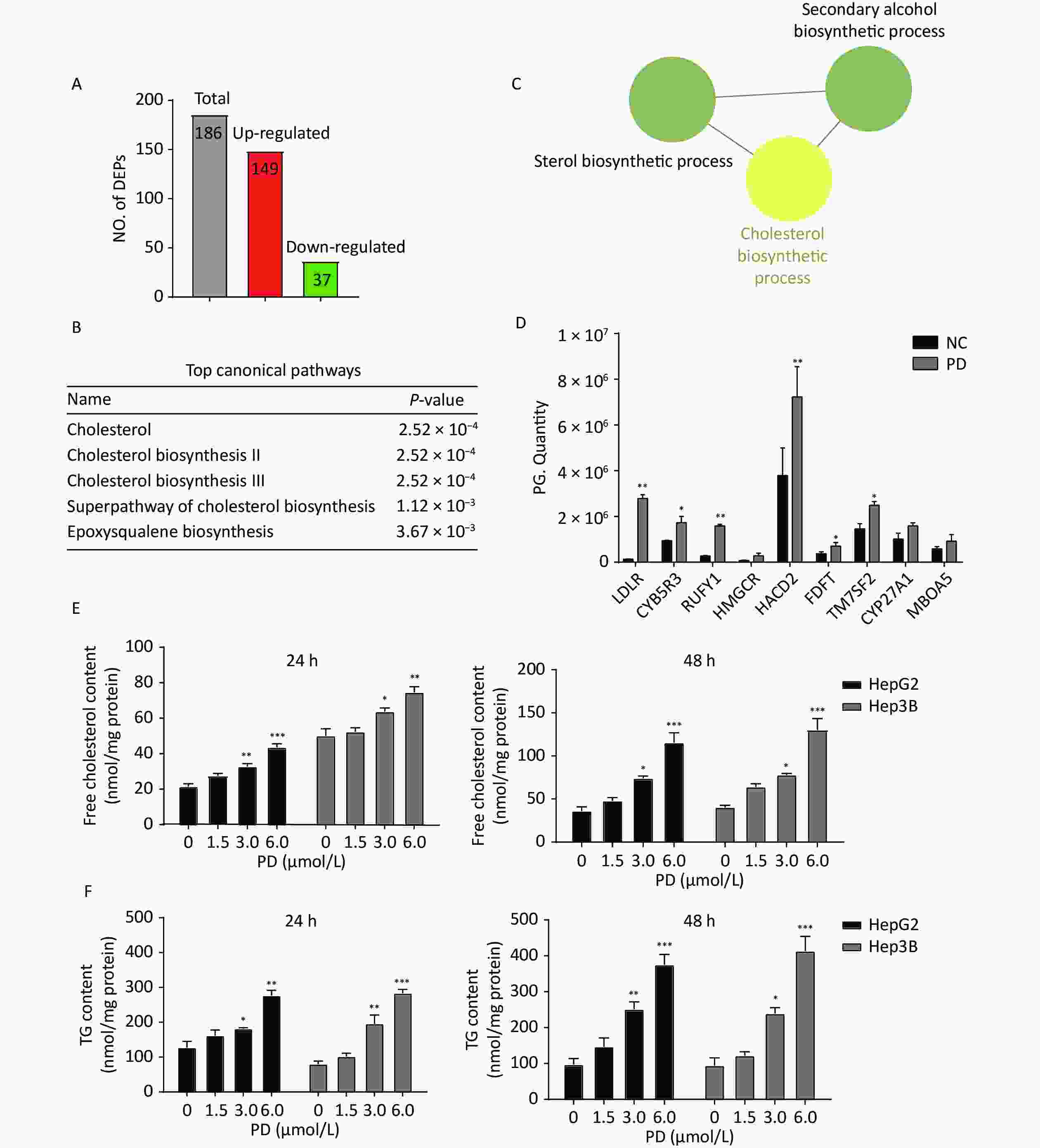
Figure 3. Quantitative proteomics identifies the deregulation of cholesterol biosynthesis induced by PD. (A–C) Differentially expressed proteins with fold change ≥ 1.5 and a P-value ≤ 0.05 were subjected to IPA analysis based on the PLGEM model (A), and the top canonical pathways are listed (B). Note that a cluster of PD-regulated proteins is involved in cholesterol biosynthesis (C). (D) Proteomics quantitative expression of SREBP associated-proteins is presented. (E–F) PD induces free cholesterol (D), and TG (E) increases markedly in HepG2 and Hep3B cells after 24 h and 48 h. Data (n = 3) are presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with control cells.
Table 1. Upregulated cholesterol biosynthesis proteins
Proteins Fold change LDLR 22.17 RUFY1 5.92 PPAL 4.61 HMGCR 4.07 EF1A2 3.12 TMEM97 2.32 HACD2 1.91 FDFT 1.87 RAB14 1.87 UB2G2 1.86 CYB5R3 1.84 GOLP3 1.73 NEUR1 1.72 FBP1L 1.72 TM7SF2 1.71 APP 1.60 PCYOX 1.57 CYP27A1 1.56 MBOA5 1.56 Cholesterol is necessary for cell cycle progression, and dysfunction causes arrest at different steps during the cycle. Previous studies have reported that an SREBP binding site in the promoter of the p21 gene has been detected, and the transient expression of nuclear SREBP causes the accumulation of p21, further reducing the expression of the CDK2 and CDK4 proteins and inducing the cell cycle in the G2/M phase[9]. Berberine-modulated lipogenesis through the SCAP/SREBP-1 pathway induces cell cycle arrest and inhibits colon cancer cell growth[10]. In the current study, we combined proteomics with bioinformatics analysis to determine whether PD induced G2/M phase arrest and suppressed the growth of liver cells via dysfunction of cholesterol biosynthesis. PD may be an effective method to disrupt cholesterol homeostasis during cancer treatment and should be further explored in the future.
The authors declare no conflicts of interest.
CHEN Yan Yan and WEN Shi Yuan conceived the study; CHEN Yan Yan performed the experiments; REN Cai Fang participated in the scientific discussion; CHEN Yan Yan and WEN Shi Yuan performed the data analyses and wrote the manuscript; CHEN Yan Yan revised the manuscript. All authors have contributed to the article and approved the submitted version.
doi: 10.3967/bes2023.009
Polyphyllin D induces G2/M Cell Cycle Arrest via Dysfunction of Cholesterol Biosynthesis in Liver Cancer Cells
-
-
S1. PD inhibits liver cancer cell proliferation. (A) Chemical structure of PD. (B) HepG2 and Hep3B cells were incubated with various concentrations (up to 6 μmol/L) of PD for 24 and 48 h, respectively, and cell viability was determined by the MTT assay. (C) PD did not exert any toxic effects on the normal LO2 hepatocyte cell line. Data (n = 3) are presented as mean ± SEM. ***, P < 0.001 compared with control cells.
Figure 1. PD causes G2/M arrest in liver cancer cells. (A) HepG2 and Hep3B cells were treated with PD (0, 1.5, 3, and 6 μmol/L) for 48 h, and the cell cycle distribution was determined by flow cytometry. (B) The percentages of cells in the G0/G1, S, and G2/M phases are presented. Data (n = 3) are presented as mean ± SEM. PD, Polyphyllin D.***, P < 0.001 compared with control cells. K, Kilocalorie; M, Million.
Figure 3. Quantitative proteomics identifies the deregulation of cholesterol biosynthesis induced by PD. (A–C) Differentially expressed proteins with fold change ≥ 1.5 and a P-value ≤ 0.05 were subjected to IPA analysis based on the PLGEM model (A), and the top canonical pathways are listed (B). Note that a cluster of PD-regulated proteins is involved in cholesterol biosynthesis (C). (D) Proteomics quantitative expression of SREBP associated-proteins is presented. (E–F) PD induces free cholesterol (D), and TG (E) increases markedly in HepG2 and Hep3B cells after 24 h and 48 h. Data (n = 3) are presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with control cells.
Table 1. Upregulated cholesterol biosynthesis proteins
Proteins Fold change LDLR 22.17 RUFY1 5.92 PPAL 4.61 HMGCR 4.07 EF1A2 3.12 TMEM97 2.32 HACD2 1.91 FDFT 1.87 RAB14 1.87 UB2G2 1.86 CYB5R3 1.84 GOLP3 1.73 NEUR1 1.72 FBP1L 1.72 TM7SF2 1.71 APP 1.60 PCYOX 1.57 CYP27A1 1.56 MBOA5 1.56 -
[1] Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2021; 71, 209−49. doi: 10.3322/caac.21660 [2] Chen B, Wu JX, Cheng SH, et al. Phase 2 study of adjuvant radiotherapy following narrow-margin hepatectomy in patients with HCC. Hepatology, 2021; 74, 2595−604. doi: 10.1002/hep.31993 [3] Tian Y, Gong GY, Ma LL, et al. Anti-cancer effects of Polyphyllin I: An update in 5 years. Chem Biol Interact, 2020; 316, 108936. doi: 10.1016/j.cbi.2019.108936 [4] Liu JZ, Liu YZ, Li HC, et al. Polyphyllin D induces apoptosis and protective autophagy in breast cancer cells through JNK1-Bcl-2 pathway. J Ethnopharmacol, 2022; 282, 114591. doi: 10.1016/j.jep.2021.114591 [5] Bisteau X, Caldez MJ, Kaldis P. The complex relationship between liver cancer and the cell cycle: A story of multiple regulations. Cancers, 2014; 6, 79−111. doi: 10.3390/cancers6010079 [6] Badal S, Delgoda R. Role of the modulation of CYP1A1 expression and activity in chemoprevention. J Appl Toxicol, 2014; 34, 743−53. doi: 10.1002/jat.2968 [7] Suski JM, Braun M, Strmiska V, et al. Targeting cell-cycle machinery in cancer. Cancer Cell, 2021; 39, 759−78. doi: 10.1016/j.ccell.2021.03.010 [8] Lasunción MA, Martínez-Botas J, Martín-Sánchez C, et al. Cell cycle dependence on the mevalonate pathway: Role of cholesterol and non-sterol isoprenoids. Biochem Pharmacol, 2022; 196, 114623. doi: 10.1016/j.bcp.2021.114623 [9] Nakakuki M, Shimano H, Inoue N, et al. A transcription factor of lipid synthesis, sterol regulatory element-binding protein (SREBP)-1a causes G1 cell-cycle arrest after accumulation of cyclin-dependent kinase (cdk) inhibitors. FEBS J, 2007; 274, 4440−52. doi: 10.1111/j.1742-4658.2007.05973.x [10] Liu YX, Hua WW, Li Y, et al. Berberine suppresses colon cancer cell proliferation by inhibiting the SCAP/SREBP-1 signaling pathway-mediated lipogenesis. Biochem Pharmacol, 2020; 174, 113776. doi: 10.1016/j.bcp.2019.113776 -
 22251Supplementary Materials.pdf
22251Supplementary Materials.pdf

-




 下载:
下载:





 Quick Links
Quick Links