-
Feline calicivirus (FCV) was a highly prevalent RNA virus causing upper respiratory tract infections in cats through the mouth and nose around the world, result in the death of cats especially under 1 year old with upper respiratory tract disease (URTD) and virulent systemic disease (FCV-VSD)[1]. Infected cats usually presented with mouth ulcers, salivation and gingivitis—stomatitis. Furthermore, both recovered cats and asymptomatic carriers can still carry and spread the FCV virus for extended periods of time, contributing to the potential for outbreak. This presented a significant threat to the health and lives of cats, and rare wildlife and took a heavy financial loss on the pet industry, making the prevention and control of the virus difficult. Hence, rapid identification of FCV in clinical samples was necessary to prevent the spread of FCV and avoid financial loss.
A variety of molecular detection method for FCV has been developed, including quantitative polymerase chain reaction (qPCR), enzymatic recombinase amplification- lateral flow dipstick (ERA-LFD) and Clustered Regularly Interspaced Short Palindromic Repeats-associated protein 13a (CRISPR-cas13a). The molecular detection method of qPCR not only depends on the specialized and expensive equipment, but also has strict requirement for various reaction temperature, not suitable for onsite detection, particularly in remote areas. In recent years, the development of isothermal amplification technology has made it possible for FCV rapid detection onsite, such as ERA-LFD and CRISPR-cas13a. Though ERA-LFD method could achieve the onsite visual detection, it was susceptible to aerosol contamination that resulting in false positive results because of the exposed lateral flow dipstick. The CRISPR-cas13a could reach to a lower detection limit of 5.5 copies/µL FCV in 60 min, but it needed targeted sequences accumulation of recombinase polymerase amplification (RPA) reaction for crRNAs to recognize and bind with, that increased operational complexity and reaction time[2].
RPA was an isothermal amplification technique that allows for rapid amplification of target genes within 20–30 minutes at 35–42 °C. There were three essential enzymes including a recombinase, a single-stranded DNA-binding protein (SSB) and a strand-displacing polymerase were employed in RPA system to achieve rapid amplification. It has been shown to be a rapid, sensitive and highly specific diagnostic method. Exo-probe was a detection probe of fluorescent for improving assay specificity and visualized result. There was a fluorophore group (FAM) and a corresponding quencher (BHQ1) group on the 5’ end and 3’ end of the tetrahydrofuran residue (THF) in exo-probe, respectively. When the exo-probe bound to the target sequence during reaction, exonuclease III cleaved the THF site, causing the separation of the fluorescent group from the quencher group, which in turn caused fluorescence to be produced. In combination with an exo-probe, aerosol contamination can be avoided, reducing the risk of false positive signals. Besides, the generated fluorescence signal could be measured by the portable fluorescence detector, which was suitable for onsite detection. The exo probe-based reverse transcription (RT)-RPA method has been applied in various pathogens. For examples, RT-RPA method combined exo-probe can detect porcine epidemic diarrhea virus at 40 °C in 20 min with detection limit of 101 copies/reaction[3]. Based on VP2 gene designed RT-RPA with exo-probe reaction achieved the detection of porcine parvovirus at 39 °C in 20 min with detection limit of 102 copies/reaction[4]. Acute hepatopancreatic necrosis disease in shrimp can be detected by RPA combing with exo probe to achieve the detection limit of 1 CFU per reaction[5]. However, this detection method applied on FCV was still not developed.
The choose of target gene was important for detection, and the RNA genome of the virus was highly susceptible to mutation. In FCV’s RNA genome, there were three open reading frames (ORF), ORF1, ORF2 and ORF3, of which ORF1 has the highest homology, and our subsequent experiments would be based on ORF1 for the design of primers and probe for the further assay.
In our study, we established a user-friendly detection platform of RT-RPA combined with exo-probe for FCV (Figure 1A). It exhibited fast, sensitive, and portable characteristics, making it suitable for onsite detection, especially in underdeveloped and remote areas.
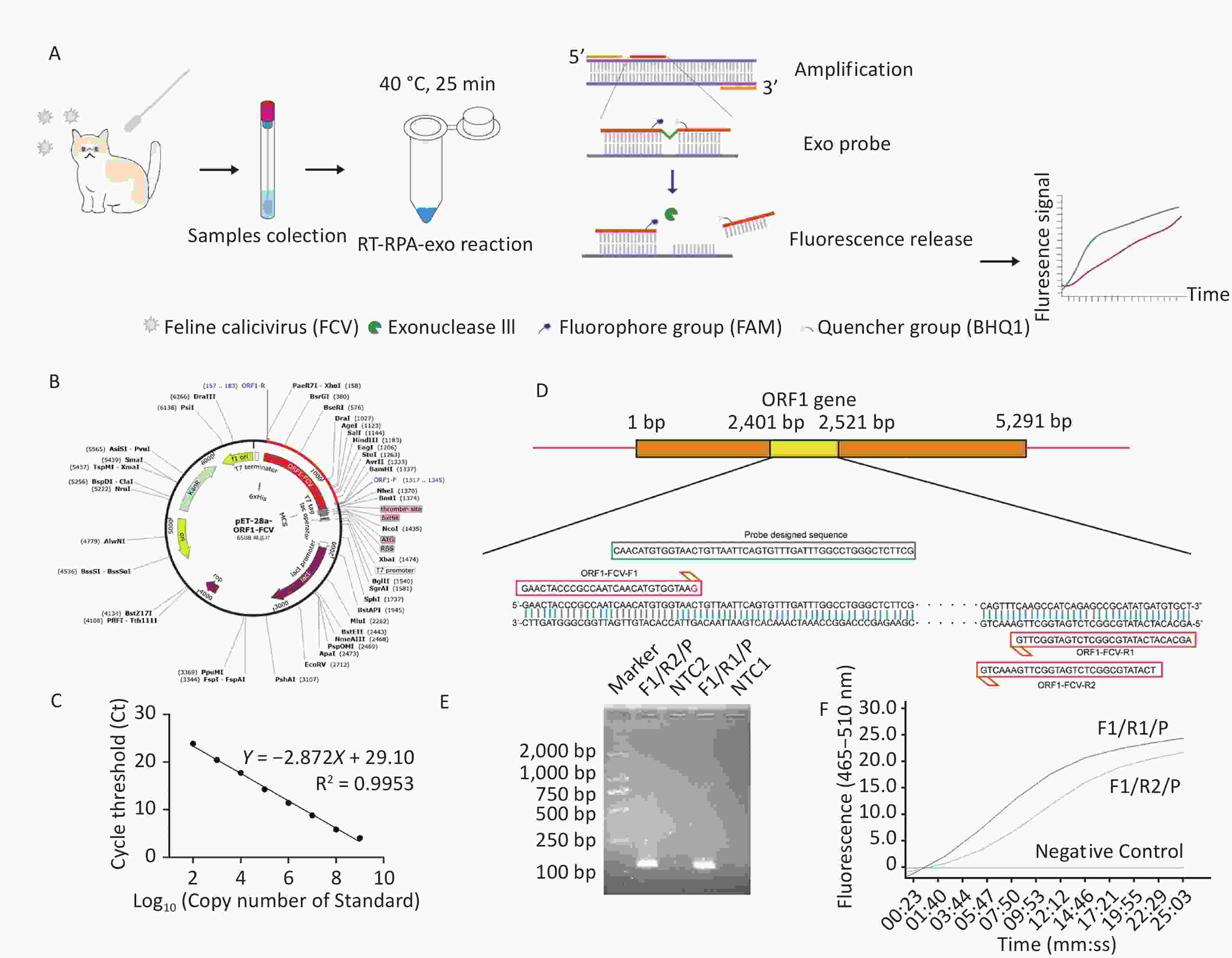
Figure 1. Schematic diagram for RT-RPA-exo and primers-probe pairs screen. (A) The Schematic diagram of established RT-RPA-exo assay. The samples of nasal and oral swabs were collected for RNA/DNA extraction, following by performing the RT-RPA-exo assay at 40 °C for 25 min. During the reaction, the THF site of exo probe was cut by exonuclease (exo) after probe recognized and bound to target dsDNA. Then, the quencher (BHQ1) group was released from dsDNA that contributed to the persistent fluorescence signal from fluorophore group (FAM) detected by fluorescence detection equipment. (B) The recombinant plasmid mapping of pET-28a (+)-ORF1-FCV. (C) The standard curve of the recombinant plasmid was obtained by the RT-qPCR assay. The template concentrations that dilutions of standard plasmid (Log10) from 102–109 copies/μL were indicated on the x-axis, whereas the corresponding cycle threshold (Ct) values were presented on the y-axis. Each dot represented the result of three duplicate amplifications of each dilution. (D) The schematic diagram of designation for ORF1-FCV-F1, ORF1-FCV-R1 and ORF1-FCV-R2 and exo probe, that were based on the highly conservative sequence (yellow rectangular box) within ORF1 gene of FCV. (E) RPA amplification combined with 1.5% agarose gel electrophoresis for preliminary primer screening of F1/R1/P and F1/R2/P, in which and RNA of FCV (ATCC VR-782) was 104 copies/µL. The lane NTC1 and NTC2 were set as negative control group without template. (F) Two primer-probe pairs of F1/R1/P and F1/R2/P were used for RT-RPA-exo reaction, in which and RNA of FCV (ATCC VR-782) was 104 copies/µL. The intensity of the fluorescent signal was detected by qPCR mechanism, and deionized water was added into the reaction system to replace various prime-probe pairs to set as negative control.
Virus’s nucleic acid preparation and clinical samples. FCV (ATCC VR-782), feline herpesvirus (FHV, ATCC VR-636), feline coronavirus (FCoV, ATCC VR-989), feline parvovirus (FPV, ATCC VR-648) and Staphylococcus aureus (S. aureus, ATCC 6538) were stored in our lab. The viral nucleic acid sample were prepared using TIANamp Virus DNA/RNA Kit (Tiangen, Beijing, China) following the manufacturer’s instruction, whereas the S. aureus genomic DNA was extracted using TIANamp Bacteria DNA Kit. There were 30 clinical samples of nasal and oral swabs from suspected cases of FCV infected pet cats, and 5 samples from healthy cats. All 35 clinical samples were obtained from local pet hospital in Taizhou. The collected samples were diluted three times with sterile PBS, and underwent repeated freeze-thawing at –80 °C for three cycles. After thorough vortexing and mixing with virus preservation solution, samples were centrifuged at 12,000 rpm for 10 min, and the supernatants were used for viral RNA/DNA extraction. The concentration of extracted nucleic acid was quantified by NanoDrop Lite spectrophotometer. And all DNA and RNA from various pathogens were stored in –80 °C for the following assay.
FCV ORF1 RNA standard preparation. The FCV virus RNA was reverse-transcribed into cDNA with Hifair AdvanceFast 1st Strand cDNA Synthesis Kit (YEASEN, Shanghai, China). PCR primer sets for highly conservative FCV-ORF1 (GenBank: OP626901.1) fragments of 1,191 bp were synthesized. The fragment details were ORF1-F (5’-CGCGGATCCTAGGAATTTGATGTCAACAT-3’) and ORF1-R (5’-CCGCTCGAGAGCACATCATATGCGGCTCT-3’) and contained two cleavage sites of BamHI and XhoI. FCV-ORF1 fragments were amplified using PCR method with pfu DNA polymerase (Tiangen) using FCV cDNA as a template, cleaved with restriction endonucleases of BamHI and XhoI, and purified with TIANquick Midi Purification Kit (Tiangen). The concentration of FCV-ORF1 DNA fragment was determined and ligated to the pET-28a (+) vector (Takara, Dalian, China) (Figure 1B). Then, the recombinant plasmid of pET-28a (+)-FCV-ORF1 was transferred into E. coli Trans5α chemically competent cells (TRANS, Beijing, China) for expansion and identified by DNA sequencing. The positive recombinant pET-28a (+)-FCV-ORF1 plasmid was transcribed into FCV ORF1 RNA standard using T7 High Efficiency Transcription kit (TRANS) and purified with EasyPure RNA Purification Kit (TRANS). The FCV ORF1 RNA standard was quantified by NanoDrop Lite spectrophotometer, and the formula for copies number was as following: copies/µL = [6.02 × 1023 × (RNA concentration) ng/uL × 10-9] / [(Sequence length + 282]) × 660][6]. The RNA standard was diluted by 10-fold gradient and detected using RT-qPCR method as described below. A standard curve was drawled based on the Ct values under different template copies and then the FCV RNA was quantified (Figure 1C).
Design and embellish of RT-RPA primer sets and probe for FCV-ORF1. Nucleic acid sequences of 10 strains of FCV virus derived from cats of various time and location (Supplementary Table S1, available in www.besjournal.com) (460-20-1, 460-20-2, 9284-5-2020-ITA, 9341-1-2020-ITA, GXLZ01-19, GXLZ02-19, GXNN01-19, GXNN02-19, GXNN05-20, GXNN06-20) were compared to obtain three highly conservative regions for primers design (Figure 1D; Supplementary Figure S1, available in www.besjournal.com). NCBI Primer blast online design tool was used to design RT-RPA primer sets and probe for this conserved FCV-ORF1. The primer lengths ranged from 28 to 35, and species were limited to FCV. Other parameters were referred to the previously published literature[7]. Based on the inter nucleotide sequence of the RT-RPA Primer, the probe was designed using Primer Premier 5 software. The probe length should be between 46 and 52 bp. Other parameters were referred to the previously published literature[7]. The THF group was inserted at the 31th base of the probe and its adjacent T-bases were replaced by FAM (6-Corboxy-DT) and BHQ1 (Black Hole Quencher 1)-dT, respectively. A C3 spacer was also needed to add at the 3' end of the probe. Two pairs of primers (FCV-ORF1-F1/R1 and FCV-ORF1-F1/R2) and one FCV-ORF1-exo-probe were designed based on the ORF1 gene for RT-RPA reaction. We conducted blast validation based on primer-designed sequences, indicating that it cannot match the genomes of other species. At the same time, structural analysis of the primer pairs and probe showed that there were no strong interfering dimers or hairpins. The detailed sequences information of primers was in Table 1.
Table 1. The sequence of primer pairs and probe for RT-RPA-exo
Assay Name Sequence (5’–3’) RT-RPA-exo ORF1-FCV-F1 5’-GAACTACCCGCCAATCAACATGTGGTAAG-3’ ORF1-FCV-R1 5’-AGCACATCATATGCGGCTCTGATGGCTTG-3’ ORF1-FCV-R2 5’-TCATATGCGGCTCTGATGGCTTGAAACTG-3’ Exo probe 5’-CAACATGTGGTAACTGTTAATTCAGTGTT[FAM-dT](THF)A[BHQ1-dT]TTGGCCTGGGCTCTTCG-C3 spacer-3’ Table S1. The acquisition time and origin of 10 FCV isolated strains
Strain name GenbanK Acquisition time (year/month) Location 460-20-1 OP626900.1 2020/11 Andria, Apulia, Italy 460-20-2 OK428795.1 2020/11 Andria, Apulia, Italy 9284-5-2020-ITA OP750454.1 2021/07 Teramo, Italy 9341-1-2020-ITA OP750455.1 2021/07 Teramo, Italy GXLZ01-19 MZ712025.1 2019/05 Liuzhou, Guangxi, China GXLZ02-19 MZ712024.1 2019/05 Liuzhou, Guangxi, China GXNN01-19 OL512761.1 2019/06 Nanning, Guangxi, China GXNN02-19 MZ712022 2019/02 Nanning, Guangxi, China GXNN05-20 MZ712019 2020/08 Nanning, Guangxi, China GXNN06-20 MZ712018 2020/08 Nanning, Guangxi, China 
Figure S1. Gene homology comparison of ORF1 in various FCV strains. Sequences from 10 strains of FCV (460-20-1, 460-20-2, 9284-5-2020-ITA, 9341-1-2020-ITA, GXLZ01-19, GXLZ02-19, GXNN01-19, GXNN02-19, GXNN05-20 and GXNN06-20) were compared. The comparison results were presented above and three highly homologous fragments between 2,401 bp to 2,521 bp were labeled with red rectangular boxes in Orf1 gene sequence.
Exo probe-based RT-RPA reaction system and program setup for FCV-ORF1. The initial reaction system for exo probe-based RT-RPA has a total volume of 50 µL as described in the protocol of Zhongce® RT-RAA Nucleic Acid Amplification Reagent (Hangzhou, S004ZC). The reaction system consisted of a tube containing reaction dry powder, 2 µL of each primer (10 µmol/L), 0.6 µL of probe (10 µmol/L), 1 µL RNA samples, 25 µL buffer A, and 2.5 µL buffer B. The remaining volume was filled with deionized water, and the mixture was thoroughly mixed before adding it to the tube containing the reaction dry powder. The experiment was performed using a qPCR equipment (LightCycler 480 II, Roche), maintaining a temperature of 40 °C for 25 min to capture the fluorescence signal from the FAM channel.
Screen of the optimal primer sets and exo probe-based RT-RPA. To determine the optimal primer sets and probe combination for exo probe-based RT-RPA. Primer sets of FCV-ORF1-F1/R1/P and FCV-ORF1-F1/R2/P were used for RPA reaction and the results were analyzed by agarose gel electrophoresis. Both primers pairs showed the obvious DNA bands at approximately 120 bp (Figure 1E). In order to further explore the amplification efficiency after adding probes, primer sets of FCV-ORF1-F1/R1 and FCV-ORF1-F1/R2 were respectively combined with FCV-ORF1-probe and used for exo RT-RPA assay (Figure 1F). The results indicated that FCV-ORF1-F1/R1/P exhibited significant higher fluorescence values compared to the FCV-ORF1-F1/R2/P within 25 minutes. Therefore, F1/R1/P was selected for further specificity and sensitivity evaluation assay.
Specificity evaluation for FCV-ORF1-F1/R1/P in exo RT-RPA. In order to explore the specificity of RT-RPA and exo RT-RPA reaction for FCV, different upper respiratory pathogens of cat containing FHV, FCoV, FPV and other no-related pathogen S. aureus were selected for test. The result showed that only FCV showed a strong fluorescent signal in 25 min and there were no generated signals in other pathogens (Figure 2A). It was worth mention that the RT-RPA was also suitable for DNA template, which has been widely verified in our lab. Hence, our established RT-RPA-system exhibited a perfect specificity.

Figure 2. The performance of established RT-RPA-exo reaction system. (A) The fluorescence history diagram exhibiting the results of RT-RPA-exo reaction for FCV and other pathogens including feline herpesvirus (FHV), feline coronavirus (FCoV), feline parvovirus (FPV) and Staphylococcus aureus (S. aureus). The red fluorescent lines represent FCV, and the green fluorescent lines represent other pathogens, which overlap with each other. (B) Detection limit of established RPA-exo-detection. Fluorescence development over time using a dilution range of 10-1 to 104 copies/reaction of FCV RNA. The amounts tested are indicated with different colors. NTC was no template control. (C) The semi-logarithmic regression of the data collected from the eight real-time RPA repeats (D) using GraphPad Prism 8.0. The reaction time was between 5–13 min for the templates at 101 to 104 copies/reaction. (E) The probit regression analysis was conducted using SPSS software based on collected eight repeats of RT-RPA-exo reaction. The limit of detection at 95% probability (7.674 copies/reaction) was depicted by a red triangle. (E) RT-qPCR and established RT-RPA-exo for clinical suspected samples (Number 1 to 30) and 5 healthy (Number 31 to 35) individuals’ detection. Orange cells represented positive results and the white cells represent negative results. (F) The clinical detection performance of RT-qPCR method and developed RT-RPA-exo method was compared based on the 29 positive clinical samples. Linear regression analysis was performed using GraphPad Prism 8.0 to determine the relationship between RT-RPA threshold time (Tt) values (y-axis) and real-time RT-PCR cycle threshold (Ct) values (x-axis). The analysis yielded an R2 value of 0.9086.
Sensitivity evaluation for FCV-ORF1-F1/R1/P in RT-RPA-exo. To investigate the sensitivity of the RT-RPA-exo reaction for FCV, FCV RNA was diluted into different concentration gradients ranging from 10-1 copies/reaction to 104 copies/reaction as templates for the reaction (Figure 2B). Each concentration was repeated eight times in the assay. The results suggested that fluorescence could be detected above a concentration of 101 copies/reaction, but no fluorescence signal was generated when the concentration was less than 101 copies/reaction. Semi-log regression analysis was performed on the data from the eight independent repeats using GraphPad Prism 8.0, showing a detection range of 5 min for 104 copies/reaction to 13 min for 101 copies/reaction (Figure 2C). In addition, probit regression analysis was conducted on the eight repeat results using SPSS software, revealing that the detection limit of the RT-RPA-exo reaction was 7.674 copies/reaction at a 95% probability (Figure 2D).
RT-qPCR assay and application of FCV exo RT-RPA detection system in clinical cat nasal and oral swabs. The clinical detection performance of the established RT-RPA-exo system was evaluated using 35 clinical samples of nasal and oral swabs obtained from diseased and healthy cats, and the results were compared with those of RT-qPCR[8] and a sample was considered positive when the Ct value was less than 40. Out of the 35 samples tested with RT-RPA-exo, 29 were positive, while RT-qPCR detected 30 positive samples. Among the 35 samples, 29 were positive in both detection methods, and 5 samples from healthy cats were negative (Figure 2E; Supplementary Table S2, available in www.besjournal.com). The coincidence rate for positive samples between RT-RPA-exo and RT-qPCR was 96.67% (29/30). Furthermore, for the detected positive samples, the RT-RPA-exo reaction provided results within 5-13 minutes, whereas RT-qPCR took approximately 60 minutes, yielding Ct values ranging from 14.01 to 28.21 (Supplementary Table S3, available in www.besjournal.com). We compared the threshold time (Tt) of the established RT-RPA-exo method recorded in qPCR instrument with the cycle threshold (Ct) values of RT-qPCR (Figure 2F), and obtained a correlation coefficient (R2) of 0.907. These findings indicated that the established RT-RPA-exo method exhibited high degree of consistency for FCV detection.
Table S2. Detection of Feline calicivirus in clinical samples
Number Sample type Sample source Status Results RT-RPA (Tt, min) RT-qPCR
(n = 3, Ct)1 Cat Oral swab Disease 5.8 14.43 ± 0.42 2 Cat Oral swab Disease 5.75 15.74 ± 0.27 3 Cat Oral swab Disease 7.25 18.50 ± 0.50 4 Cat Oral swab Disease − 25.01 ± 0.20 5 Cat Oral swab Disease 10.52 21.2 ± 0.26 6 Cat Oral swab Disease 5 16.50 ± 0.42 7 Cat Oral swabs Disease 7.2 18.22 ± 0.34 8 Cat Oral swab Disease 12.5 26.56 ± 0.31 9 Cat Oral swab Disease 6.5 17.50 ± 0.22 10 Cat Oral swab Disease 9.25 21.8 ± 0.36 11 Cat Oral swab Disease 5.86 16.5 ± 0.45 12 Cat Oral swab Disease 8.7 20.8 ± 0.40 13 Cat Oral swab Disease 8.36 20.22 ± 0.38 14 Cat Oral swab Disease 9.2 20.61 ± 0.35 15 Cat Oral swab Disease 5.62 14.65 ± 0.24 16 Cat Oral swabs Disease 5.98 16.64 ± 0.35 17 Cat Nasal swab Disease 9.48 21.84 ± 0.48 18 Cat Nasal swab Disease 5.74 16.28 ± 0.37 19 Cat Nasal swab Disease 8.95 21.69 ± 0.28 20 Cat Nasal swab Disease 7.62 17.95 ± 0.46 21 Cat Nasal swab Disease 7.65 16.65 ± 0.11 22 Cat Nasal swab Disease 10.65 22.32 ± 0.48 23 Cat Nasal swab Disease 11.75 25.38 ± 0.42 24 Cat Nasal swab Disease 11.20 22.48 ± 0.18 25 Cat Nasal swab Disease 6.62 17.89 ± 0.28 26 Cat Nasal swab Disease 9.2 25.54 ± 0.21 27 Cat Nasal swab Disease 8.45 16.67 ± 0.17 28 Cat Nasal swab Disease 7.15 19.94 ± 0.28 29 Cat Nasal swab Disease 12.15 27.9 ± 0.31 30 Cat Nasal swab Disease 5.24 15.62 ± 0.19 31 Cat Oral swab Healthy − − 32 Cat Oral swab Healthy − − 33 Cat Oral swab Healthy − − 34 Cat Nasal swab Healthy − − 35 Cat Nasal swab Healthy − − Table S3. Comparation of RT-RPA-exo and RT-qPCR
Item RT-RPA-exo RT-qPCR S2 [1] Time 15–30 min 1 h Specificity 100% 100% Limit of detection 7.674 copies/reaction 70 copies/reaction Clinical performance 96.7 % (29/30) 100% (30/30) Note. The data of time, specificity, limit of detection of RT-qPCR S2 was referenced by Abd-Eldaim MM’s work[1]. Reference: Abd-Eldaim MM, Wilkes RP, Thomas KV, et al. Development and validation of a TaqMan real-time reverse transcription-PCR for rapiddetection of feline calicivirus. Arch Virol, 2009; 154, 555−60. RPA-combined with exo probe for fluorescence visualization detection has been applied in detection of Yersinia pestis[9], Vibrio Cholerae and Vibrio Parahaemolyticus[10] and so on. Nowadays, new combination for RPA combined with CRISPR/Cas12a or Cas13a has applied. Though it can introduce an optimum detection limit, it extended detection time to 1 h and increased complexity of detection because of the increase in operational steps, which bring difficult for fast and point of care detection. RPA-combined with exo probe still has its advantage of relatively less detection time and avoiding aerosol pollution. Besides, the cost for RT-RPA-exo reaction was about $2 and was suitable for testing of individual samples.
In summary, we have successfully developed a rapid, accurate, and reliable detection method called RT-RPA-exo for detecting FCV. This innovative method allowed for the detection of FCV within just 25 minutes at a temperature of 40 °C, with a detection limit of 101 copies per reaction. Compared to the commonly used RT-qPCR method, our RT-RPA-exo method has demonstrated an impressive consistency rate of 96.67% in terms of positive results. Furthermore, our method offered significant advantages in terms of detection time and portability. By integrating it with a portable fluorescence detector like the Genie III tube scanner[4], we effectively shortened the overall detection time to 25 min and eliminated the reliance on laboratory equipment. This breakthrough enabled on-site detection of FCV, making it particularly useful for small laboratories, resource-poor areas, and remote regions where access to sophisticated equipment may be limited.
The authors declare that they have no competing interests.
MU Hong Yun conceived and designed the project. MU Hong Yun and ZHOU Hong Lei performed the assay and wrote manuscript. JING Zheng and LU Wei participated in preparation of samples. JING Zheng was the leader of the project and revised the manuscript. All authors approved the final manuscript.
doi: 10.3967/bes2024.036
An exo Probe-based Reverse Transcription and Recombinase Polymerase Amplification for Rapid Detection of Feline Calicivirus in Cats
-
-
Figure 1. Schematic diagram for RT-RPA-exo and primers-probe pairs screen. (A) The Schematic diagram of established RT-RPA-exo assay. The samples of nasal and oral swabs were collected for RNA/DNA extraction, following by performing the RT-RPA-exo assay at 40 °C for 25 min. During the reaction, the THF site of exo probe was cut by exonuclease (exo) after probe recognized and bound to target dsDNA. Then, the quencher (BHQ1) group was released from dsDNA that contributed to the persistent fluorescence signal from fluorophore group (FAM) detected by fluorescence detection equipment. (B) The recombinant plasmid mapping of pET-28a (+)-ORF1-FCV. (C) The standard curve of the recombinant plasmid was obtained by the RT-qPCR assay. The template concentrations that dilutions of standard plasmid (Log10) from 102–109 copies/μL were indicated on the x-axis, whereas the corresponding cycle threshold (Ct) values were presented on the y-axis. Each dot represented the result of three duplicate amplifications of each dilution. (D) The schematic diagram of designation for ORF1-FCV-F1, ORF1-FCV-R1 and ORF1-FCV-R2 and exo probe, that were based on the highly conservative sequence (yellow rectangular box) within ORF1 gene of FCV. (E) RPA amplification combined with 1.5% agarose gel electrophoresis for preliminary primer screening of F1/R1/P and F1/R2/P, in which and RNA of FCV (ATCC VR-782) was 104 copies/µL. The lane NTC1 and NTC2 were set as negative control group without template. (F) Two primer-probe pairs of F1/R1/P and F1/R2/P were used for RT-RPA-exo reaction, in which and RNA of FCV (ATCC VR-782) was 104 copies/µL. The intensity of the fluorescent signal was detected by qPCR mechanism, and deionized water was added into the reaction system to replace various prime-probe pairs to set as negative control.
S1. Gene homology comparison of ORF1 in various FCV strains. Sequences from 10 strains of FCV (460-20-1, 460-20-2, 9284-5-2020-ITA, 9341-1-2020-ITA, GXLZ01-19, GXLZ02-19, GXNN01-19, GXNN02-19, GXNN05-20 and GXNN06-20) were compared. The comparison results were presented above and three highly homologous fragments between 2,401 bp to 2,521 bp were labeled with red rectangular boxes in Orf1 gene sequence.
Figure 2. The performance of established RT-RPA-exo reaction system. (A) The fluorescence history diagram exhibiting the results of RT-RPA-exo reaction for FCV and other pathogens including feline herpesvirus (FHV), feline coronavirus (FCoV), feline parvovirus (FPV) and Staphylococcus aureus (S. aureus). The red fluorescent lines represent FCV, and the green fluorescent lines represent other pathogens, which overlap with each other. (B) Detection limit of established RPA-exo-detection. Fluorescence development over time using a dilution range of 10-1 to 104 copies/reaction of FCV RNA. The amounts tested are indicated with different colors. NTC was no template control. (C) The semi-logarithmic regression of the data collected from the eight real-time RPA repeats (D) using GraphPad Prism 8.0. The reaction time was between 5–13 min for the templates at 101 to 104 copies/reaction. (E) The probit regression analysis was conducted using SPSS software based on collected eight repeats of RT-RPA-exo reaction. The limit of detection at 95% probability (7.674 copies/reaction) was depicted by a red triangle. (E) RT-qPCR and established RT-RPA-exo for clinical suspected samples (Number 1 to 30) and 5 healthy (Number 31 to 35) individuals’ detection. Orange cells represented positive results and the white cells represent negative results. (F) The clinical detection performance of RT-qPCR method and developed RT-RPA-exo method was compared based on the 29 positive clinical samples. Linear regression analysis was performed using GraphPad Prism 8.0 to determine the relationship between RT-RPA threshold time (Tt) values (y-axis) and real-time RT-PCR cycle threshold (Ct) values (x-axis). The analysis yielded an R2 value of 0.9086.
Table 1. The sequence of primer pairs and probe for RT-RPA-exo
Assay Name Sequence (5’–3’) RT-RPA-exo ORF1-FCV-F1 5’-GAACTACCCGCCAATCAACATGTGGTAAG-3’ ORF1-FCV-R1 5’-AGCACATCATATGCGGCTCTGATGGCTTG-3’ ORF1-FCV-R2 5’-TCATATGCGGCTCTGATGGCTTGAAACTG-3’ Exo probe 5’-CAACATGTGGTAACTGTTAATTCAGTGTT[FAM-dT](THF)A[BHQ1-dT]TTGGCCTGGGCTCTTCG-C3 spacer-3’ S1. The acquisition time and origin of 10 FCV isolated strains
Strain name GenbanK Acquisition time (year/month) Location 460-20-1 OP626900.1 2020/11 Andria, Apulia, Italy 460-20-2 OK428795.1 2020/11 Andria, Apulia, Italy 9284-5-2020-ITA OP750454.1 2021/07 Teramo, Italy 9341-1-2020-ITA OP750455.1 2021/07 Teramo, Italy GXLZ01-19 MZ712025.1 2019/05 Liuzhou, Guangxi, China GXLZ02-19 MZ712024.1 2019/05 Liuzhou, Guangxi, China GXNN01-19 OL512761.1 2019/06 Nanning, Guangxi, China GXNN02-19 MZ712022 2019/02 Nanning, Guangxi, China GXNN05-20 MZ712019 2020/08 Nanning, Guangxi, China GXNN06-20 MZ712018 2020/08 Nanning, Guangxi, China S2. Detection of Feline calicivirus in clinical samples
Number Sample type Sample source Status Results RT-RPA (Tt, min) RT-qPCR
(n = 3, Ct)1 Cat Oral swab Disease 5.8 14.43 ± 0.42 2 Cat Oral swab Disease 5.75 15.74 ± 0.27 3 Cat Oral swab Disease 7.25 18.50 ± 0.50 4 Cat Oral swab Disease − 25.01 ± 0.20 5 Cat Oral swab Disease 10.52 21.2 ± 0.26 6 Cat Oral swab Disease 5 16.50 ± 0.42 7 Cat Oral swabs Disease 7.2 18.22 ± 0.34 8 Cat Oral swab Disease 12.5 26.56 ± 0.31 9 Cat Oral swab Disease 6.5 17.50 ± 0.22 10 Cat Oral swab Disease 9.25 21.8 ± 0.36 11 Cat Oral swab Disease 5.86 16.5 ± 0.45 12 Cat Oral swab Disease 8.7 20.8 ± 0.40 13 Cat Oral swab Disease 8.36 20.22 ± 0.38 14 Cat Oral swab Disease 9.2 20.61 ± 0.35 15 Cat Oral swab Disease 5.62 14.65 ± 0.24 16 Cat Oral swabs Disease 5.98 16.64 ± 0.35 17 Cat Nasal swab Disease 9.48 21.84 ± 0.48 18 Cat Nasal swab Disease 5.74 16.28 ± 0.37 19 Cat Nasal swab Disease 8.95 21.69 ± 0.28 20 Cat Nasal swab Disease 7.62 17.95 ± 0.46 21 Cat Nasal swab Disease 7.65 16.65 ± 0.11 22 Cat Nasal swab Disease 10.65 22.32 ± 0.48 23 Cat Nasal swab Disease 11.75 25.38 ± 0.42 24 Cat Nasal swab Disease 11.20 22.48 ± 0.18 25 Cat Nasal swab Disease 6.62 17.89 ± 0.28 26 Cat Nasal swab Disease 9.2 25.54 ± 0.21 27 Cat Nasal swab Disease 8.45 16.67 ± 0.17 28 Cat Nasal swab Disease 7.15 19.94 ± 0.28 29 Cat Nasal swab Disease 12.15 27.9 ± 0.31 30 Cat Nasal swab Disease 5.24 15.62 ± 0.19 31 Cat Oral swab Healthy − − 32 Cat Oral swab Healthy − − 33 Cat Oral swab Healthy − − 34 Cat Nasal swab Healthy − − 35 Cat Nasal swab Healthy − − S3. Comparation of RT-RPA-exo and RT-qPCR
Item RT-RPA-exo RT-qPCR S2 [1] Time 15–30 min 1 h Specificity 100% 100% Limit of detection 7.674 copies/reaction 70 copies/reaction Clinical performance 96.7 % (29/30) 100% (30/30) Note. The data of time, specificity, limit of detection of RT-qPCR S2 was referenced by Abd-Eldaim MM’s work[1]. Reference: Abd-Eldaim MM, Wilkes RP, Thomas KV, et al. Development and validation of a TaqMan real-time reverse transcription-PCR for rapiddetection of feline calicivirus. Arch Virol, 2009; 154, 555−60. -
[1] Bordicchia M, Fumian TM, Van Brussel K, et al. Feline calicivirus virulent systemic disease: clinical epidemiology, analysis of viral isolates and in vitro efficacy of novel antivirals in australian outbreaks. Viruses, 2021; 13, 2040. doi: 10.3390/v13102040 [2] Huang J, Liu YJ, He YW, et al. CRISPR-Cas13a based visual detection assays for feline calicivirus circulating in southwest China. Front Vet Sci, 2022; 9, 913780. doi: 10.3389/fvets.2022.913780 [3] Wang JC, Zhang RX, Wang JF, et al. Real-time reverse transcription recombinase polymerase amplification assay for rapid detection of porcine epidemic diarrhea virus. J Virol Methods, 2018; 253, 49−52. doi: 10.1016/j.jviromet.2018.01.001 [4] Wang JC, Liu LB, Han QA, et al. An exo probe-based recombinase polymerase amplification assay for the rapid detection of porcine parvovirus. J Virol Methods, 2017; 248, 145−47. doi: 10.1016/j.jviromet.2017.06.011 [5] Yu HY, Yang XH, Zhang J, et al. A rapid real-time recombinase polymerase amplification assay for diagnosis of acute hepatopancreatic necrosis disease in shrimp. Acta Biochim Biophys Sin (Shanghai), 2021; 53, 381−84. doi: 10.1093/abbs/gmaa175 [6] Wang P, Zhao CJ, Lu QW, et al. RPA-ligation-qPCR combined method for genotyping the SARS-CoV-2 key mutation E484Q. Acta Biochim Biophys Sin (Shanghai), 2022; 54, 1924−27. doi: 10.3724/abbs.2022152 [7] Yang XH, Zhang X, Wang Y, et al. A real-time recombinase polymerase amplification method for rapid detection of vibrio vulnificus in seafood. Front Microbiol, 2020; 11, 586981. doi: 10.3389/fmicb.2020.586981 [8] Meli ML, Berger A, Willi B, et al. Molecular detection of feline calicivirus in clinical samples: A study comparing its detection by RT-qPCR directly from swabs and after virus isolation. J Virol Methods, 2018; 251, 54−60. doi: 10.1016/j.jviromet.2017.10.001 [9] Wei X, Li Y, Lu X, et al. Rapid detection of Yersinia pestis by real-time recombinase-aided amplification. Biomed Environ Sci, 2021; 34, 309−13. [10] Liao L, Shi W, Ma C, et al. Duplex detection of Vibrio cholerae and Vibrio parahaemolyticus by real-time recombinase polymerase amplification. Biomed Environ Sci, 2022; 35, 1161−65. -
 23308+Supplementary Materials.pdf
23308+Supplementary Materials.pdf

-




 下载:
下载:






 Quick Links
Quick Links