-
Dengue virus (DENV) is a mosquito-borne, infectious viral agent that comprises four distinct serotypes (DENV-1–4). Infection with any serotype of DENV causes asymptomatic disease, dengue fever (DF), or severe dengue[1,2]. DF is primarily circulated in tropical and subtropical regions located between 25° latitudes in the north and south worldwide. Traditionally, the endemic regions of DF include Africa, America, the eastern Mediterranean, Southeast Asia, and the Western Pacific[2]. In recent decades, due to complex reasons, including economy and trade globalization, climate change, environmental deterioration, and social factors, the geographic distribution of DF had expanded continuously, and about 3.9 billion people living in 128 countries are at risk of infection with DENV[3]. Meanwhile, the incidence of DF has increased dramatically, with 390 million DENV infections being reported each year and 96 million with apparent clinical manifestations[4]. Thus, DF has become a global public health problem.
Like the global prevalence of DF, the epidemic regions of DF in China have expanded gradually and the incidence of DF has increased dramatically in recent years. For instance, most of the DF cases and outbreaks had occurred in the coastal provinces[5] in the past; however, the first outbreak of DF in central China was reported in Henan province in 2013[6]. DF epidemic has become a public health concern in China. Fujian is a coastal province located in southeastern China and had no DF case reported in history until 1999 when an outbreak of DF occurred in Fuzhou city. Since then, the import of cases has been reported almost every year in the province and local outbreaks occurred intermittently[7]. We had previously characterized the epidemiological and etiological of DF in Fujian province[8]. However, it is noteworthy that the import of DF cases and local outbreaks were more frequent and some changes were observed regarding the situation of the DF epidemic in recent years. For a better understanding of the changes in the epidemic situation and for providing novel insights for disease control, we conducted a descriptive epidemiological and phylogenetic investigation in this study.
-
Epidemiological Data Epidemiological data supplemented with field investigations of patients with DF were collected from the ‘China Information System for Disease Control and Prevention’[9].
Case Definition DF diagnosis was in conformity with the ‘National diagnostic criteria and principle of management of dengue fever (WS 216–2001)’[10] and the ‘Diagnostic criteria for dengue fever (WS 216–2008)’[11]. Combined with the clinical manifestation and the epidemiological evidence, a laboratory-confirmed case was defined as one with a specific nucleotide acid or NS1 antigen of DENV being tested positive in the acute stage or a specific IgG antibody positively converted in convalescence, whereas a clinically diagnosed case was defined as one with a specific IgM antibody being determined positive. Imported and indigenous DF cases were defined epidemiologically according to whether the patients traveled to DF endemic regions and exposed to mosquito bite within 14 d before the onset of illness. A local DF outbreak was defined as more than two indigenous DF cases found in a relatively small area, such as a community, village, school, or other crowded unit, during the longest incubation period of 14 d of the illness.
Specimens and Viral Isolates Serum specimens were collected from patients for laboratory analysis. The need for informed consent was waived for mandatory measures for disease control. Specimens with the specific nucleotide acid being positive[12] were selected and inoculated (1:20) into a monolayer of mosquito C6/36 cells and blind-passed two passages. When the obvious cellular pathogenic effect appeared, the supernatants were collected and stored at −80 °C for further etiological study.
Reagents QIAamp Viral RNA Mini Kit, QIAGEN One-Step RT-PCR Kit, and QIAquick Gel Extraction Kit were purchased from Qiagen (Hilden, Germany). Commercially available real-time RT-PCR reagents for viral serotyping were purchased from Liferiver (Shanghai, China). Other reagents were routinely prepared, and the specific primers (Supplementary Table S1, available in www.besjournal.com) for amplification of the viral complete envelope (E) gene were synthesized by BioSune Biotechnology Co., Ltd (Shanghai, China)[13].
Primer name Sequence (5’-3’) Length of product (bp) DENV-1E F ATAGGAACATCCATCAC 1,609 DENV-1E R TGCCRCTTCCACATTTGAG DENV-2E F TTGAGACATCCAGGCTTCACC 1,610 DENV-2E R CCACTATCGGCCTGCACCAT DENV-3E F CCATCCATGACAATGAGATG 1,591 DENV-3E R ATTTGTATTGCTCTGTCC Table S1. RT-PCR primers used for amplification of full length envelope gene of DENV
-
Epidemiological Analysis The epidemiological data were sorted and statistically analyzed using the Excel software. The geographical distribution of cases was mapped using the ArcGIS software.
Viral RNA Extraction and Serotyping Viral RNA was extracted from the viral stock using the QIAamp Viral RNA Mini Kit, and serotyping was performed using the real-time RT-PCR Kits according to the manufacturer’s instructions.
E Gene Amplification and Sequencing Full-length sequences of E genes were amplified using the QIAGEN One-Step RT-PCR Kit with the corresponding serotype-specific primers (Supplementary Table S1). To 25 μL reaction mixture, 5 μL viral RNA was added as the template. The reaction mixture was incubated at 50 °C for 30 min and 94 °C for 15 min, followed by 30 cycles of 94 °C for 30 s, 45 °C for 30 s, and 72 °C for 30 s, then maintained at 72 °C for 5 min, and finally kept at 4 °C. The RT-PCR products were purified using the QIAquick Gel Extraction Kit and subsequently sequenced using the same primers on the automated capillary 3730xl DNA Analyzer (Applied Biosystems, USA) by BioSune Biotechnology Co., Ltd. Complete E genes were assembled using the SeqMan program from the Lasergene software package (DNASTAR, Madison, USA).
Phylogenetic Analysis Complete E gene sequences of DENV isolates of each serotype, together with the sequences of the corresponding serotype available from the GenBank database, were aligned independently. Phylogenetic analyses of the E genes of each serotype were performed independently using the neighbor-joining (NJ) method with 1,000 bootstrap replications and Kimura 2-parameter substitution model using the Mega software 6.0.
-
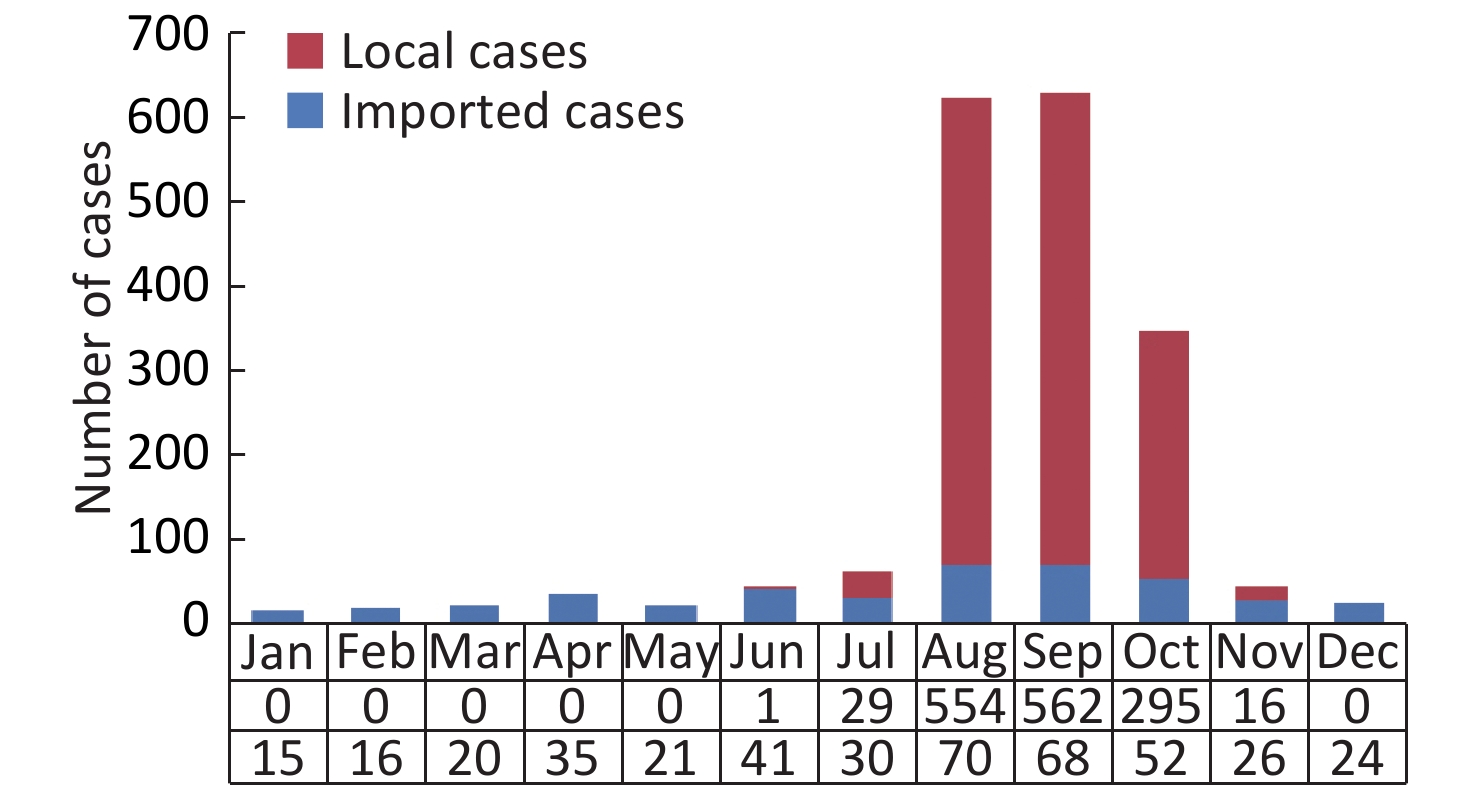
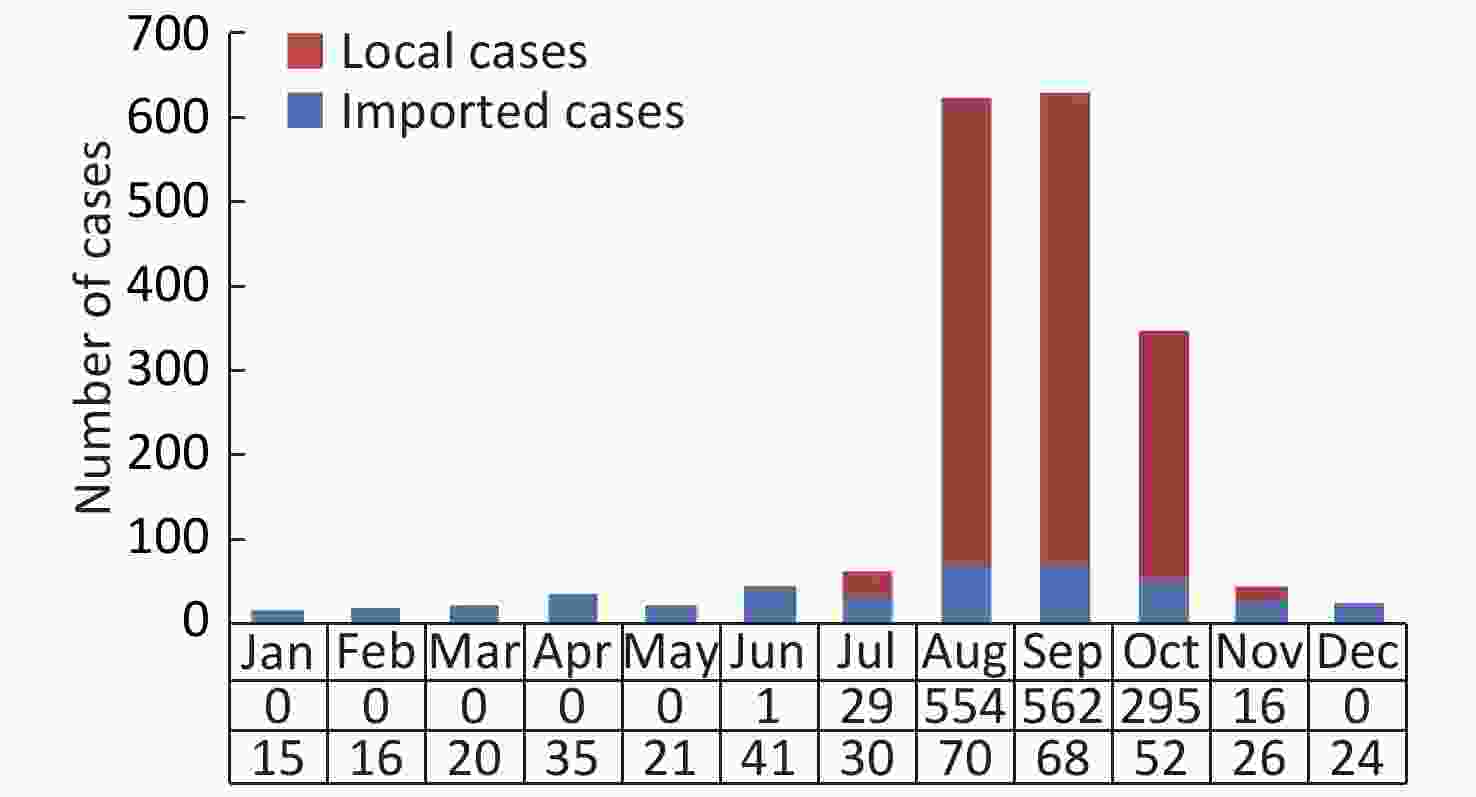
A total of 1,876 DF cases, including 418 imported and 1,458 indigenous, were reported in Fujian province during 2004–2017. All the cases presented mild or typical symptoms of DF, and no deaths or severe clinical outcomes were found. The sex ratio (male/female) of imported cases (1:0.54, 272/146) was higher than that of indigenous cases (1:1.21, 659/799); however, the median age of imported cases (35 years; IQR: 26–44 years) was lower than that of indigenous cases (48 years; IQR: 31–62 years). Therefore, it appears that male individuals in coastal cities responded primarily to the virus imported into Fujian. The spatial and temporal distribution revealed that the import of cases occurred throughout the year (Supplementary Figure S1 available in www.besjournal.com) and in cities throughout the entire province; however, 92.3% (386/418) of the cases were reported in seven coastal cities, including Fuzhou (123), Quanzhou (130), Putian (76), Xiamen (29), Ningde (18), Zhangzhou (6), and Pingtan (4). The indigenous cases were reported in the summer season (Supplementary Figure S1) in five coastal cities and one inland city (Nanping) (Table 1). According to field epidemiological investigations, the imported cases primarily originated from Southeast Asia (309/418), followed by other endemic regions of DF (80/418) worldwide and the surrounding provinces in southern China (29/418) (Table 1).
Type or origin* 2004 2005 2006 2007 2008 2009 2010 2011 2012 2013 2014 2015 2016 2017 Total Indigenous Fuzhou 93 1 1 5 33 907 13 1,053 Putian 103 9 147 8 267 Nanping 122 3 125 Quanzhou 1 5 6 Zhangzhou 6 6 Ningde 1 1 Subtotal 93 0 0 103 9 0 1 0 0 1 276 41 921 13 1,458 Imported Southeast Asia 9 14 3 9 16 13 21 17 23 36 29 42 32 45 309 America 1 1 1 2 1 6 Oceania 1 2 6 3 5 14 3 34 South Asia 1 9 4 5 4 22 Africa 9 2 2 4 17 Yunnan 1 1 Guangdong 22 1 1 24 Hainan 1 1 Taiwan 2 2 Zhejiang 1 1 Subtotal 9 14 3 9 16 14 23 18 26 61 57 54 55 59 418 Total 102 14 3 112 25 14 24 18 26 62 333 95 976 72 1,876 Table 1. Summary of indigenous and imported DF cases reported in Fujian during 2004–2017
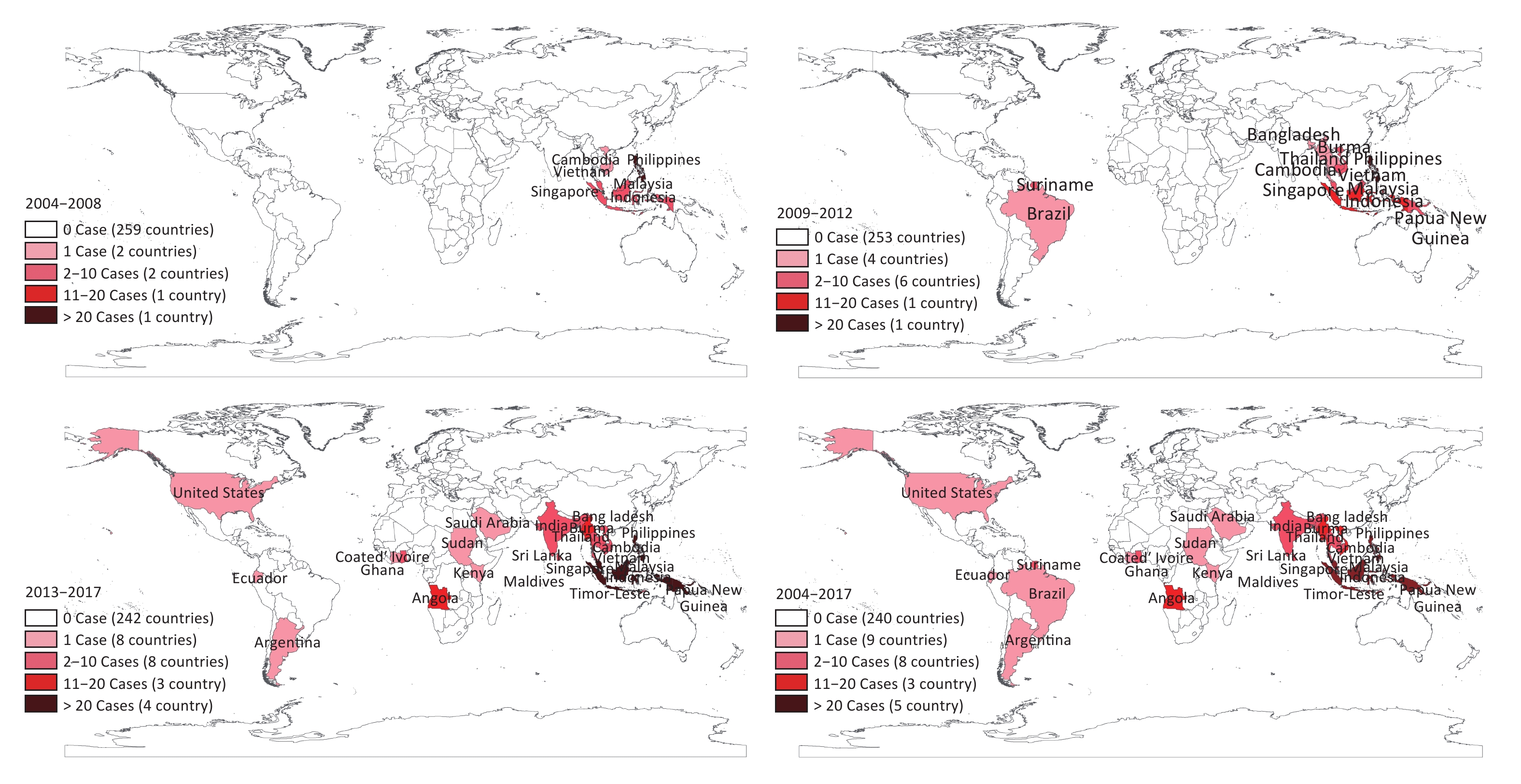
From 2013 onward, the number of imported DF cases increased significantly (Figure 1). Compared with 132 cases imported during 2004–2012, there were 286 cases imported into Fujian province during the period 2013–2017. Furthermore, the original region from where the cases were imported expanded significantly. The imported cases unexceptionally originated from Southeast Asia before 2009; however, the regions had expanded thereafter to America, Oceania, South Asia, and Africa (Figure 2). In addition to the cases imported from abroad, 29 cases were imported from the surrounding provinces of Guangdong, Yunnan, Hainan, Taiwan, and Zhejiang in China since 2013.
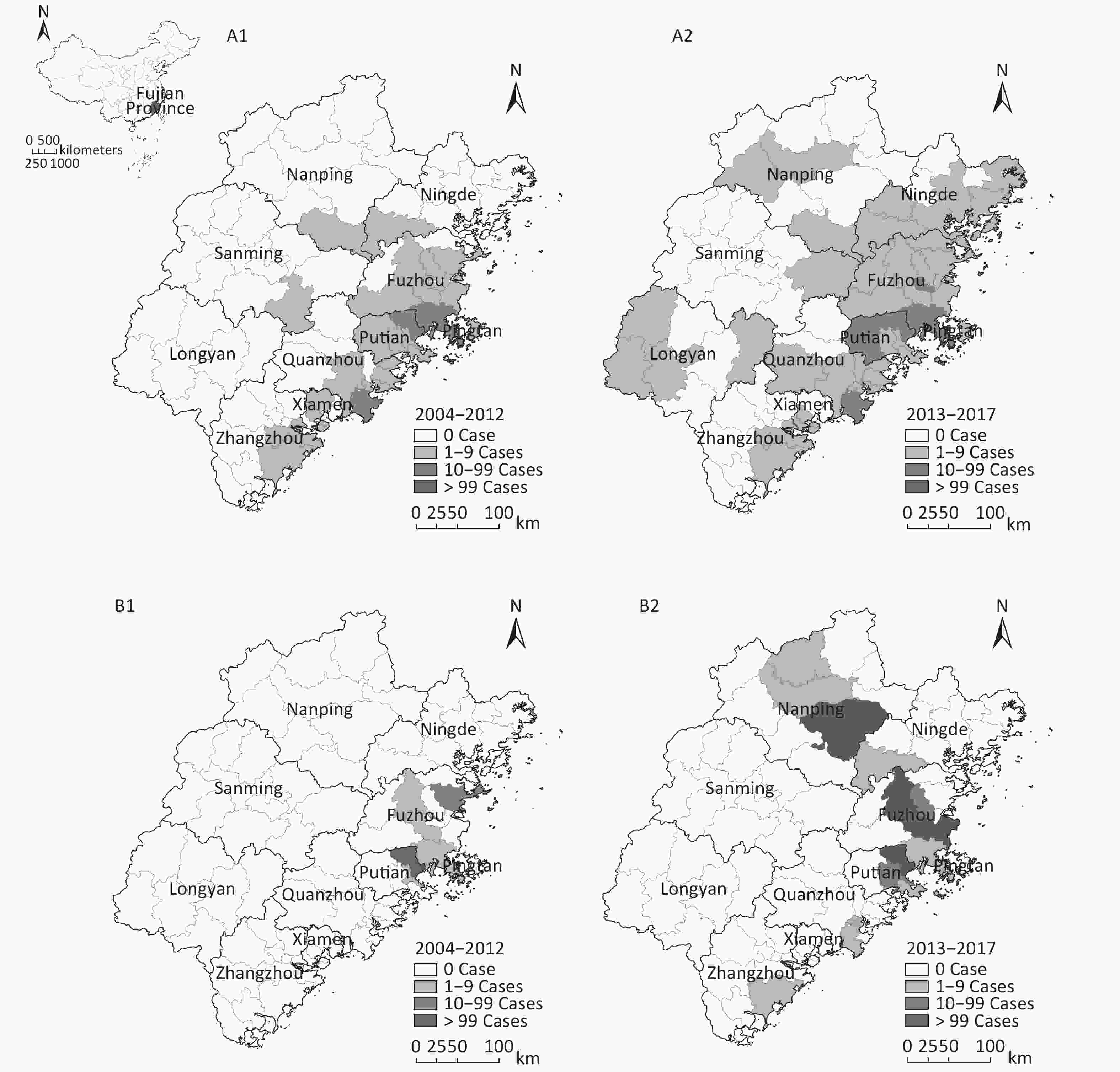
Figure 1. Distribution of DF cases in Fujian during 2004–2017. (A) Distribution of imported DF cases. (B) Distribution of indigenous DF cases. The number of DF cases is marked on the map according to counties in Fujian province.
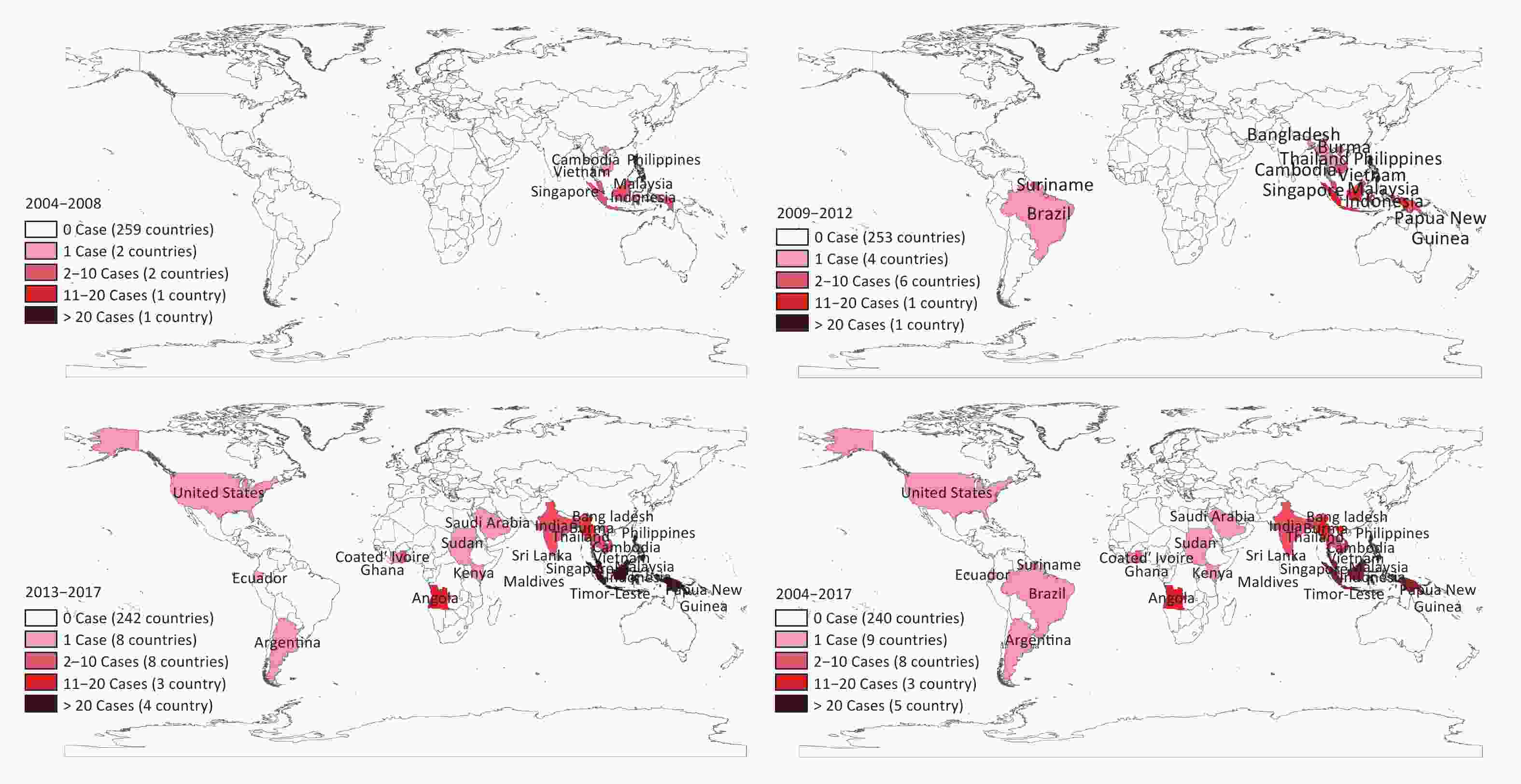
Figure 2. Expansion of original regions from which the DF cases were imported into Fujian province during 2004–2017.
With the increasing number of imported cases, local DF outbreaks occurred more frequently and the incidence of indigenous DF cases increased significantly in Fujian province (Figure 1). Of a total of 13 local DF outbreak events reported during 2004–2017, 10 had occurred in the past 5 years. Of the 1,458 indigenous cases, 1,252 (85.9%) were reported from 2013 onward. In addition to the increasing incidence of indigenous cases, the DF-affected areas in Fujian extended. Only two coastal cities (Fuzhou and Putian) reported indigenous cases before 2014; however, six cities had reported indigenous cases and four coastal cities (Fuzhou, Putian, Zhangzhou, and Quanzhou) and one inland city (Nanping) experienced local outbreaks of DF since 2014. Nonetheless, the majority of indigenous cases (1,445/1,458) were distributed in Fuzhou, Putian, and Nanping cities (Figure 1) in which most of the local DF outbreaks had occurred.
-
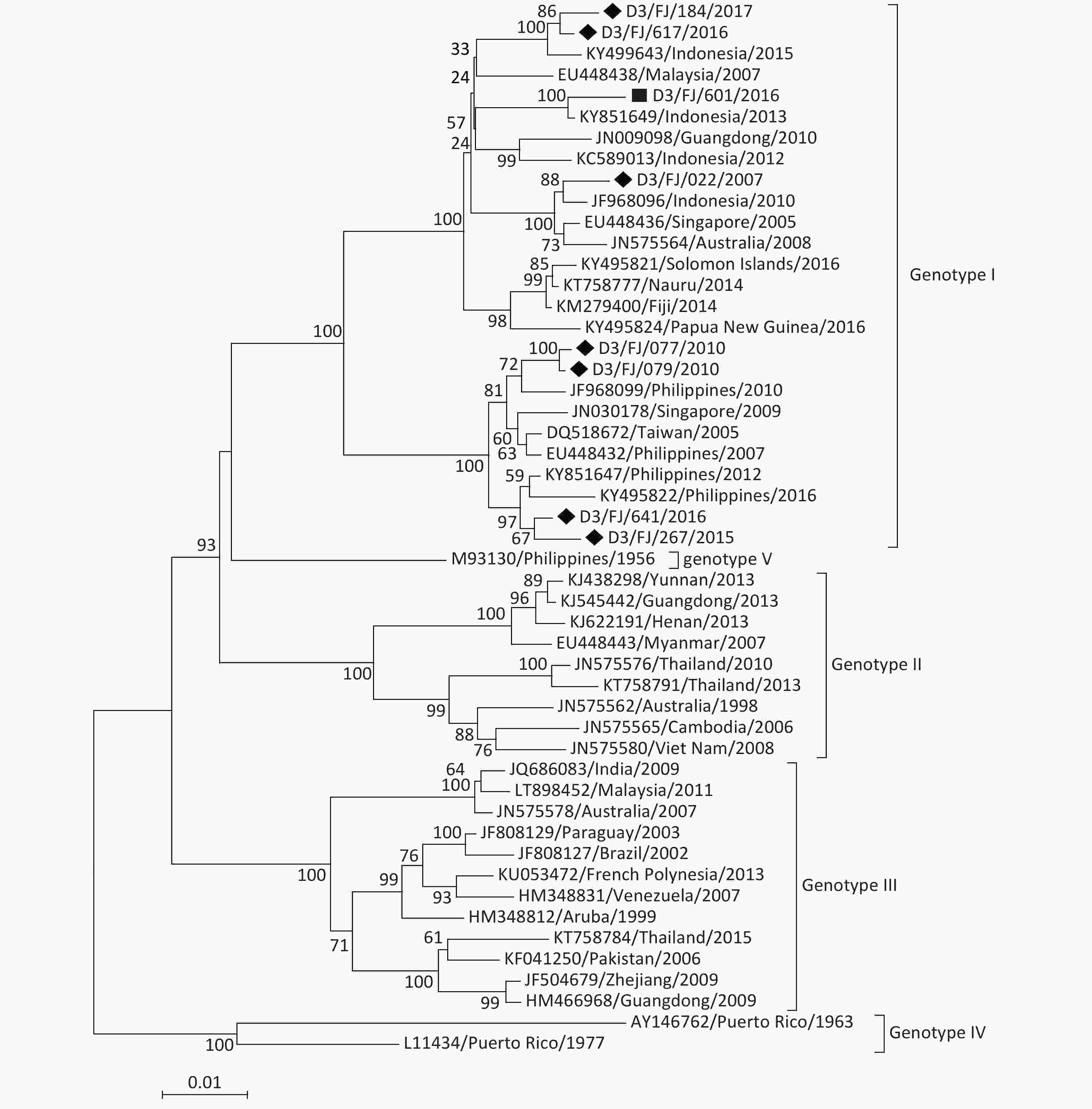
A total of 66 strains of DENV, including 31 DENV-1, 27 DENV-2, and 8 DENV-3, were isolated from patients with DF in Fujian province during 2004–2017. Of these isolates, 30 strains were isolated from imported cases and the remaining 36 strains were isolated from indigenous cases. The viral isolates were designated as ‘Serotype/Province/Serial number/Year’, and all their E gene sequences were deposited in GenBank (Supplementary Table S2, available in www.besjournal.com). Complete E gene sequences of DENV-1, -2, and -3 were aligned respectively with reference sequences downloaded from GenBank and expressed as ‘Accession No./Origin/Year’. The respective NJ trees of distinct serotypes were constructed, and the results of the phylogenetic analysis of DENVs of distinct serotypes are described in detail below.
Isolate Origin* Case type Acc. No. Isolate Origin* Case type Acc. No. D1/FJ/397/2004 Fuzhou Indigenous KU570091 D2/FJ/368/2007 Putian Indigenous KU570120 D1/FJ/412/2004 Fuzhou Indigenous KU570092 D2/FJ/102/2008 Putian Indigenous KU570123 D1/FJ/431/2004 Fuzhou Indigenous KU570093 D2/FJ/161/2013 Indonesia Indigenous KU570124 D1/FJ/003/2012 Suriname Imported KC759167 D2/FJ/162/2013 Indonesia Imported KU570125 D1/FJ/189/2012 Viet Nam Imported KU570095 D2/FJ/163/2013 Indonesia Imported KU570126 D1/FJ/031/2013 Angola Imported KU570098 D2/FJ/229/2014 Putian Indigenous KU570127 D1/FJ/011/2013 Angola Imported KU570096 D2/FJ/232/2014 Putian Indigenous KU570128 D1/FJ/023/2013 Angola Imported KU570097 D2/FJ/236/2014 Putian Indigenous KU570129 D1/FJ/072/2013 Singapore Imported KU570099 D2/FJ/269/2014 Nanping Indigenous KU570132 D1/FJ/189/2013 Thailand Imported KU570100 D2/FJ/270/2014 Nanping Indigenous KU570133 D1/FJ/202/2013 Philippine Imported KU570101 D2/FJ/196/2015 Philippine Imported MH729972 D1/FJ/006/2014 Viet Nam Imported KU570102 D2/FJ/269/2015 Philippine Imported MH729973 D1/FJ/211/2014 Nanping Indigenous KU570104 D2/FJ/303/2015 Putian Indigenous MH729974 D1/FJ/222/2014 Nanping Indigenous KU570111 D2/FJ/304/2015 Putian Indigenous MH729975 D1/FJ/224/2014 Nanping Indigenous KU570112 D2/FJ/305/2015 Putian Indigenous MH729976 D1/FJ/346/2014 Guangdong Imported KU570115 D2/FJ/336/2015 Fuzhou Indigenous MH729977 D1/FJ/353/2014 Guangdong Imported KU570116 D2/FJ/361/2015 Fuzhou Indigenous MH729978 D1/FJ/463/2014 Indonesia Imported KU570117 D2/FJ/409/2015 Fuzhou Indigenous MH729979 D1/FJ/466/2014 Guangdong Imported KU570118 D2/FJ/408/2015 Singapore Imported MH729980 D1/FJ/328/2015 Fuzhou Indigenous MH729960 D2/FJ/386/2016 Fuzhou Indigenous MH729981 D1/FJ/364/2015 Fuzhou Indigenous MH729961 D2/FJ/387/2016 Fuzhou Indigenous MH729982 D1/FJ/365/2015 Fuzhou Indigenous MH729962 D2/FJ/388/2016 Fuzhou Indigenous MH729983 D1/FJ/366/2015 Fuzhou Indigenous MH729963 D2/FJ/621/2016 PNG* Imported MH729984 D1/FJ/424/2016 Nanping Indigenous MH729964 D2/FJ/185/2017 Coated'lvoire Imported MH729985 D1/FJ/629/2016 Nanping Indigenous MH729965 D2/FJ/188/2017 Zhejiang Imported MH729986 D1/FJ/630/2016 Nanping Indigenous MH729966 D3/FJ/022/2007 Indonesia Imported KU570088 D1/FJ/186/2017 Myanmar Imported MH729967 D3/FJ/077/2010 Philippine Imported KU570089 D1/FJ/198/2017 Viet Nam Imported MH729968 D3/FJ/079/2010 Philippine Imported KU570090 D1/FJ/221/2017 Fuzhou Indigenous MH729969 D3/FJ/267/2015 Philippine Imported MH729987 D1/FJ/222/2017 Fuzhou Indigenous MH729970 D3/FJ/601/2016 Zhangzhou Indigenous MH729988 D1/FJ/226/2017 Fuzhou Indigenous MH729971 D3/FJ/617/2016 Indonesia Imported MH729989 D2/FJ/091/2007 Putian Indigenous KU570119 D3/FJ/641/2016 Philippine Imported MH729990 D2/FJ/099/2007 Putian Indigenous EU249523 D3/FJ/184/2017 Indonesia Imported MH729991 Note. *The origins of imported DF cases were referred according to field epidemiological investigations. Abrreviation: PNG, Papua New Guinea. Table S2. Summary of sixty-six DENV strains isolated and sequenced in Fujian in 2004–2017 for phylogenetic analysis
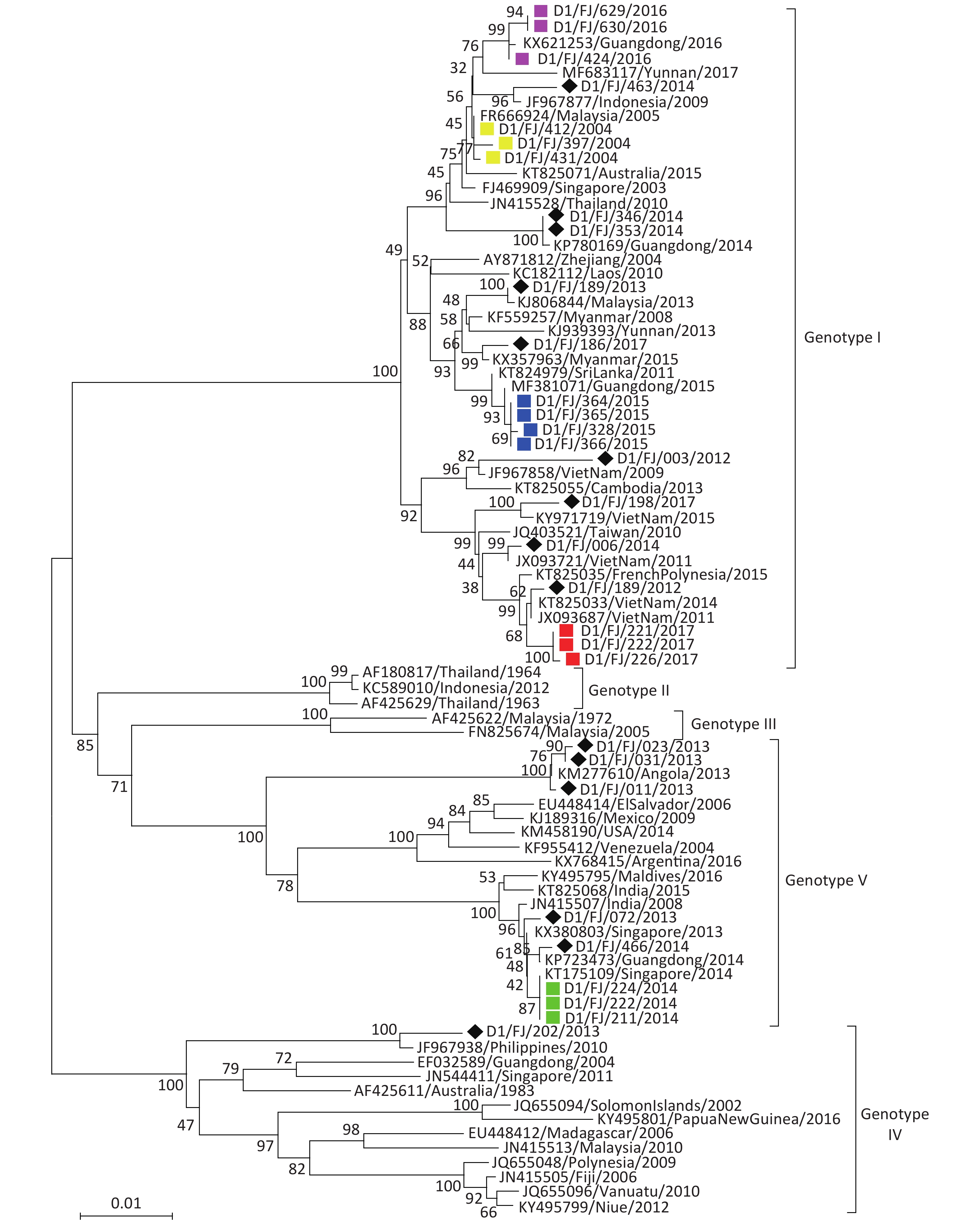
DENV-1 The phylogenetic tree (Figure 3) revealed that the 31 isolates of DENV-1 obtained in this study fall into three genotypes (I, IV, and V). Of the isolates, 22 strains fall into genotype I, 8 into genotype V, and 1 into genotype IV. The tree also indicated that the isolates collected from imported DF cases with DENV-1 infection primarily originated from the countries in Southeast Asia, despite three (D1/FJ/011/2013, D1/FJ/023/2013, and D1/FJ/031/2013) from Angola in Africa and three (D1/FJ/346/2014, D1/FJ/353/2014, and D1/FJ/466/2014) from the adjacent Guangdong province. The results were consistent with the field epidemiological investigations, except the strain D1/FJ/003/2012 that clustered evolutionarily with the JF967858/Viet Nam/2009 strain, but the field investigation suggested that the patient came from Suriname in South America[14]. The speculated reason for this finding was that a similar virus found in Vietnam had transmitted into Suriname and caused the human infection before.
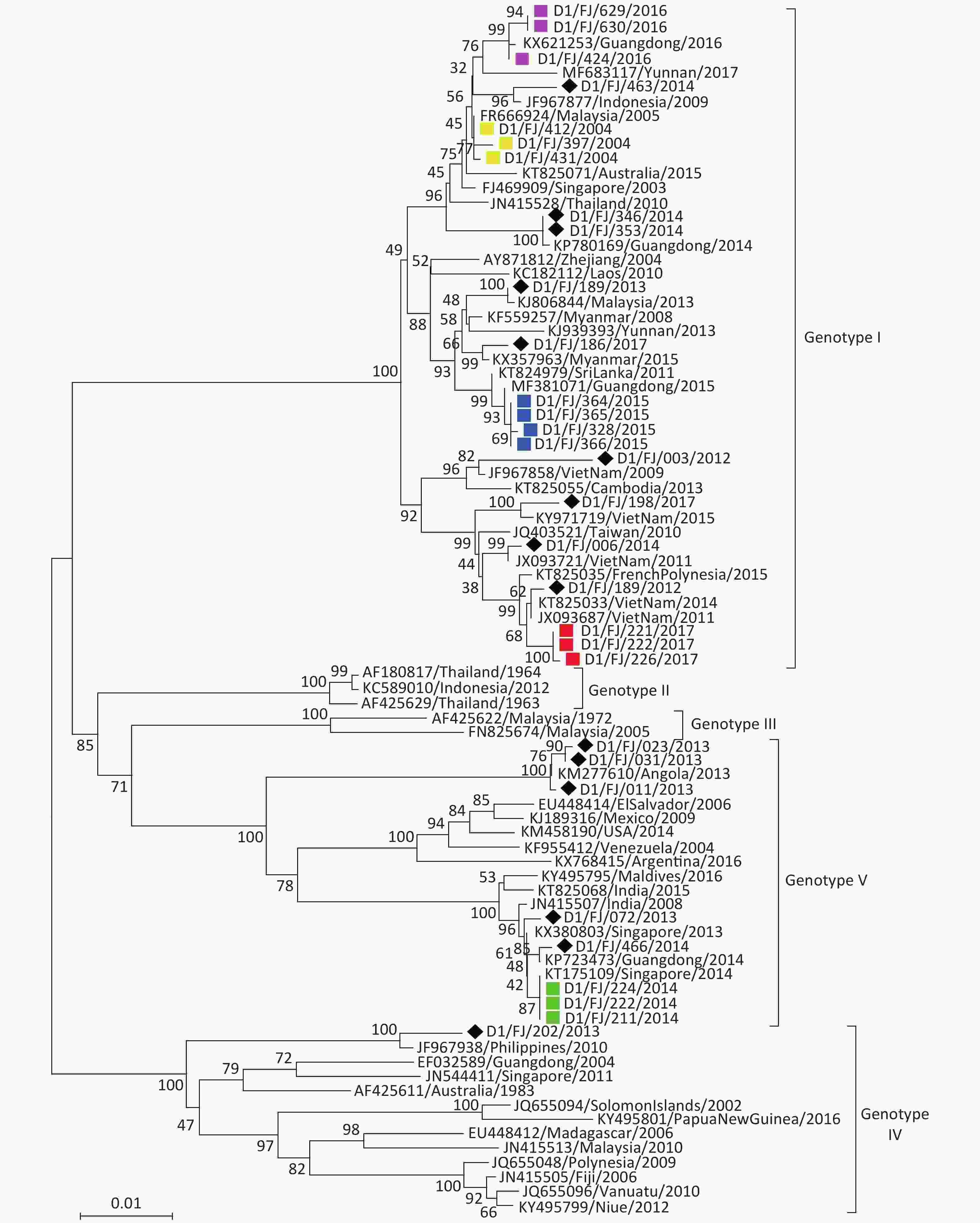
Figure 3. Envelope gene-based phylogenetic tree of DENV-1 isolated in Fujian during 2004–2017. Solid squares in yellow, blue, red, green, and purple indicate the isolates from the outbreaks in Fuzhou (2004, 2015, and 2017) and Nanping (2014 and 2016), respectively. The imported strains are marked with black diamonds.
Of the 16 DENV-1 isolates from indigenous patients in five local outbreaks, 13 belonged to genotype I and the remaining 3 belonged to genotype V. There was less genetic similarity between the isolates from different outbreak events. Instead, the viruses shared a high genetic similarity with the strains found in regions outside of Fujian province. For instance, the DENV-1 isolates (Figure 3) from the local outbreak in Nanping city in 2014 shared 100% sequence identity with the KT175109/Singapore/2014 strain, and the isolates from the outbreak in Fuzhou in 2004 shared 99.7% sequence identity with the strain FR666924/Malaysia/2005. Therefore, it is speculated that the import of viral agents from Singapore and Malaysia caused the local dissemination and outbreak in Nanping and Fuzhou, respectively. It is worth noting that the DENV-1 isolated from the outbreak in Fuzhou in 2015 clustered with the MF381071/Guangdong/2015 strain and the isolates from the outbreak in Nanping in 2016 clustered with the KX821263/Guangdong/2016 strain. As DENV-1 has continually circulated in Guangdong province for at least 18 years since 1997[15] and outbreaks of DENV-1 had occurred in the same year in Guangdong, these two local outbreaks were therefore caused by the DENV-1 imported from the adjacent Guangdong province.
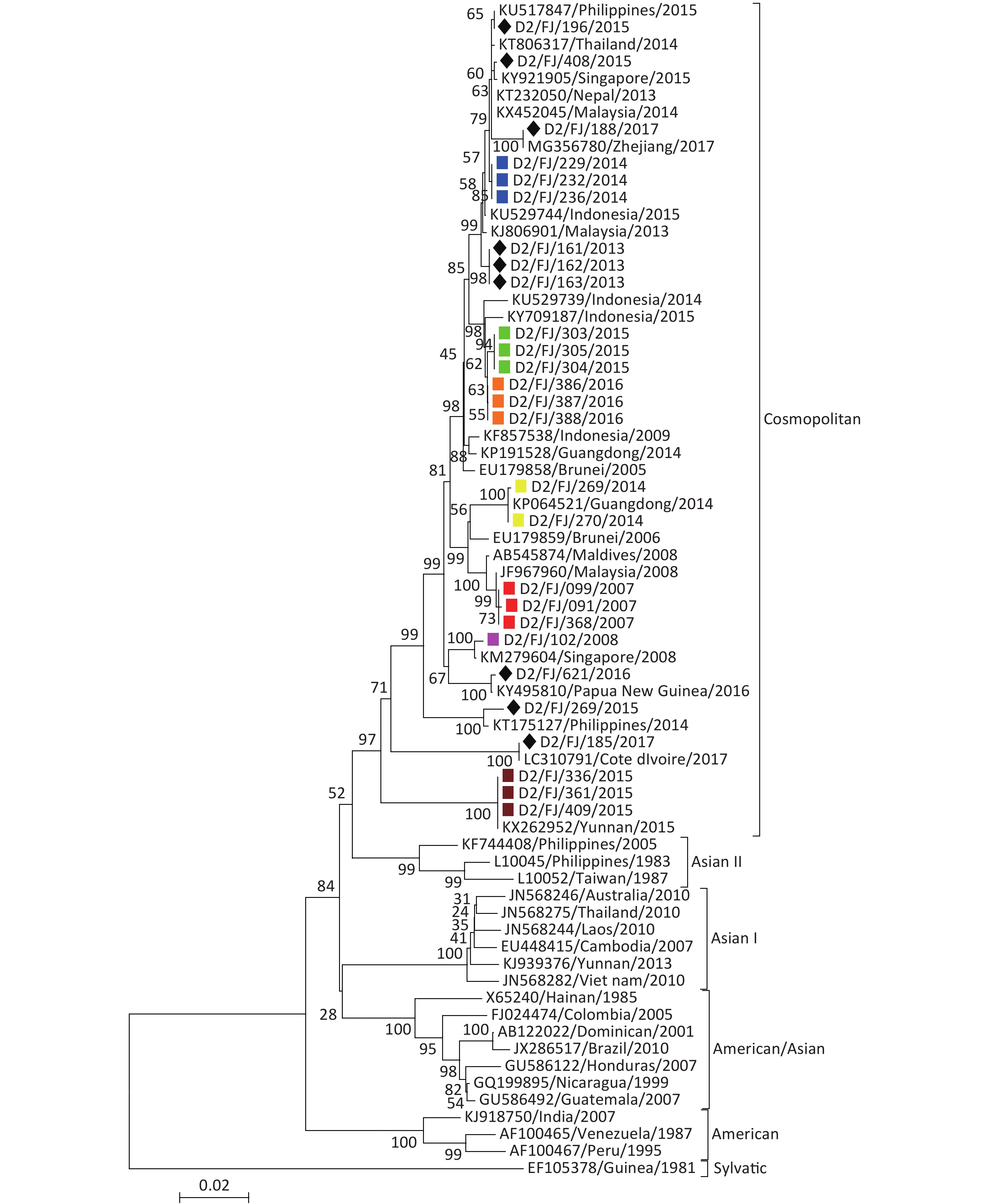
DENV-2 The phylogenetic tree (Figure 4) revealed that all the 27 isolates of DENV-2 belonged to the cosmopolitan genotype. Of the 12 isolates collected from patients with imported DF, 10 clustered with DENV-2 found in Southeast Asia or Oceania, one with the virus in Coate d'lvoire in Africa and one with the virus found in the adjacent Zhejiang province[16]. These results suggested that the DENV-2 imported into Fujian, consistent with the field investigations, primarily originated from Southeast Asia. There were seven local DENV-2 outbreaks in 2007, 2008, 2014, 2015, and 2016 involving three cities (Putian, Fuzhou, and Nanping). Based on the phylogenetic analysis and the sequence similarity, it appears that the strains from the five local outbreak events (Putian in 2007, 2008, 2014, and 2015 and Fuzhou in 2015) originated from Malaysia, Singapore, and Indonesia, respectively. However, the isolates from the local outbreaks in Nanping in 2014 and Fuzhou in 2015 were believed to have been originated from Guangdong and Yunnan provinces, respectively, as the isolates shared 100% sequence identity respectively with the strains found in the two provinces in the same years [15,17].
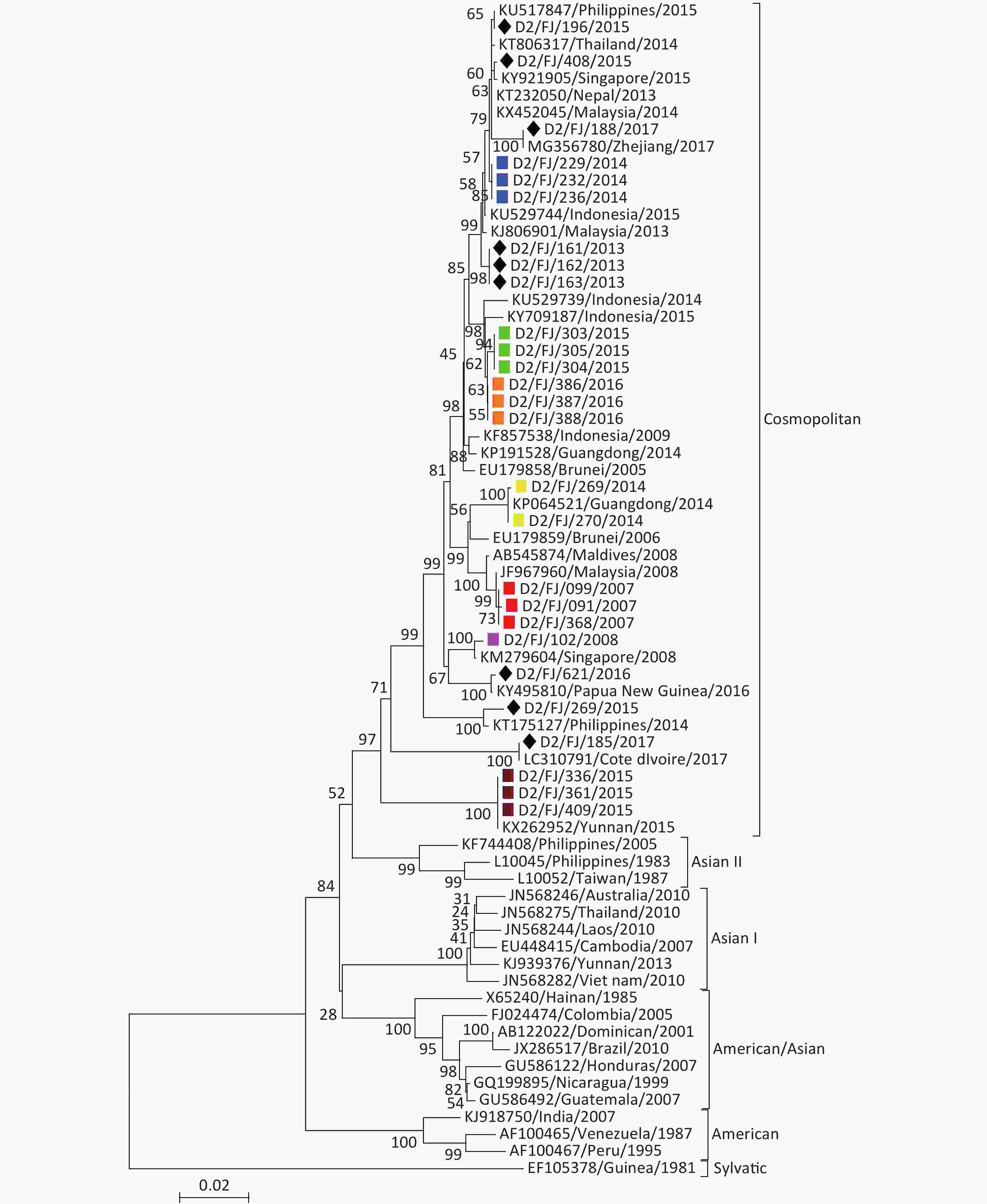
Figure 4. Envelope gene-based phylogenetic tree of DENV-2 isolated in Fujian during 2004–2017. Solid squares in red, purple, blue, green, brown, orange, and yellow indicate the isolates from the outbreaks in Putian (2007, 2008, 2014, and 2015), Fuzhou (2015 and 2016), and Nanping (2014), respectively. The imported strains are marked with a black diamond.
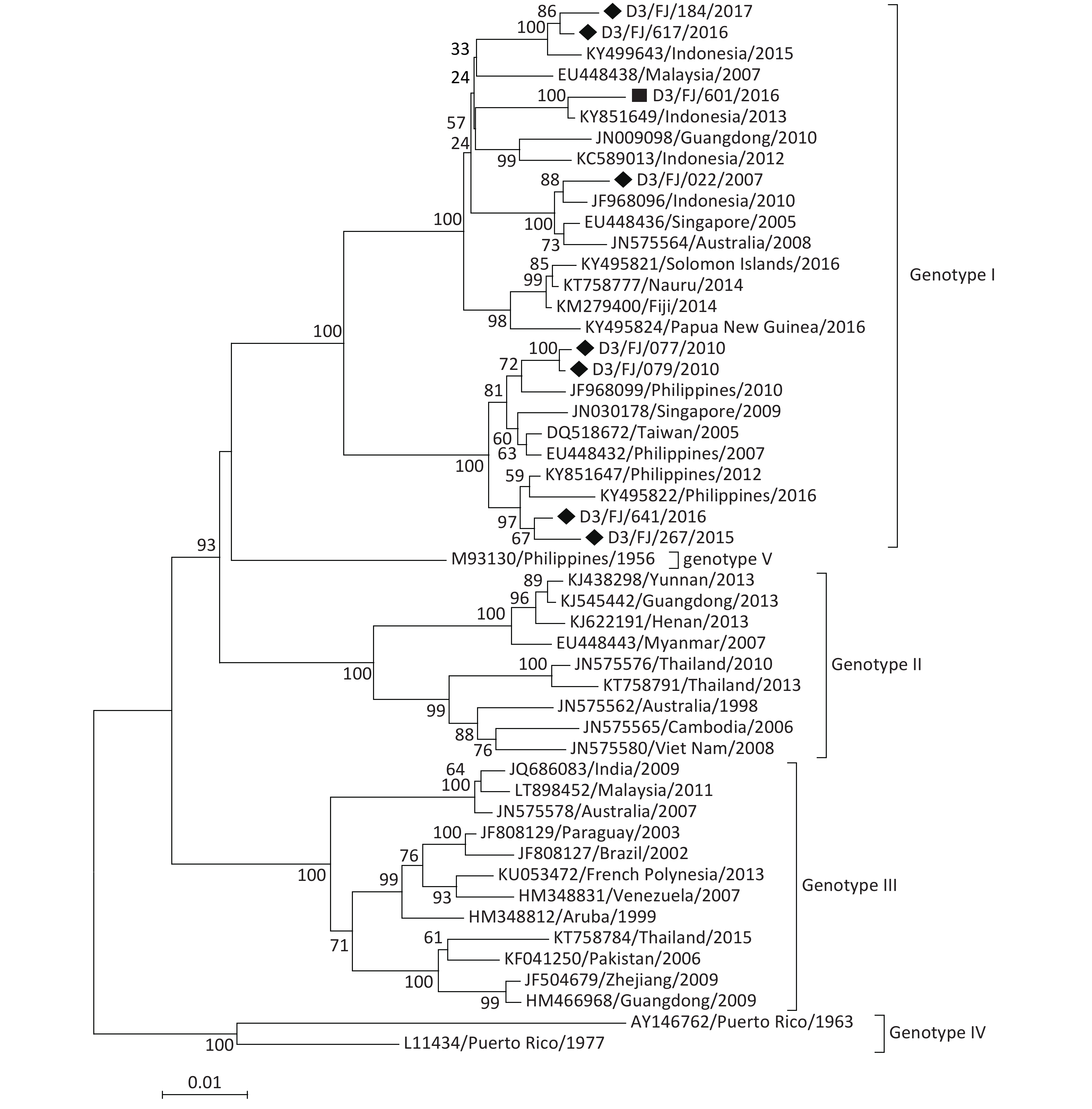
DENV-3 Of the eight DENV-3 isolates, seven originated from the sporadic imported cases and only one originated from the indigenous patient in the local outbreak in Zhangzhou in 2016. The phylogenetic tree (Figure 5) revealed that all the DENV-3 strains belonged to genotype I. Based on the phylogenetic analysis, the isolates originated most probably from the Philippines and Indonesia in Southeast Asia, which was consistent with the field investigations. The only D3/FJ/601/2016 isolate collected from the indigenous patient was believed to have been originated from Indonesia because it shared the highest sequence identity (99.3%) with the strain KY851649/Indonesia/2013.
-
In this study, we have updated the epidemic situation and described the changes in the epidemic status of DF in Fujian province. First, the number of DF cases, either imported or indigenous, reported in Fujian had increased dramatically over the recent years. Especially in 2013–2017, the number of reported DF cases had increased by six-fold compared with that in the past 9 years (2004–2012). Second, despite the fact that most of the imported cases originated from the traditional Southeast Asian regions, the original regions from where the DF cases were imported had gradually expanded. Combined with the results of the phylogenetic analysis, the field investigations demonstrated that an increasing number of cases originated from the regions of South Asia, Africa, America, and Oceania. In addition, since 2013, the imported cases originated more frequently from the surrounding provinces in China where the local DF outbreaks occurred[15-17]. Third, sporadic indigenous cases and local outbreaks of DF in Fujian had occurred frequently, especially in 2013–2017, and the DF-affected areas had extended gradually, a situation similar to that in Mainland China[18]. The local outbreaks occurred every year from 2013 onward, and as of 2017, six cities in Fujian had reported indigenous cases and five cities had experience local DF outbreaks. In 2014, Nanping, an inland city, had experienced a large outbreak of DF, which suggested that the disease began to affect inland areas in Fujian province.
Phylogenetic analysis is an useful approach for investigating viral evolution and in classification studies[19,20] and is widely used in pathogen tracking in the transmission of infectious diseases[5] to overcome insufficient information and memorial bias by field investigations. In this study, the phylogenetic analysis revealed the genetic diversity of the DENV strains (DENVs) in Fujian province and provided molecular evidence indicating that the DENVs isolated from either imported or indigenous cases in Fujian were genetically similar to the DENVs isolated from regions outside of the province. In addition, the genetic heterogeneity of indigenous isolates revealed that DF had not been endemic so far in Fujian province. It is reasonable to state that the continuous import of DF cases not only from abroad but also from the surrounding regions in China responded to the ascent of indigenous cases and the frequent local outbreaks in the province in recent years.
Contributed by the continuous and frequent import of DF, the incidence of DF cases has increased with a wider geographic distribution in Fujian in recent years. With the implementation of the Belt and Road strategy, Fujian will confront with an increasing number of imported DF cases. In addition, in the expanding regions, the cases that are imported easily could result in diversity of the viral agent, which would threaten the clinical outcome of the patients. Therefore, one important measure for the prevention of DF is to focus on the early recognition and clinical management of imported cases not only from abroad but also from domestic regions. As the mosquito vector of Aedes albopictus is distributed widely in Fujian due to the suitable climate[21], it risks Fujian the potential endemic of DF and easily results in large outbreaks similar to the episode that occurred in Guangdong province in 2014[22]. Therefore, effective mosquito control is another important measure for DF control in the province.
-
DF is still considered as an imported epidemic disease in Fujian province. The frequent importation of DF cases from not only abroad, especially Southeast Asian countries, but also from the domestic surrounding provinces had resulted in the increased incidence, frequent local outbreaks, and expansion of distribution in Fujian in recent years. The new situation of the epidemic has challenged the control of the disease in the province. Early recognition and management of imported cases and effective mosquito control measures are crucial for DF control in Fujian province.
-
No conflict of interest to declare.
-
WANG Jin Zhang participated in study design, experiment performance, data analysis, and manuscript writing. YOU Li Bin, KAN Nai Peng, and LIN Qi assisted in experiment performance and data processing. WENG Yu Wei and ZHENG Kui Cheng were involved in study design, discussion of results, and manuscript revision. All authors had read and approved the final manuscript.
Frequent Import and Multiple Sources of Dengue Fever have Changed the Epidemic Situation of the Disease in Fujian Province, China
doi: 10.3967/bes2020.016
- Received Date: 2019-04-16
- Accepted Date: 2019-09-06
-
Key words:
- Dengue fever /
- Epidemic /
- Dengue virus /
- Phylogenetic analysis
Abstract:
| Citation: | WANG Jin Zhang, YOU Li Bin, KAN Nai Peng, LIN Qi, WENG Yu Wei, ZHENG Kui Cheng. Frequent Import and Multiple Sources of Dengue Fever have Changed the Epidemic Situation of the Disease in Fujian Province, China[J]. Biomedical and Environmental Sciences, 2020, 33(2): 123-132. doi: 10.3967/bes2020.016 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: