-
Respiratory infections are associated with high morbidity and mortality and are a major global health problem[1]. Acute respiratory infections are caused by multiple respiratory pathogens, including viruses and bacteria. Viral-bacterial co-infections, which have become increasingly common and a global concern, can lead to substantial complications, causing higher morbidity and adverse prognosis[2]. Previous studies have reported low positive detection rates of targeted pathogens related to acute respiratory infections, owing to the limited number of detected pathogens and variations in the sensitivity of diagnostic methods[3-4]. Low positive detection rates may impede our understanding of respiratory pathogen characteristics and hamper the development of precise treatment and prevention strategies.
In China, surveillance of severe acute respiratory infections (SARIs) is used to track influenza activity and identify pathogens in hospitalized patients[5-6]. To understand the etiological and epidemiological characteristics of SARIs and inform treatment and prevention strategies, we conducted a retrospective analysis during a period of increased respiratory illnesses in Beijing, China, in the autumn and winter of 2023, from 10 sentinel hospitals across 10 districts in Beijing (Supplementary Table S1). We examined the etiology and co-infection patterns of patients with SARI using targeted next-generation sequencing (tNGS). Oropharyngeal swab samples were collected to detect multiple pathogens using tNGS. A total of 107 pathogens were detected in the swab samples, including 74 viral and 33 bacterial pathogens (consisting of eight species of Mycoplasma and Chlamydia), which was higher than that found in previous studies[7]. Detailed descriptions of the pathogen tests are provided in Supplementary Table S2.
During the autumn and winter of 2023, 635 hospitalized patients were enrolled in the study, and their samples were tested using the SARI surveillance system. The median age of the enrolled patients was 41 years (interquartile range: 0.5–95.0 years) and age groups were created as follows: children (≤ 5 years old; 12.3%, 78/635), adolescents (6–17 years old; 27.2%, 173/635), adults (18–59 years old; 18.6%, 118/635), and older adults (≥ 60 years old; 41.9%, 266/635)[7]. Most patients were male (53.7%), and 572 (90.1%) were tested positive for at least one pathogen. The positive pathogen detection rate was 89.7% (70/78) among children, 92.5% (160/173) among adolescents, 89.0% (105/118) among adults, and 89.1% (237/266) among older adults, demonstrating a significantly improved detection rate compared to previous studies[3-4], which were not statistically significant (P > 0.05). The positive pathogen detection rate did not significantly differ between sexes, and was 53.0% (303/572) among males and 47.0% (269/572) among females (P > 0.05). The rate of resistance to macrolide antibiotics was 18.5% (106/572) among all patients, with 17.1% (12/70) among children, 52.5% (84/160) among adolescents, 3.8% (4/105) among adults, and 2.5% (6/237) among older adults. No pertussis resistance genes were detected in any of these cases.
Of the 572 patients who were detected positive for at least one bacterial or viral pathogen, 1,113 pathogens were detected, indicating the presence of a virus, bacteria, or co-infection. Sixteen bacterial species were identified. Stenotrophomonas maltophilia had the highest positivity rate (30.8%, 223/724), followed by Mycoplasma pneumoniae (19.6%, 142/724), Acinetobacter baumannii (9.7%, 70/724), Haemophilus influenzae (8.4%, 61/724), Klebsiella pneumoniae (6.9%, 50/724), and Pseudomonas aeruginosa (6.9%, 50/724) (Figure 1). In total, 30 viral species were identified. The most frequently identified viral pathogens were the Epstein-Barr (EB) virus (20.6%, 80/389), followed by HSV-1 (16.2%, 63/389), IFV-A (10.5%, 41/389), RV (7.2%, 28/389), HAdV-3 (5.4%, 21/389), and RSV-B (5.4%, 21/389).
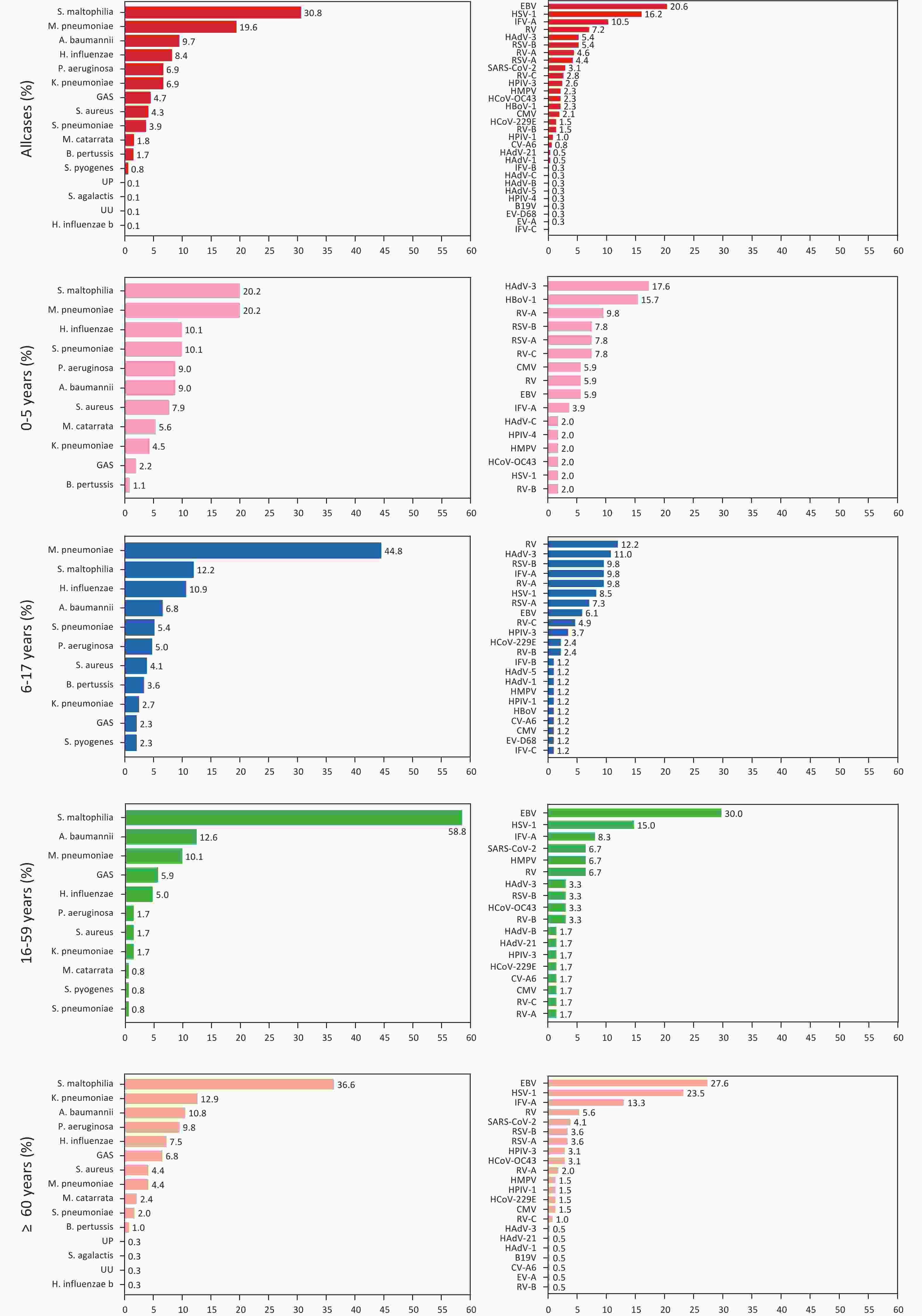
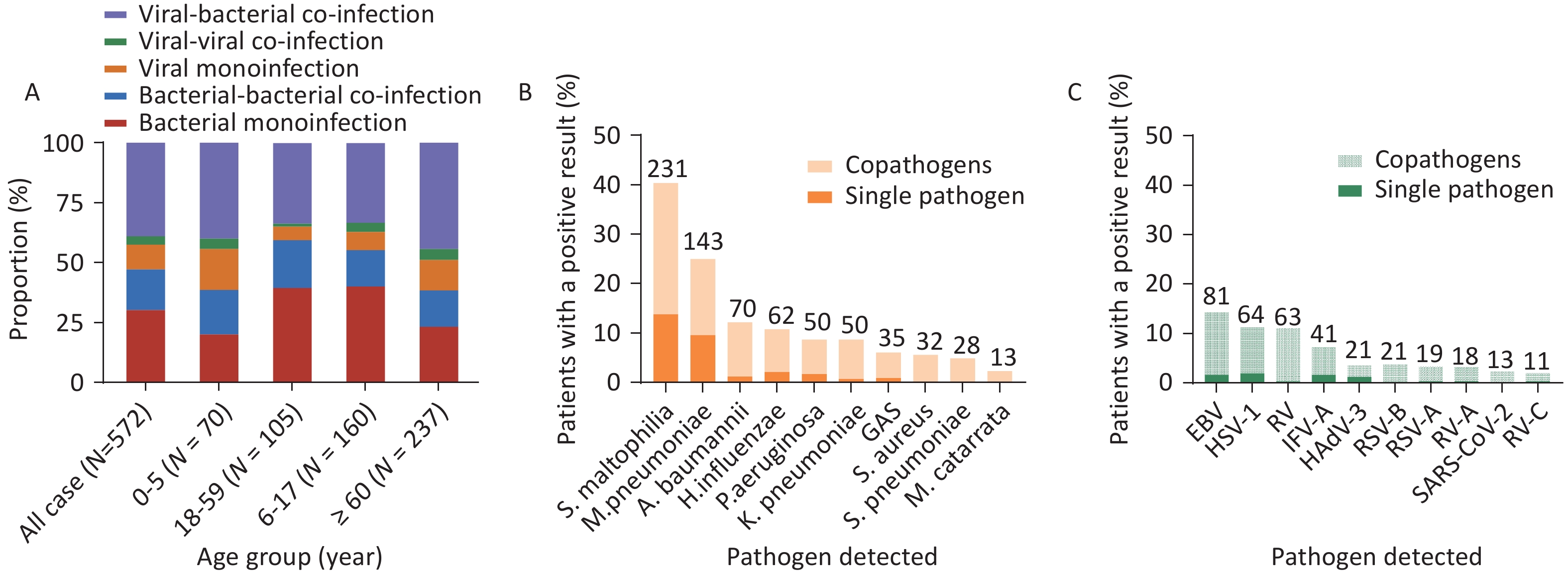
Figure 1. Composition of bacterial and viral pathogens in patients with severe acute respiratory infections (SARIs) across different age groups.
Eleven bacterial and 16 viral species were observed in the 0–5 year age group. The pathogens with the highest positivity rates were S. maltophilia (20.2%, 18/89) and M. pneumoniae (20.2%, 18/89), followed by Streptococcus pneumoniae (10.1%, 9/89), H. influenzae (10.1%, 9/89), A. baumannii (9.0%, 8/89), and P. aeruginosa (9.0%, 8/89). The most commonly detected virus was HAdV-3 (17.6%, 9/51), followed by HBoV-1 (15.7%, 8/51), RV-A (9.8%, 5/51), RSV-B (7.8%, 4/51), RSV-A (7.8%, 4/51), and RV-C (7.8%, 4/51) (Figure 1). Among adolescents, 11 bacterial and 22 viral species were detected. The pathogen with the highest positivity rate was M. pneumoniae (44.8%, 99/221), which was significantly higher than that of other pathogens. Furthermore, S. maltophilia had a positivity rate of 12.2% (27/221), followed by H. influenzae (10.9%, 24/221), A. baumannii (6.8%, 15/221), S. pneumoniae (5.4%, 12/221), and P. aeruginosa (5.0%, 11/221). The most commonly detected viral species was the human rhinovirus (12.2%, 10/82), followed by HAdV-3 (11.0%, 9/82), RSV-B (9.8%, 8/82), IFV-A (9.8%, 8/82), RV-A (9.8%), and HSV-1 (8.5%, 7/82) (Figure 1). Among adults, 11 bacterial and 18 viral species were detected. The pathogen with the highest positivity rate was S. pneumoniae (58.8%; 70/119), followed by A. baumannii (12.6%; 15/119), M. pneumoniae (10.1%; 12/119), Group A streptococcus (GAS) (5.9%; 7/119), H. influenzae (5.0%; 6/119), and P. aeruginosa (1.7%; 2/119). The most commonly detected viral species was the EB virus (30.0%, 18/60), followed by HSV-1 (15%, 9/60), IFV-A (8.3%, 5/60), SARS-CoV-2 (6.7%, 4/60), HMPV (6.7%, 4/60), and RV (6.7%, 4/60) (Figure 1). Among older adults, 15 bacterial and 22 viral species were detected. The pathogens with the highest positivity rate was S. maltophilia (36.6%, 108/295), followed by K. pneumoniae (12.9%, 38/295), A. baumannii (10.8%, 32/295), P. aeruginosa (9.8%, 29/295), H. influenzae (7.5%, 22/295), GAS (6.8%, 20/295), and Staphylococcus aureus (4.4%, 13/295). EB was the most commonly detected virus (27.6%, 54/196), followed by HSV-1 (23.5%, 46/196), IFV-A (13.3%, 26/196), RV (5.6%, 11/196), SARS-CoV-2 (4.1%, 8/196), and RSV-B (3.6%, 7/196) (Figure 1). Among children and adolescents, the higher number of respiratory illnesses were attributed to known pathogens, which is consistent with the results of previous studies[9].
To identify the infection and co-infection patterns of patients with SARI, we analyzed bacterial and viral single and co-infections in all participants and across all age groups. Among the 572 patients, a pathogen was detected in 232 patients (40.5%), including 173 patients with single bacterial infections (30.2%) and 59 patients with single viral infections (10.3%). Co-infections were identified in 340 patients (59.4%), with 223 (39.0%) exhibiting viral-bacterial co-infections, 97 (17.0%) exhibiting bacterial-bacterial co-infections, and 20 (3.5%) exhibiting viral-viral co-infections (Figure 2A). The proportion of viral-viral co-infections was the lowest in the different age groups, followed by single viral infections. Specifically, the proportions of viral-viral co-infections were 4.3% (3/70), 1.3% (2/160), 3.8% (4/105), and 4.6% (11/237) in the four increasing age groups, respectively (Figure 2A). Within the 0–5-year and ≥ 60-year age groups, viral-bacterial co-infections were predominant, accounting for 40.0% (28/70) and 44.3% (105/237), respectively (Figure 2A). However, among the 6–17-year and 18–59-year age groups, single bacterial infection rates were predominant, accounting for 39.4% (63/160) and 40.0% (42/105), respectively. The rates of viral single infections in the 0–5-year (17.1%, 12/70) and the ≥60-year age groups (12.7%, 30/237) were higher than those in the 6–17-year (5.6%, 9/160) and 18–59-year age groups (7.6%, 8/105) (Figure 2A). Viral or bacterial infections increase the likelihood of viral-bacterial co-infections due to the presence of existing infections in the respiratory tract[10].
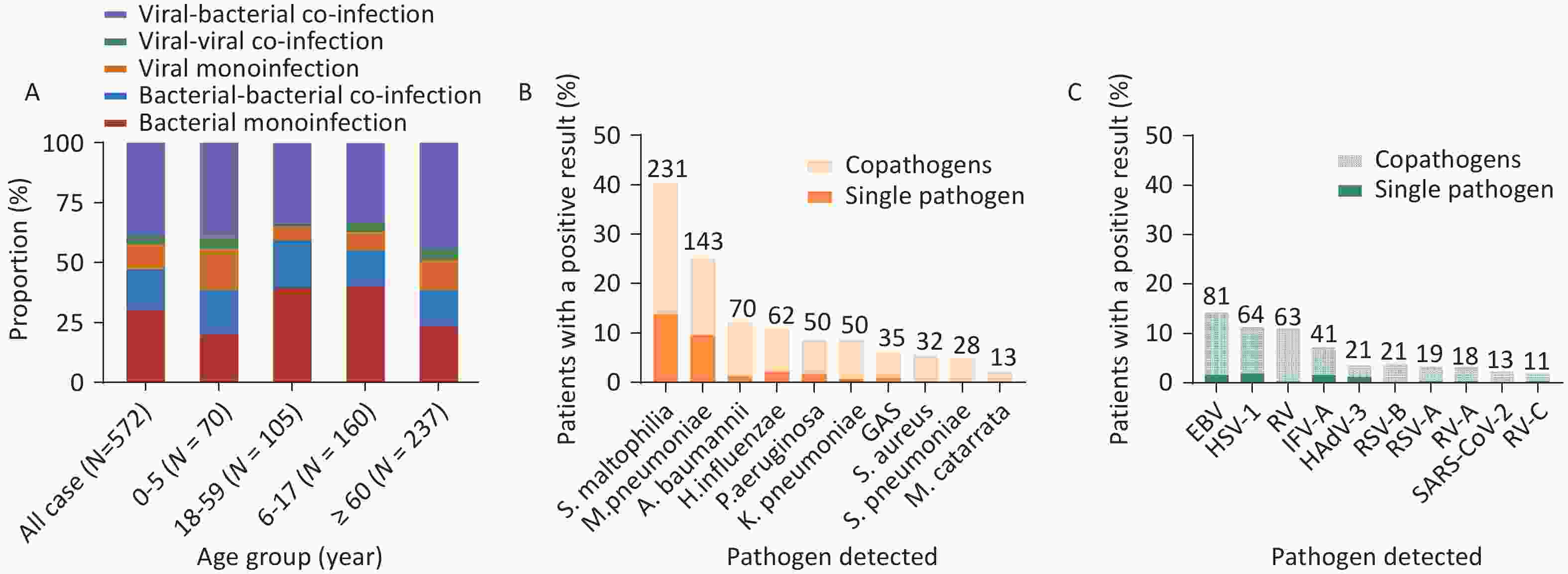
Figure 2. (A) Patterns of single and co-infections in patients with SARIs across different age groups. (B) Specific bacterial pathogens detected as single and co-pathogens. (C) Specific viral pathogens detected as single and co-pathogens.
To analyze single and co-infected pathogens, we selected the top 10 pathogens, including both bacteria and viruses. Among the 572 patients, the most frequently detected pathogen was S. maltophilia, which was detected in 231 (40.4%) patients, with 79 cases of single-pathogen infection (13.8%) and 152 cases of co-pathogen infection (26.6%), which was higher than that of other bacteria. The next most frequently detected pathogen was M. pneumoniae, which was detected in 143 patients (25%), with 55 exhibiting a single-pathogen infection (9.6%) and 88 with co-infections (15.4%) (Figure 2B). Furthermore, A. baumannii ranked third, with 70 infected patients (12.2%), with 1.2% and 11.0% single-pathogen infections and co-infections detected, respectively. Notably, S. aureus (n = 32, 5.6%), S. pneumoniae (n = 28, 4.9%), and Moraxella catarrhalis (n = 13, 2.3%) were detected only in co-infections with other pathogens (Figure 2B).
The EB virus was most frequently detected in both single infections (n = 9, 1.6%) and co-infections (n = 72, 12.6%), totaling 81 cases (14.2%). HSV-1 was detected in 11.2% (n = 64) of the patients, with 11 single-pathogen infections (1.9%) and 53 co-infections (9.3%). RV accounted for 11.0% (n = 63) of the patients, with two single-pathogen infections (0.3%) and 61 co-infections (10.7%). IFV-A accounted for 7.2% (n = 41) of the patients, including nine single-pathogen infections (1.6%) and 32 co-infections (5.6%). All human SARS-CoV-2 infections (n = 13, 2.3%) were co-infections. Co-infections were predominant among the viral pathogens, especially the EB virus, HSV-1, and RV (Figure 2C).
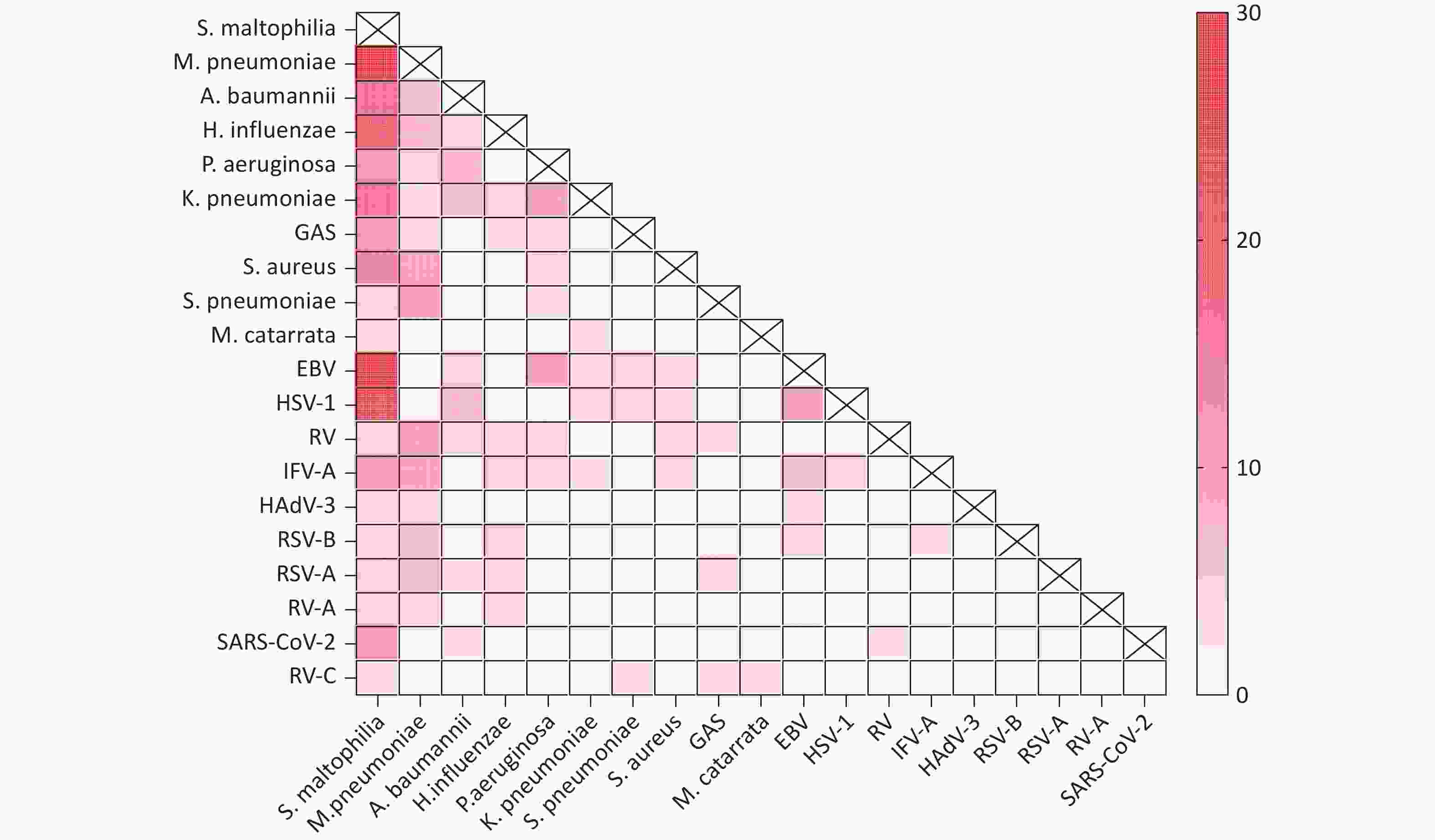
The most common bacterial co-infections were S. maltophilia and M. pneumoniae (n = 27), S. maltophilia and H. influenzae (n = 16), S. maltophilia and A. baumannii (n = 14), and S. maltophilia and K. pneumoniae (n = 14). The most common viral-bacterial co-infections were S. maltophilia, EB virus, and HSV-1. The number of co-infections was the highest for S. maltophilia and the EB virus (n = 25), followed by S. maltophilia and HSV-1 (n = 21), S. maltophilia and IFV-A (n = 9), and S. maltophilia and SARS-CoV-2 (n = 9). Viral-viral co-infections were primarily detected for the EB virus and HSV-1 (n = 8), the EB virus and IFV-A (n = 5), and HSV-1 and IFV-A (n = 4) (Figure 3).
Our study had some limitations. First, the surveillance interval was short and our pathogen samples were limited to those collected between October and November 2023. Second, the number of patients with SARI was relatively small, which could have introduced a selection bias. Third, the sample characteristics at baseline, such as vaccination status, drug use, and treatment outcomes, had some missing data. Fourth, tNGS may have caused sample contamination and potential false positives.
In conclusion, we assessed the spectrum of respiratory infectious pathogens in patients with SARI and the co-infection patterns among different age groups. These findings could help identify the predominant respiratory pathogens and improve diagnosis, treatment, and prevention.
Co-Circulation of Respiratory Pathogens that Cause Severe Acute Respiratory Infections during the Autumn and Winter of 2023 in Beijing, China
doi: 10.3967/bes2025.050
- Received Date: 2024-12-22
- Accepted Date: 2025-02-25
The authors declare no competing interests.
&These authors contributed equally to this work.
| Citation: | Jingzhi Li, Da Huo, Daitao Zhang, Jiachen Zhao, Chunna Ma, Dan Wu, Peng Yang, Quanyi Wang, Zhaomin Feng. Co-Circulation of Respiratory Pathogens that Cause Severe Acute Respiratory Infections during the Autumn and Winter of 2023 in Beijing, China[J]. Biomedical and Environmental Sciences. doi: 10.3967/bes2025.050 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: