-
Cancer is a major threat to human health worldwide. Colorectal cancer (CRC), a highly prevalent malignant tumor, poses a significant public health challenge. Therefore, the identification of effective biomarkers is of great significance[1]. The NFKBIE gene encodes an inhibitor of nuclear factor κBε (IkBε). IκBε, a key regulator of the NF-κB signaling pathway, is closely associated with tumorigenesis. However, their roles in CRC remain unclear[2]. Pan-cancer research is crucial for accelerating the identification of biomarkers and translational medical research, as it can reveal molecular commonalities and differences among different tumor types[3]. Thus, this study aimed to explore the role of NFKBIE in CRC using pan-cancer analysis to provide a basis for the diagnosis and treatment of CRC.
In this study, based on CRC RNA-sequencing data from The Cancer Genome Atlas (TCGA) database, we used the DESeq2 software package to screen for differentially expressed genes (DEGs) between tumor and normal tissues. Subsequently, the “survival” package was used to obtain prognosis-related genes of CRC, and immune-related genes were retrieved from the ImmPort database. By integrating upregulated DEGs, prognosis-related genes, and immune-related genes, and conducting a literature-based screening, NFKBIE was ultimately determined as the research target. Next, we analyzed NFKBIE expression in normal and pan-cancer tissues using the Human Protein Atlas (HPA), TCGA, and Genotype-Tissue Expression (GTEx) databases. The relationships between NFKBIE expression and pan-cancer diagnosis, prognosis, and tumor immunity were analyzed using receiver operating characteristic (ROC) curves, Kaplan–Meier curves, and correlation analyses. Finally, we analyzed the expression of NFKBIE in CRC using data from TCGA, Gene Expression Profiling Interactive Analysis 2 (GEPIA2), HPA, and University of Alabama at Birmingham Cancer Data Analysis Portal (UALCAN) databases, along with clinical samples, and explored its association with the clinicopathological features of patients with CRC. Using functional enrichment, immune correlation, and single-cell analyses, we elucidated the specific functions of NFKBIE in CRC.
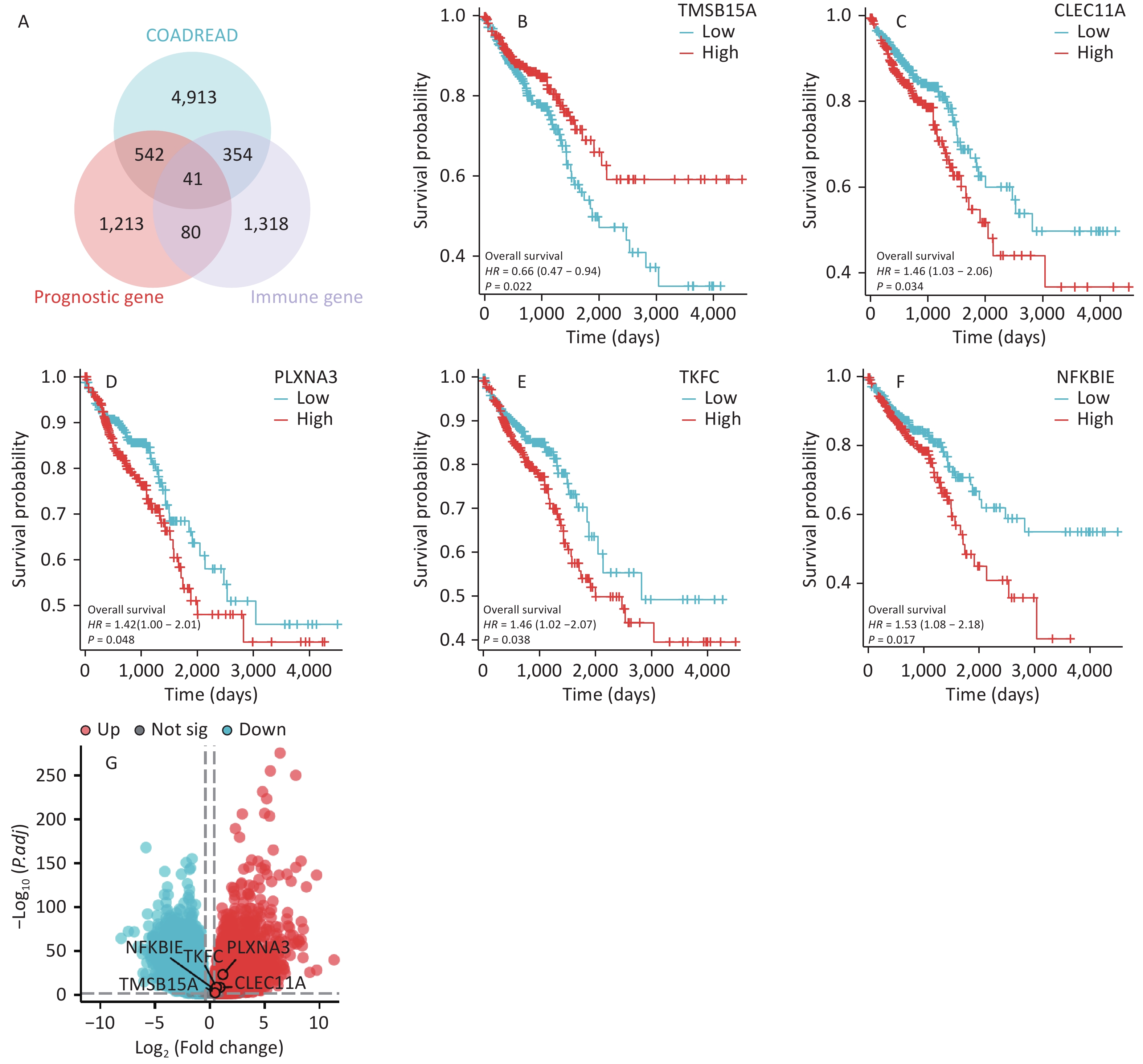
Based on the DEGs, prognosis-related genes, and immune-related genes in CRC, 41 overlapping genes were identified (Figure 1A). Through a literature review, five genes that have not yet been studied in CRC were identified. We conducted survival prognosis and differential volcano plot analyses for the five genes in patients with CRC. The results showed that high expression of TMSB15A was beneficial for the prognosis of patients with CRC (P < 0.05) (Figure 1B), whereas high expression of CLEC11A, PLXNA3, TKFC, and NFKBIE was unfavorable for the prognosis of patients with CRC (P < 0.05) (Figure 1C–F). In addition, all five genes were upregulated in CRC (Figure 1G). Comprehensive analysis indicated that NFKBIE was related to the prognosis of CRC and was closely associated with cancer development. Therefore, NFKBIE was selected as the research target.
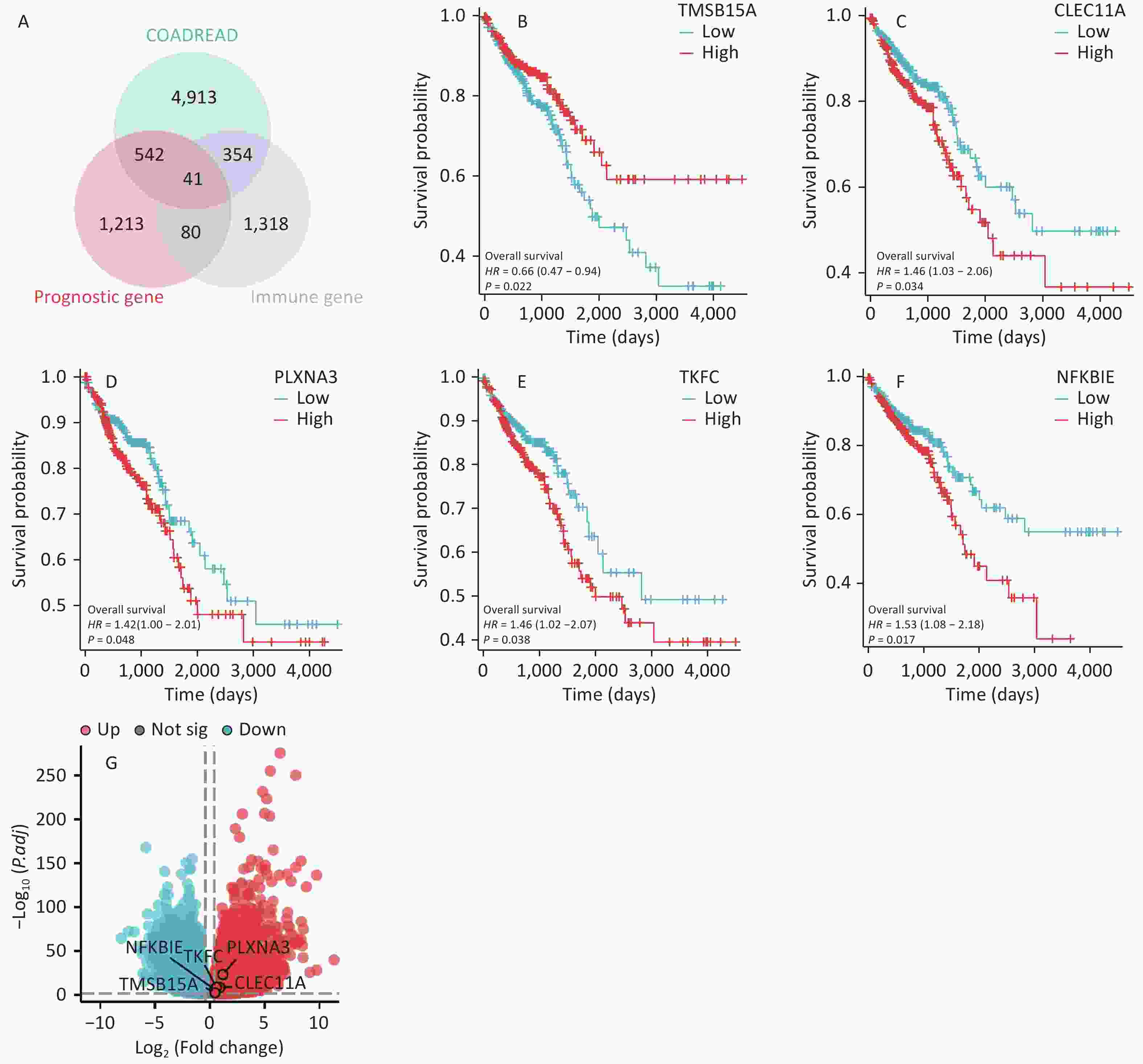
Figure 1. Screening of target genes in CRC. (A) Venn diagram of upregulated DEGs, prognosis-related genes, and immune-related genes in CRC. (B–F) Prognostic analysis of the five target genes in CRC. (G) Volcano plot of the five target genes in CRC. P < 0.05 is considered statistically significant. CRC, colorectal cancer; DEGs, differentially expressed genes.
Pan-cancer analysis revealed that NFKBIE mRNA and protein were mainly expressed in the bone marrow, lymphoid tissues, and digestive tract (Supplementary Figure S1A). Based on pan-cancer expression data from TCGA and GTEx databases, we observed that NFKBIE was abnormally upregulated in most tumors, especially in colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) (P < 0.05) (Supplementary Figure S1B–D). The ROC analysis results showed that the area under the curve (AUC) of NFKBIE was > 0.7 in 19 types of cancers, including COAD and READ (Supplementary Figure S2A–S). This indicates that NFKBIE is a promising diagnostic biomarker for many cancers. In addition, survival analysis results showed that increased NFKBIE expression was associated with poor prognosis in patients with COAD, glioblastoma multiforme, kidney renal papillary cell carcinoma (KIRP), and liver hepatocellular carcinoma, but with good prognosis in patients with breast invasive carcinoma and skin cutaneous melanoma (P < 0.05) (Supplementary Figure S3A, D–I). High NFKBIE expression was a risk factor for disease-specific survival in patients with COAD and KIRP (P < 0.05) (Supplementary Figure S3B). Increased NFKBIE expression was closely associated with poor progression-free intervals) in kidney renal clear cell carcinoma and KIRP (P < 0.05) (Supplementary Figure S3C). These findings suggest that NFKBIE has a good prognostic predictive value in multiple cancers. Furthermore, immune correlation analysis indicated that NFKBIE was positively correlated with the expression of most immune-infiltrating cells and immune checkpoints in pan-cancer, especially B cells and macrophages (Supplementary Figure S4A–D). This suggests that NFKBIE expression is closely related to immune response in tumors.
Therefore, we conducted a systematic analysis to explore the role of NFKBIE in CRC. Based on TCGA data and paired and unpaired differential analyses, we observed that NFKBIE was highly expressed in CRC (P < 0.001) (Figure 2A–B). Analyses of the GEPIA2 database and qPCR experiments demonstrated that NFKBIE was highly expressed in CRC (P < 0.05) (Figure 2C–D). In addition, analyses of the HPA and UALCAN databases revealed that NFKBIE was highly expressed in CRC tissues (Figure 2E–F). Subsequently, based on the CRC data from TCGA, we plotted an ROC curve, and the results showed an AUC of 0.837 (Figure 2G), indicating that NFKBIE may be a diagnostic biomarker for CRC. Finally, survival analysis showed that high NFKBIE expression was unfavorable for the prognosis of patients with CRC (P < 0.05) (Figure 2H–I). To further analyze the relationship between NFKBIE and the clinicopathological features of patients with CRC, we used the chi-square test, two-class logistic model, Wilcoxon rank-sum test, and Kruskal–Wallis rank-sum test. The results showed that upregulated NFKBIE expression was significantly associated with clinicopathological parameters (sex, age, and lymphatic invasion) (P < 0.05) (Supplementary Figure S5A–C, Supplementary Tables S1–2). Compared with the normal control group, NFKBIE was more highly expressed at the T, N, and M stages (Supplementary Figure S5D–F), suggesting that NFKBIE may be involved in CRC progression. In the pathological stage II, lymphatic invasion N0, and N-stage N1 subgroups, the overall survival of patients with CRC decreased as NFKBIE expression increased (Supplementary Figure S5G–I), further indicating the potential value of NFKBIE as a prognostic biomarker. Moreover, univariate and multivariate regression analyses showed that NFKBIE expression, N stage, pathological stage, age, lymphatic invasion, and residual tumors were independent risk factors affecting the prognosis of patients with CRC (Supplementary Table S3).
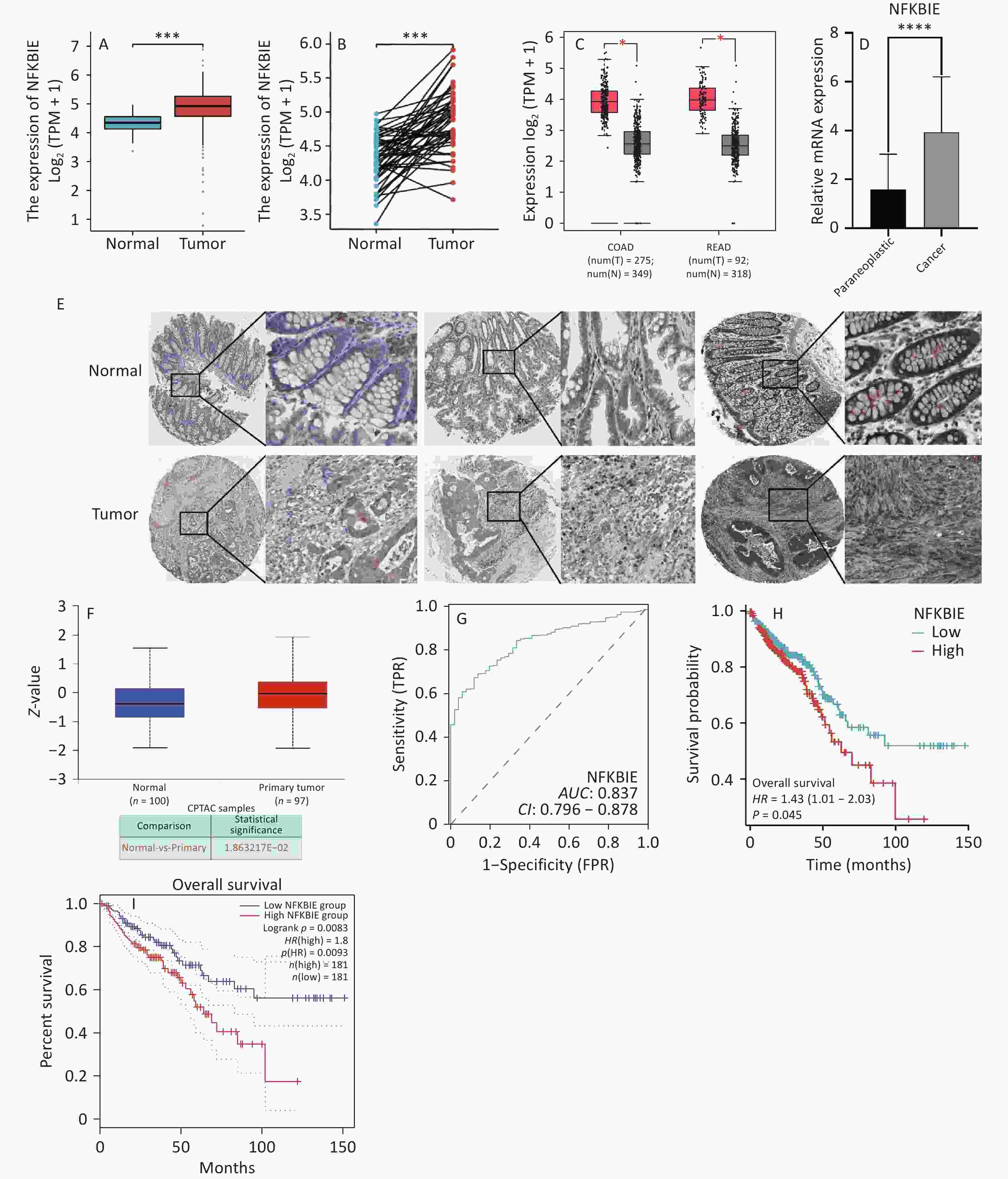
Figure 2. Expression, diagnostic efficacy, and survival prognosis of NFKBIE in CRC. (A–B) Unpaired and paired expressions of NFKBIE in TCGA. (C) Expression of NFKBIE in GEPIA2. (D) qPCR analysis of NFKBIE expression. (E) Immunohistochemical image of NFKBIE protein in HPA. (F) Expression of NFKBIE protein in UALCAN. (G) ROC curve of NFKBIE. (H) OS of NFKBIE in TCGA. (I) OS of NFKBIE in GEPIA2. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. P < 0.05 is considered statistically significant. CRC, colorectal cancer; DEGs, differentially expressed genes; TCGA, The Cancer Genome Atlas; HPA, Human Protein Atlas; UALCAN, University of Alabama at Birmingham Cancer Data Analysis Portal; GEPIA2, Gene Expression Profiling Interactive Analysis 2ROC, receiver operating characteristic; OS, overall survival.
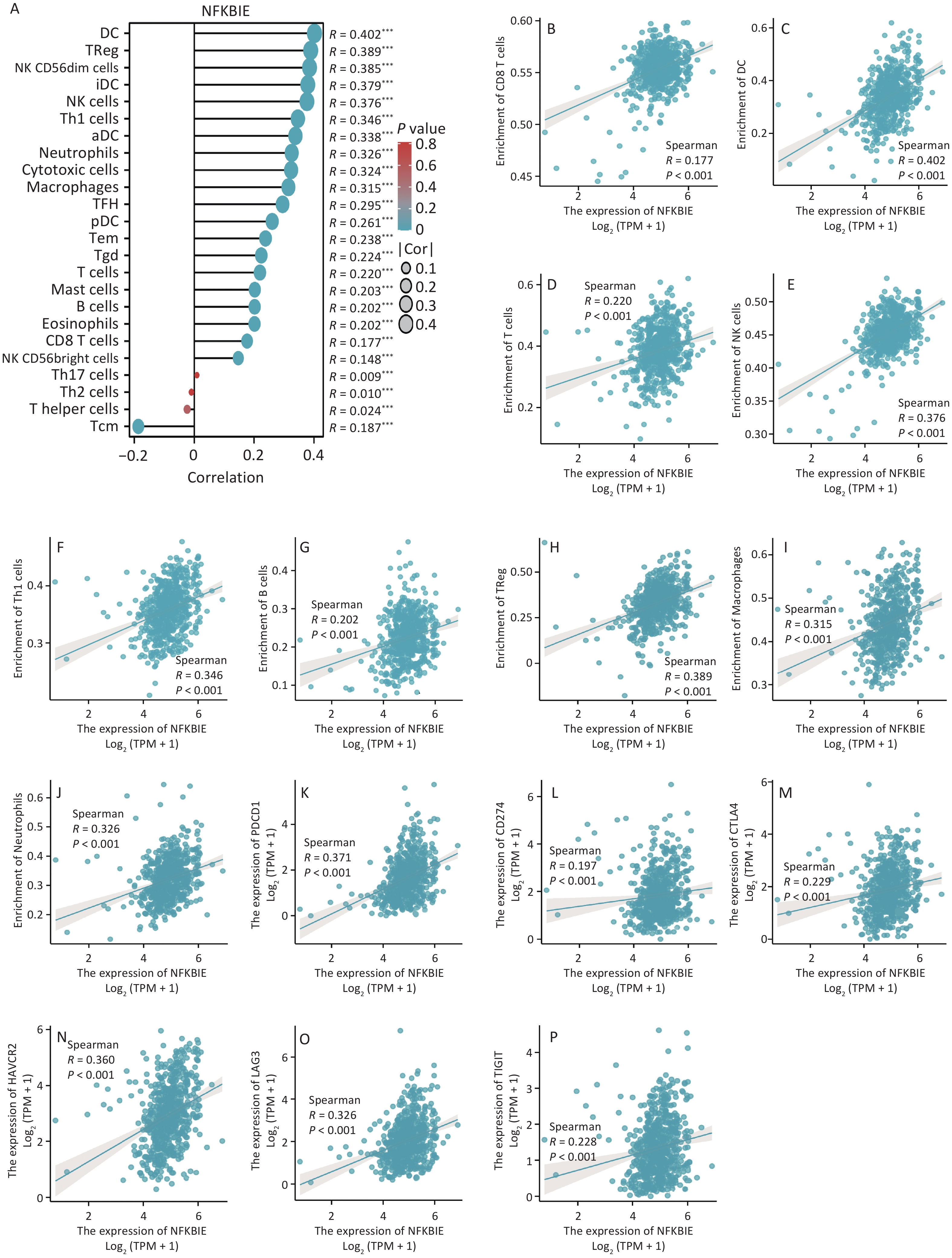
To explore the molecular mechanism of NFKBIE in CRC, we performed an enrichment analysis. First, through correlation analysis, we identified 1,282 genes that were highly correlated with NFKBIE. Subsequently, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. GO analysis revealed that NFKBIE was mainly associated with immune receptor activity (Supplementary Figure S6A). Immune receptors are protein molecules on the surface of immune cells that play crucial roles in initiating and regulating immune responses[4]. KEGG analysis indicated that NFKBIE was primarily enriched in the MAPK signaling pathway (Supplementary Figure S6B). Activation of the MAPK signaling pathway facilitates the expression of genes related to M2-type macrophages[5]. However, M2-type macrophages promote tumor angiogenesis, matrix remodeling, and immunosuppression[6]. To further explore the molecular mechanism of NFKBIE in CRC, we conducted gene set enrichment analysis using its DGEs. The results showed that NFKBIE was enriched in tumor- and immune-related pathways (Supplementary Figure S6C–E). Subsequently, when exploring the relationship between NFKBIE and immune-infiltrating cells and immune checkpoints in CRC, we observed that NFKBIE expression was positively correlated with most immune cells and checkpoints, especially macrophages (Figure 3A–P). Moreover, in samples with high NFKBIE expression, we observed an increase in the number of macrophages (Supplementary Figure S7A–I). Finally, single-cell sequencing demonstrated that NFKBIE was mainly distributed in the CRC macrophages (Supplementary Figure S8). Through comprehensive analysis, we observed that NFKBIE may promote the polarization of macrophages to the M2 type and assist tumor cells in immune escape through multiple pathways, thereby exacerbating CRC progression.
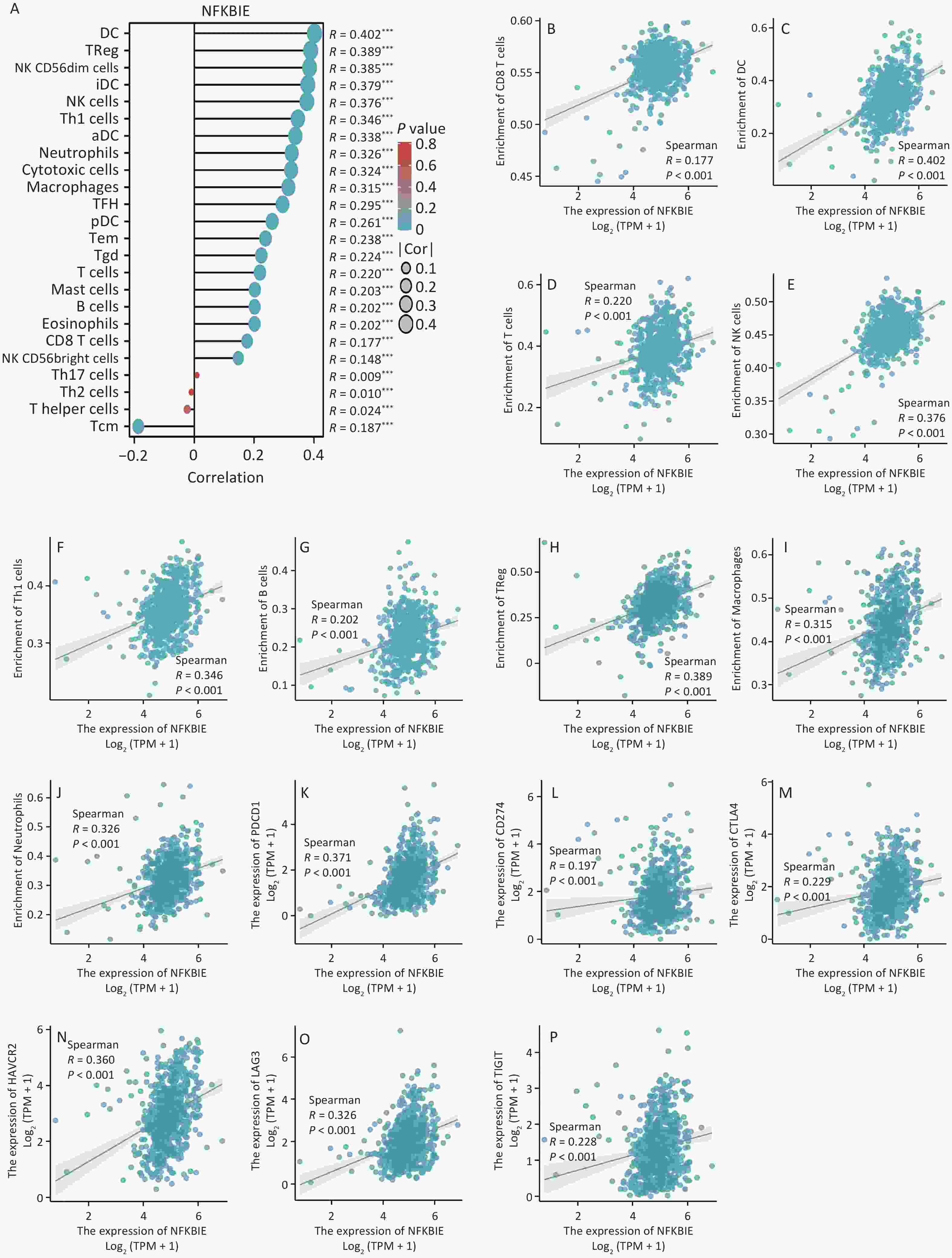
Figure 3. Immune correlation analysis of NFKBIE. (A) Correlation between NFKBIE expression and 24 types of immune-infiltrating cells. (B–J) Scatter plots of the correlation between NFKBIE expression and the number of immune-infiltrating cells. (K–P) Scatter plots showing the correlation between NFKBIE expression and immune checkpoints. *P < 0.05, **P < 0.01, ***P < 0.001. NS: P ≥ 0.05. P < 0.05 is considered statistically significant.
In summary, NFKBIE plays important diagnostic, prognostic, and immunological roles in pan-cancer and holds promise as a novel biomarker related to CRC diagnosis, prognosis, and immunity. This study relied mainly on bioinformatics analysis of public databases and lacked functional verification in in vitro and in vivo. Thus, we were unable to directly confirm the regulatory effect of NFKBIE on immune cell infiltration or its specific molecular mechanism. Future research should verify the impact of NFKBIE on macrophage polarization using CRC cell models with NFKBIE knockdown or overexpression. These findings provide a solid basis for the translational application of NFKBIE as an immunotherapeutic target.
NFKBIE: Novel Biomarkers for Diagnosis, Prognosis, and Immunity in Colorectal Cancer: Insights from Pan-cancer Analysis
doi: 10.3967/bes2025.124
- Received Date: 2025-05-08
- Accepted Date: 2025-09-11
The authors declare that they have no competing interests.
This study was approved by the Ethics Committee of the First Affiliated Hospital of Hebei North University (Approval No. K2024147).
&These authors contributed equally to this work.
| Citation: | Chenyang Hou, Peng Wang, Fengxu Yan, Yanyan Bo, Zhenpeng Zhu, Xiran Wang, Shan Liu, Dandan Xu, JiaJia Xiao, Jun Xue, Fei Guo, Qingxue Meng, Rensen Ran, Weizheng Liang. NFKBIE: Novel Biomarkers for Diagnosis, Prognosis, and Immunity in Colorectal Cancer: Insights from Pan-cancer Analysis[J]. Biomedical and Environmental Sciences. doi: 10.3967/bes2025.124 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: