-
Skin cutaneous melanoma (SKCM), a highly invasive malignant tumor originating from skin melanocytes, poses a significant threat to public health[1,2]. Its development is closely associated with multiple factors, such as ultraviolet radiation, gene mutations, and immune escape. Among these, imbalance in the immune surveillance and clearance of tumor cells is a crucial link to disease progression[3,4]. Tripartite motif-containing 27, which belongs to the TRIM protein family and is encoded by the TRIM27 gene, contains the RING, B-box, and coiled-coil domains. It participates in biological processes such as cell-cycle regulation, signal transduction, and immune response mainly by modifying target proteins through ubiquitination. Notably, increasing evidence indicates that TRIM27 is closely associated with the tumor immune microenvironment and contributes to cancer immune escape via multiple mechanisms, thereby promoting tumor development[5]. However, the role of TRIM27 in SKCM remains unclear, thus prompting our investigation to elucidate this.
In this study, prognostic genes of SKCM were obtained from The Cancer Genome Atlas (TCGA) database using the “survival” (v3.3.1) package, whereas immunity-related genes were retrieved from the ImmPort database. The two gene sets were intersected to identify overlapping genes, which were then analyzed in terms of their differential expression, diagnostic efficiency, and survival prognosis value using the University of California, Santa Cruz XENA (UCSC XENA, https://xenabrowser.net/datapages/) database. On the basis of comprehensive comparisons and a literature review that revealed TRIM27 is closely related to cancer development, TRIM27 was selected as the research subject. Next, the Gene Expression Omnibus (GEO) and Human Protein Atlas (HPA) databases were used to analyze the mRNA and protein expression levels of TRIM27 in normal and SKCM tissues. Then, the impact of TRIM27 expression on patient survival was investigated using receiver operating characteristic (ROC) curve and survival prognosis analyses based on the SKCM data extracted from the GEO, TCGA, and Gene Expression Profiling Interactive Analysis 2 (GEPAI2, http://gepia2.cancer-pku.cn/#index) databases. Additionally, the correlations between TRIM27 expression and clinicopathological features were analyzed on the basis of TCGA-SKCM data using the stats (v.4.2.1) and car (v.3.1.0) packages of R software. Independent variables affecting the prognosis of patients with SKCM were screened using multivariate Cox regression analysis, and a prognostic nomogram model was constructed using the survival (v.3.3.1) and rms (v.8.0.0) packages. Finally, the specific functions of TRIM27 in SKCM were comprehensively explored using functional enrichment, immune correlation, single-cell, gene variation, and drug sensitivity analyses.
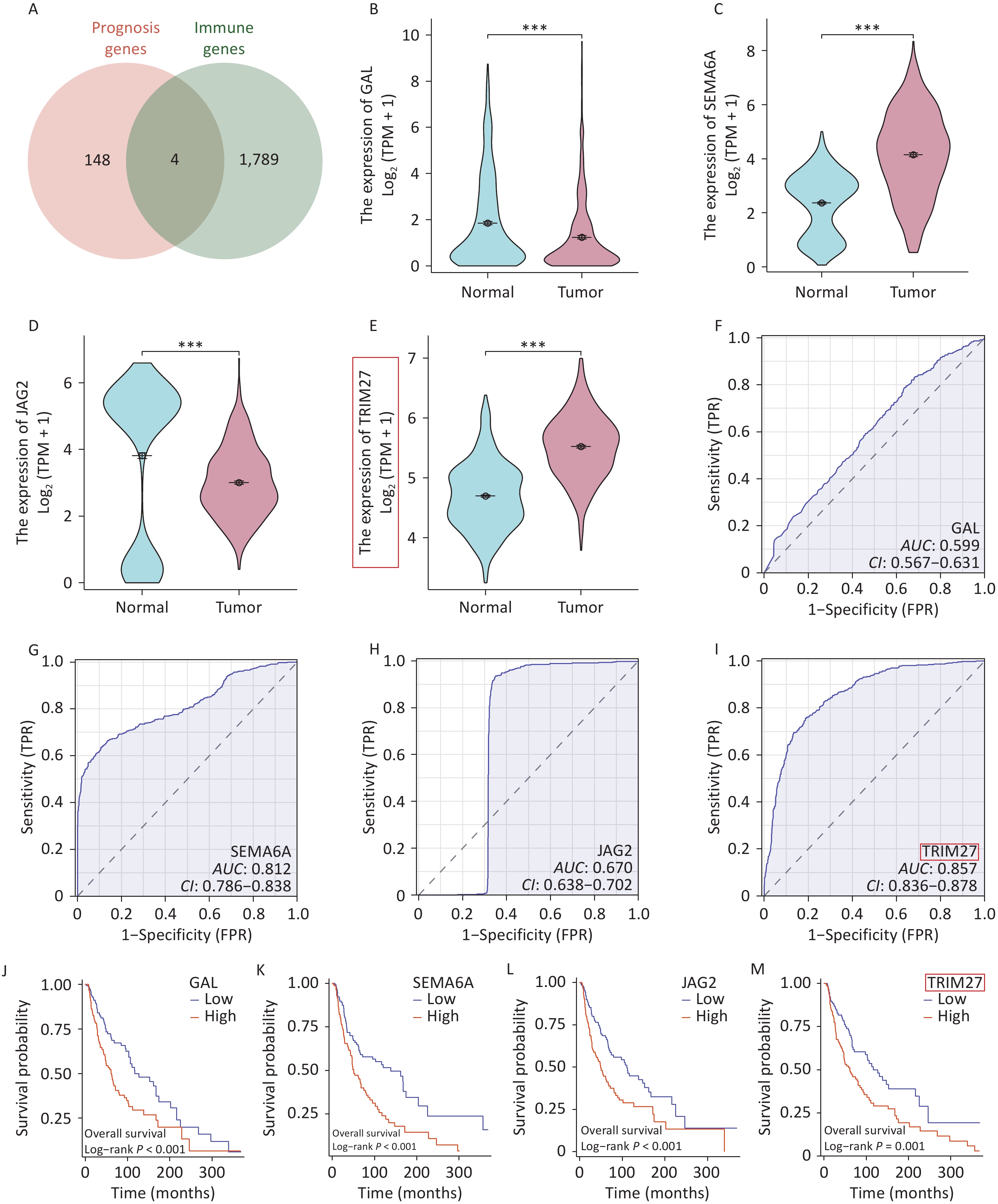
Through intersection of the ImmPort and TCGA datasets, four overlapping genes were identified: GAL, SEMA6A, JAG2, and TRIM27 (Figure 1A) (Supplementary Table S1). Intergroup differential expression, ROC, and survival prognosis analyses of these genes using UCSC XENA data showed that TRIM27 was highly expressed in SKCM, which was unfavorable for patient prognosis, and it had strong diagnostic value (Figure 1B–M). The volcano plot generated from differential expression analysis of the GEO data showed that TRIM27 was upregulated in both the GSE15605 and GSE114445 datasets (Figure 2A–B). Additionally, this gene was highly expressed in the GSE15605 and GSE114445 datasets and had high diagnostic value (P < 0.001) (Figure 2C–F). Survival analysis based on the GEPIA2 and TCGA-SKCM datasets showed that patients with high TRIM27 expression levels had a shorter overall survival (OS) time than that of patients expressing low levels of the protein (P < 0.01) (Figure 2G–H). Finally, analysis using the HPA database revealed that the TRIM27 protein was highly expressed in SKCM tissues and was mainly expressed in the cell nucleus (Figure 2I–M). In conclusion, TRIM27 may serve as both a diagnostic and prognostic biomarker in SKCM.
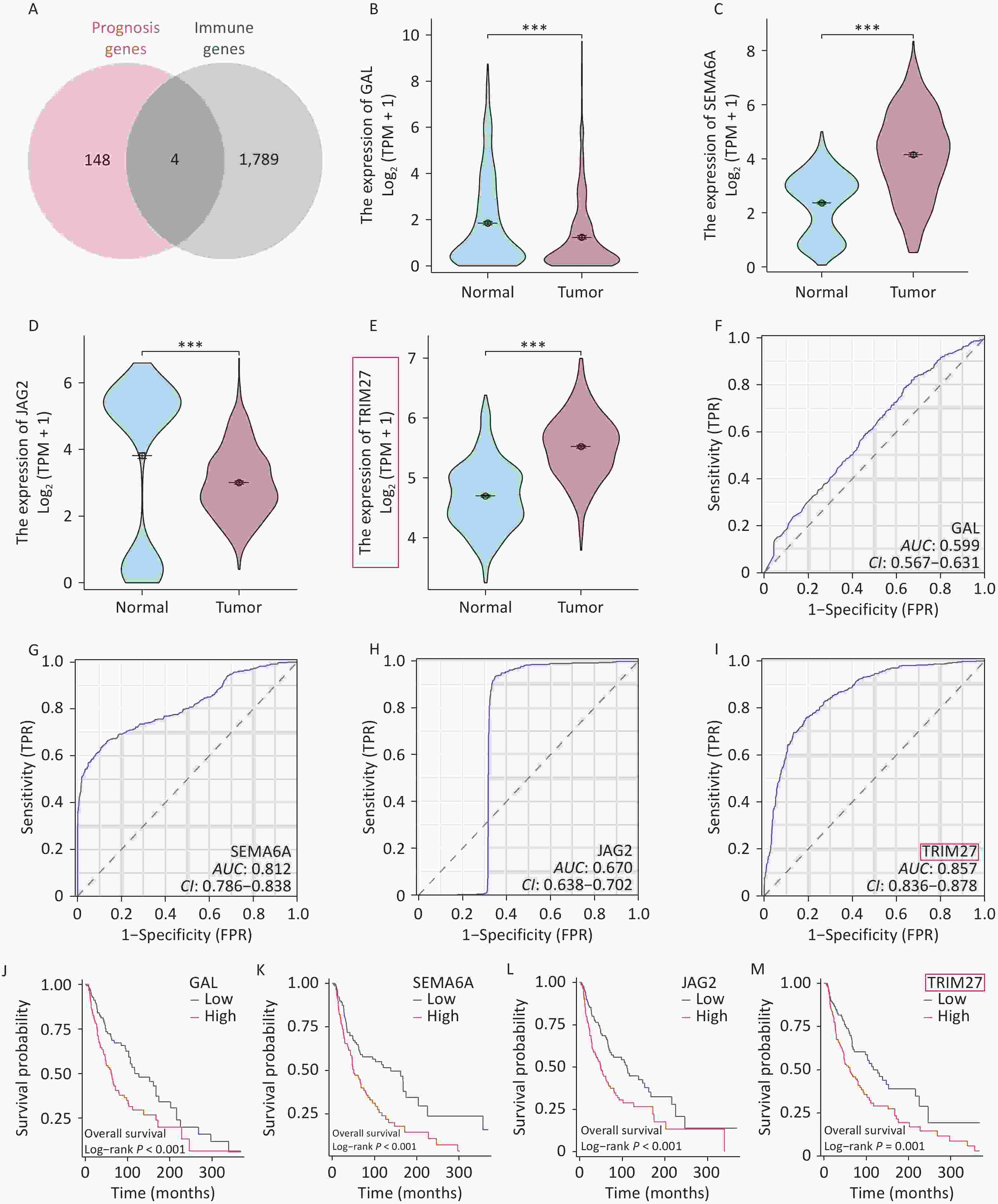
Figure 1. Screening and characterization of prognostic and immunity-related genes in skin cutaneous melanoma (SKCM). (A) Venn diagram of prognostic and immunity-related genes in SKCM. (B–E) Comparison of intergroup expression of overlapping genes in SKCM based on UCSC XENA data. (F–I) ROC curves of overlapping genes in SKCM based on UCSC XENA data. (J–M) Survival curves of overlapping genes in SKCM based on TCGA data. *P < 0.05, ***P < 0.001.
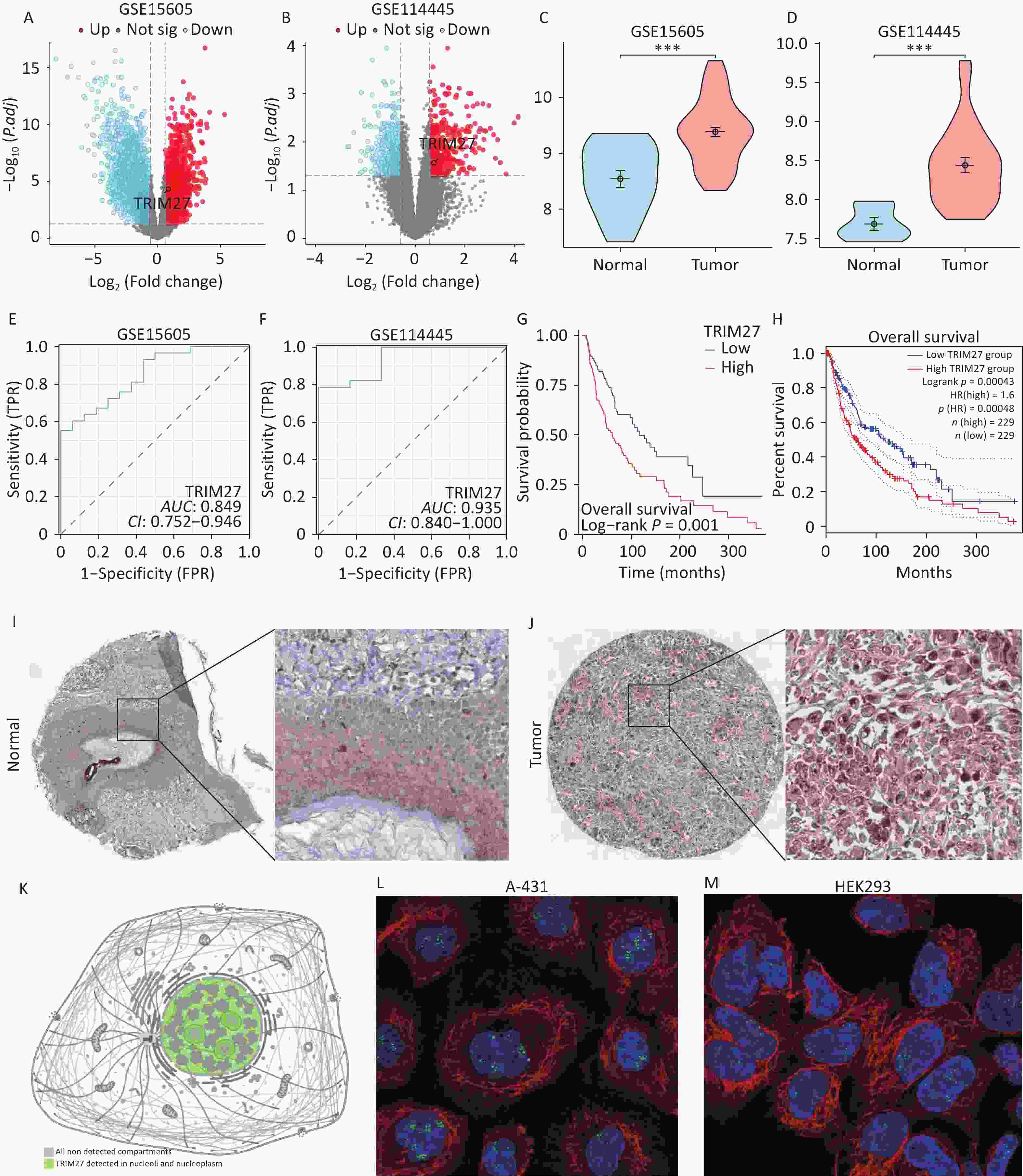
Figure 2. Analyses of the expression and subcellular localization of TRIM27 in SKCM and its diagnostic and prognostic abilities. (A–B) Volcano plots of TRIM27 in the GEO datasets. (C–D) Comparison of the intergroup expression of TRIM27 in the GEO datasets. (E–F) ROC curves of TRIM27 in the GEO datasets. (G) Kaplan-Meier curve of TRIM27 effects in SKCM based on TCGA data. (H) Kaplan-Meier curve of TRIM27 effects in SKCM based on GEPIA2 data. (I–J) Immunohistochemical staining images of TRIM27 in SKCM based on HPA data. (K) Subcellular localization map of TRIM27 based on HPA data. (L–M) Subcellular localization maps of TRIM27 in A-431 and HEK293 cells based on HPA data. *P < 0.05, ***P < 0.001.
To determine the clinical significance of TRIM27, we analyzed the association between its expression levels and patient clinicopathological features. TRIM27 expression was found to be correlated with lymphatic metastasis and the pathological stage (P < 0.05) (Supplementary Figure S1A–B). In the OS event analysis, TRIM27 expression was significantly higher in the death group (Dead) than in the survival group (Alive) (P < 0.001) (Supplementary Figure S1C). Additionally, its expression was significantly increased in patients with disease-specific survival and progression-free interval events (P < 0.001) (Supplementary Figure S1D–E). This indicates that high TRIM27 expression may drive aggressive tumor progression and is associated with malignant phenotypes, such as a reduced survival rate and increased recurrence risk. To further investigate the impacts of TRIM27 expression and clinicopathological parameters on patient survival, univariate and multivariate Cox regression analyses were performed. The results showed that the T stage, N stage, M stage, and TRIM27 expression level were independent risk factors affecting OS (P < 0.05) (Supplementary Table S2). A prognostic nomogram was then constructed on the basis of the independent prognostic factors and TCGA-SKCM data. First, we established a time-dependent survival ROC curve based on TRIM27 expression levels. The results showed that the area under the ROC curve values were higher than 0.50, indicating the suitability of the nomogram model for prognostic prediction (Supplementary Figure S2A–C). The nomogram results showed that a higher total score (obtained by adding the corresponding points of each factor) correlated with lower 1-, 3-, and 5-year survival probabilities for patients (Supplementary Figure S2D), suggesting that this model can provide a reference for individualized clinical prognostic assessment and treatment plan formulation. Next, we verified the accuracy of the prognostic nomogram model using a prognostic calibration curve. The C-index of the prognostic calibration plot was 0.679, and the calibration plot was very close to the diagonal line, indicating good calibration performance (Supplementary Figure S2E–G).
To analyze the molecular mechanism underlying the involvement of TRIM27 in SKCM pathogenesis, we performed functional enrichment, immune correlation, single-cell, gene variation, and drug-sensitivity analyses. First, analyses of the GSE15605 and TCGA datasets revealed 1018 differentially expressed genes (DEGs) between SKCM samples with high and those with low TRIM27 expression (Supplementary Figure S3A and Table S3). The DEGs were annotated using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases and gene set enrichment analysis (GSEA). The GO analysis showed that TRIM27 was mainly associated with “epidermis development” (Supplementary Figure S3B), whereas the KEGG analysis revealed that the gene was significantly enriched in the “cytokine-cytokine receptor interaction” pathway, which is a key hub in cancer immune escape (Supplementary Figure S3C). The abnormal activation of this pathway in the tumor microenvironment drives immunosuppression through multiple mechanisms: (1) Induction of the expansion of immunosuppressive cells: tumor-secreted colony-stimulating factor-1 (CSF-1) and C-C motif ligand 2 (CCL2) recruit monocytes to the tumor site, which differentiate into M2-type macrophages under the action of interleukin (IL)-6/IL-10, thereby inhibiting the function of CD8+T cells[6]. (2) Initiating the T-cell exhaustion program: persistent tumor necrosis factor-alpha (TNF-α) signals can induce T-cell functional exhaustion by activating the nuclear factor-kappa B (NF-κB) pathway[7]. GSEA analysis showed that the “matrisome” pathway was enhanced in samples expressing high TRIM27 levels, whereas the keratinization process, cornified envelope formation, and developmental biology pathways were inhibited (Supplementary Figure S3D). Comprehensive enrichment analysis indicated that TRIM27 may create an immunosuppressive microenvironment through the cytokine network and maintain the undifferentiated state of tumor cells by inhibiting keratinization-related genes. Immune-correlation analysis showed that TRIM27 expression was negatively correlated with the scores of the tumor microenvironment composition, the infiltration of most immune cells, and immune checkpoints (Figure 3A–D). This further suggests that TRIM27 may lead to an immunosuppressive microenvironment, which is consistent with the association between an immunosuppressive microenvironment and poor tumor prognosis in related studies. Moreover, single-cell analysis revealed that TRIM27 was mainly expressed in monocytes in SKCM (Supplementary Figure S4A–E). In the tumor microenvironment, monocytes can be polarized into M2-type macrophages under cytokine induction[8]. Additionally, M2-type macrophages may induce immunosuppression. Finally, we performed gene variation and drug sensitivity analyses of TRIM27. Gene amplification (4.9%) was the main form of TRIM27 variation in SKCM, and the gene was sensitive to several drugs such as BI-2536 and AT-7519 (Supplementary Figure S5A–E).
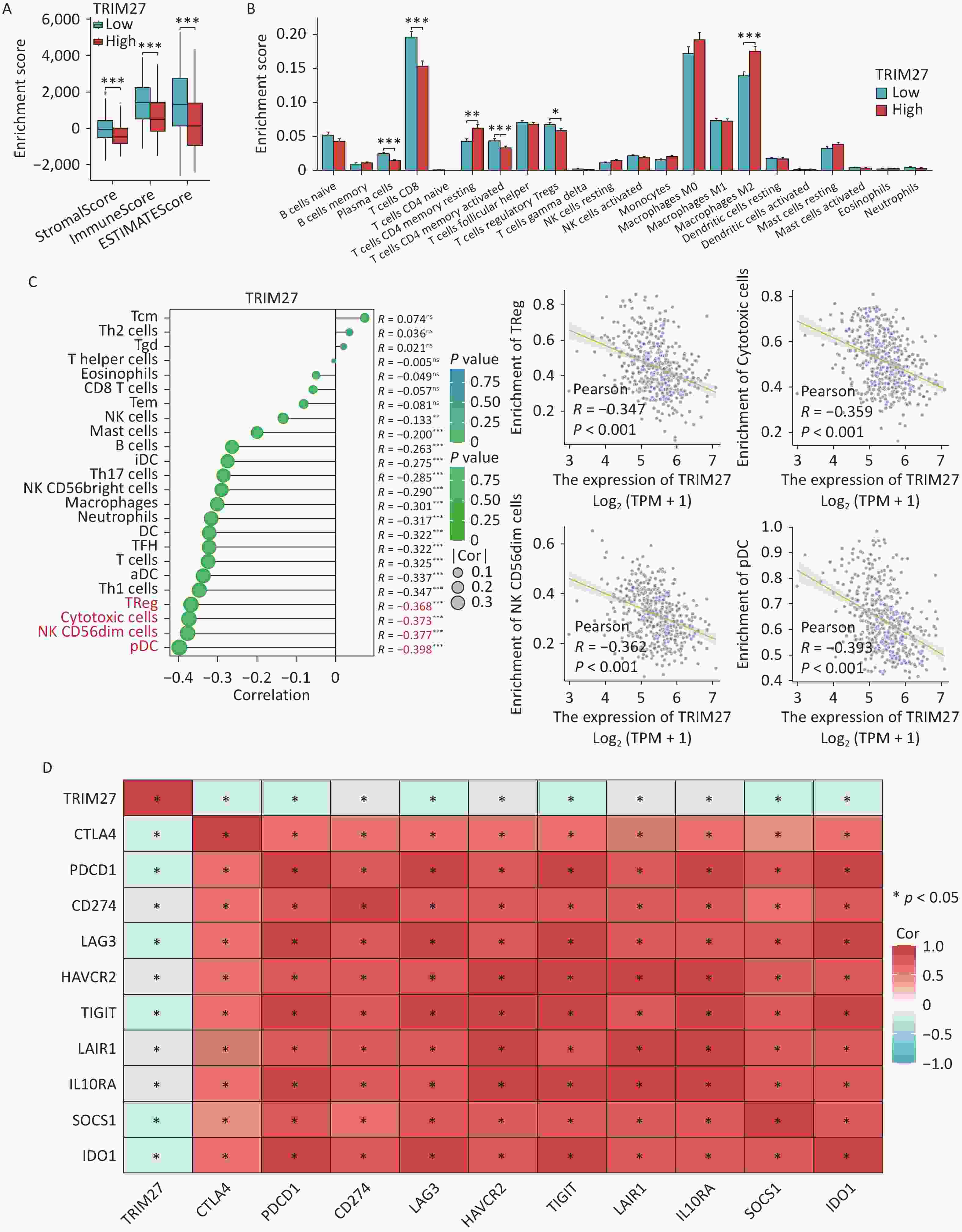
Figure 3. Relationship between TRIM27 and tumor immunity in SKCM. (A) Stromal, immune, and ESTIMATE scores in the high- and low-TRIM27-expression groups. (B) Differences in the proportions of immune cells between the high- and the low-TRIM27-expression groups. (C) Lollipop and scatter plots showing the correlation between TRIM27 expression and immune cell infiltration. (D) Heatmap showing the correlation between TRIM27 expression and immunosuppressive checkpoint genes. *P < 0.05, **P < 0.01, ***P < 0.001.
In summary, TRIM27 is highly expressed in SKCM, is associated with a poor prognosis, and has high diagnostic value. The study findings revealed that TRIM27 may drive the development of SKCM by creating an immunosuppressive microenvironment and inhibiting epidermal differentiation, ultimately leading to a poor prognosis for patients. This suggests that TRIM27 is a promising biomarker for both prognostic assessment and immune regulation and thus provides a new research direction for the precise diagnosis and treatment of SKCM.
TRIM27: A Bifunctional Biomarker for Immune Microenvironment Regulation and Prognostic Assessment in Skin Cutaneous Melanoma
doi: 10.3967/bes2025.158
- Received Date: 2025-07-14
- Accepted Date: 2025-09-19
The authors declare that they have no competing interests.
Not applicable
| Citation: | Weizheng Liang, Chenyang Hou, Fengxu Yan, Yanyan Bo, Shan Liu, Dandan Xu, JiaJia Xiao, Xiran Wang, Fei Guo, Qingxue Meng, Zhongwu Li, Rensen Ran. TRIM27: A Bifunctional Biomarker for Immune Microenvironment Regulation and Prognostic Assessment in Skin Cutaneous Melanoma[J]. Biomedical and Environmental Sciences. doi: 10.3967/bes2025.158 |







 Quick Links
Quick Links
 DownLoad:
DownLoad:

