-
Ionizing radiation is an extremely ubiquitous and significant environmental hazard that has been identified as both public health and national security risks. Exposure to ionizing radiation in daily life is usually low, but radiation accidents and incidents can cause significant exposure[1]. In a large-scale radiological event or nuclear exposure incident, thousands of individuals may be exposed. Rapidly distinguishing exposed from non-exposed individuals and estimating of the doses received by individuals are crucial to guide the appropriate trial and clinical medical treatment[2]. In the absence of physical dosimeter, biological dosimetry can be used to estimate the dose after radiation exposure[3].
In the past few years, gene expression alteration following ionizing radiation exposures has been shown to potentially function as a convenient, rapid and high-throughput radiation biodosimetry in a large-scale radiological emergency[4-6]. The expression levels of different candidate genes were regulated in a dose-and time-dependent manner after exposure to ionizing radiation, which have been identified in whole blood[7], isolated lymphocytes[8], and T cells[9] using microarrays or quantitative real-time polymerase chain reaction (qRT-PCR). Most studies have suggested that gene expression changes in human peripheral blood cells, particularly lymphocytes, can be used as a sensitive tool to assess radiation exposure. Furthermore, high-throughput platforms for peripheral blood RNA isolation combined with 384-well low-density arrays qRT-PCR or DNA microarrays techniques represent a new dimension in high sampling capacity. However, monitoring specific single gene expression to assess the absorbed dose in response to radiation exposure is limited. Therefore, developing a radiation-specific gene expression panel with high reproducibility may be necessary for covering a range of doses, dose-rates, and time-points to improve accuracy and robustness of dose estimation.
In the present study, bibliometric analysis was employed to qualitatively and quantitatively assess global research from 2000 to 2017. The radiation-responsive genes with high reproducibility studied in the inclusive literature were found to possess consistent dose-effect relationships in more than three independent studies. To determine the significant dose-effect relationship of these genes after radiation exposure, we validated the expression of radiation-responsive genes expression in human B lymphoblastoid cell line (AHH-1) at certain time points following exposure to 60Co γ-rays using qRT-PCR.
Data Retrieval Data were obtained from Pubmed/Medline, Embase, and the Web of Science databases. To improve retrieval accuracy, Medical Subject Headings (MeSH) terms were used to search both on the Pubmed/Medline and Embase database. Pubmed/Medline was searched as follows: 'gene expression' [MeSH Terms] or ('gene' [All Fields] and 'expression' [All Fields]) or 'gene expression' [All Fields], and biodosimetry [All Fields]; Embase was searched as follows: 'gene'/exp or gene and ('expression'/exp or expression) and ('biodosimetry'/exp or 'biodosimetry'). For bibliometric analysis, the Web of Science Core Collection is the most widely used source of scientific information. In this study, the terms 'gene expression' and 'biodosimetry' were used to retrieve titles, key words, author information, abstracts, and reference from the Web of Science. The period of publication was limited from 2000 to 2017.
Inclusive and Exclusive Criteria Inclusive and exclusive criteria were as follows: (1) peer-reviewed articles were published and indexed in three databases, including original research articles, meeting abstracts, and proceedings, excluding reviews, editorial material, and books. Citation databases were as follows: Science Citation Index Expanded (1900-present); Social Sciences Citation Index (1990-present); Arts and Humanities Citation Index (1990-present); Conference Proceedings Citation Index-Science (1990-present); Conference Proceedings Citation Index-Social Science and Humanities (1990-present); Emerging Sources Citation Index (2015-present). (2) Inclusion of the publications should include a summary of the information that can provide sufficient information, or wherein full-text information can be obtained. (3) Species were not limited. (4) Repeated publications from different database sources were excluded. (5) Articles that were inconsistent with the purpose of the study were also excluded.
Bibliometric Analysis Results from the Web of Science were exported and analyzed with Microsoft Excel 2010, including the type of literature, annual publication output, journal publication, distribution of publication by subject area, countries and institutions, and top published authors of studies on ionizing radiation-induced gene expression alteration. To further explore the most-cited articles, CiteSpace Ⅵ (64 bits) was used to construct co-citation references network. CiteSpace Ⅵ (64 bits) is a visualization tool that can analyze trends and patterns in scientific literature and detect critical points in the development of a field[10].
Screening the Radiation-responsive Genes All the ionizing radiation-induced differential expression genes from inclusive literature were listed and analyzed with Microsoft Excel 2010. The inclusive genes possessed an obvious dose-effect relationship after exposure to radiation in at least one independent study. The highly reproducible radiation-responsive genes studied in the inclusive literature were found to possess consistent dose-effect relationships in more than three independent studies.
Bioinformatics Analysis Gene Ontology (GO) enrichment (http://gather.genome.duke.edu/) was applied to biologically classify the genes. Then, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (http://www.kegg.jp/) was used to map the differential genes to the possible molecular pathways. STRING (http://string.embl.de/) and Cytoscape 3.4.0 (http://www.cytoscape.org/) were employed to build protein interaction networks onto which radiation-responsive genes expression can be mapped.
Cell Culture Previous studies have proved that lymphocytes are both sensitive to early radiation injury and highly responsive in terms of induced gene expression changes. In this study, AHH-1 cells were obtained from American Type Culture Collection (Manassas, VA, USA). AHH-1 cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% heat-inactivated fetal bovine serum (HyClone; GE Healthcare Life Sciences, Logan, UT, USA), 2 mmol/L L-glutamine (Thermo Fisher Scientific, Inc.), 100 U/mL penicillin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 100 μg/mL streptomycin (Sigma-Aldrich; Merck KGaA). The AHH-1 cell line is diploid and its population-doubling time ranges between 16 and 19 h. AHH-1 cells were incubated at 37 ℃ in a humidified 5% CO2 atmosphere.
Sample Irradiation Irridiation with 60Co γ-rays was performed in the Beijing Radiation Center (Beijing, China). The source radioactivity was 130 TBq. The exposure setup was calibrated by physical measurement using a tissue-equivalent ionizing chamber. The radiation dose rate was calculated using the source radioactivity and the distance between the source and sample: a dose-rate of 1 Gy/min corresponded to a source-sample distance of 73.5 cm. The homogeneous irradiation field was 30 cm × 30 cm; the samples were placed within a 5 cm radius circle and the uncertainty of the calibration was 1%. AHH-1 cells were seeded in flasks (1 × 107 cells/mL) and irradiated. Radiation doses 0, 1, 3, and 5 Gy, at a dose rate of 1 Gy/min, were used in the validation experiment; the dose point of 0 Gy was used as the negative control. Following exposure, AHH 1 cells were incubated at 37 ℃ for 4 and 24 h for further analysis. Three independent experiments were performed.
qRT-PCR Total RNA was extracted from AHH-1 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed into complementary DNA using a Reverse Transcription system kit (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. Individual PCR reactions were performed in 20 µL samples in a 96-well plate (Applied Biosystems™; Thermo Fisher Scientific, Inc.), containing 0.5 µL of each forward and reverse primer (10 µmol/L each), 2 µL cDNA, 7 µL distilled water, and 10 µL 2 × SYBR-Green PCR Master Mix (Applied Biosystems™; Thermo Fisher Scientific, Inc.). qRT-PCR was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems™; Thermo Fisher Scientific, Inc.). Amplification was performed under the following conditions: initial cycle at 95 ℃ for 10 min, followed by 40 cycles at 95 ℃ for 15 s, and at 60 ℃ for 1 min. Relative fold changes in expression were calculated using the ΔΔCt method with actin beta (ACTB) and beta-2-microglobulin (B2M) as reference controls. All experiments were performed thrice.
Statistical Methods Data were presented as the mean ± standard deviation (SD). Statistical analysis was performed by using the SPSS software version 19.0 (SPSS Inc., Chicago, IL, USA). The statistical significance of the difference between groups was assessed by one-way analysis of variance (ANOVA). P < 0.05 was considered to indicate a statistically significant difference.
Bibliometric Analysis of Publication Outputs From 2000 to 2017, the Pubmed/Medline, the Embase, and the Web of Science databases were searched for 52, 69, and 88 publications, respectively. A total of 209 studies were retrieved from the three databases. A total of 79 research articles met the literature inclusion criteria. Some fluctuations were observed with regard to the total number of publications over the past 20 years. Overall, ionizing radiation-induced differentially expressed genes research was initiated in 2001-2003, significantly increasing in 2006-2011; the number of publications in 2011 (13 articles) was six-fold higher than that in 2006 (two articles). However, the number of publications slightly decreased in 2012. The growth rate of publications has been steady over the past five years (2012-2017). The journal Radiation Research published the highest number of publications (23, 29.11%), followed by International Journal of Radiation Biology (16, 20.25%), Health Physics (6, 7.60%), and Radiation Protection Dosimetry (6, 7.60%). The USA (52 articles) and Germany (14 articles) were the top two countries engaged in ionizing radiation-induced differentially expressed genes research, accounting for 83.54% of the total number of publications. Additionally, the top research institutions for studies on ionizing radiation-induced differential expression genes were Columbia University (USA), followed by the United States Department of Defense, National Institutes of Health (NIH) USA, Ghent University (Belgium), and Julich Research Center (Germany).
The 79 articles on ionizing radiation-induced differentially expressed genes research were drafted by more than 100 authors. Amundson from Columbia University (USA), who first reported the potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation, has published 16 articles over the past 17 years, which was higher than those of other authors.
In literature metrology, citation frequency of an article is an important index to assess publication quality. In the present study, Citespace Ⅵ (64 bits) was used to construct the co-citation references network. An article that Amundson SA (2004, 145 citation) published in Cancer Research had the highest number of citations, suggesting that it is the most important article in the related researches, followed by articles published by Paul S (2008; 102 citation), Dressman HK (2007; 83 citation), Kabacik S (2011; 55 citation), and Meadows SK (2008; 55 citation) in International Journal of Radiation Oncology Biology Physics, PLoS Medicine, PLoS One, and International Journal of Radiation Biology.
Screening the Highly Reproducible Radiation-Responsive Genes Differentially expressed radiation-responsive genes were screened from 79 articles. A total of 165 genes, the expression levels of which were previously reported to be up-or down-regulated in response to ionizing radiation, have been identified and validated by microarrays or qRT-PCR. The above differential expressed radiation-responsive genes were clustered and sorted according to occurrence frequency. The top 35 radiation-responsive genes, which derived from more than three independent studies and showed significant association with doses and time after exposure to ionizing radiation, are presented in Figure 1. Cyclin-dependent kinase inhibitor 1A (CDKN1A) was identified as a candidate for dose assessment by the highest frequency studies, indicating that it is one of the most fundamental and important radiation-responsive genes, followed by damage-specific DNA-binding protein 2 (DDB2), growth arrest and DNA damage-inducible 45 (GADD45A), ferredoxin reductase (FDXR), Bcl-2 binding component 3 (BBC3), and Bcl-2 associated X protein (BAX). Additionally, mdm2 p53 binding protein homolog (MDM2), xeroderma pigmentosum complementation group C (XPC), cyclin G1 (CCNG1), and proliferating cell nuclear antigen (PCNA) were found to occure highly in most studies.
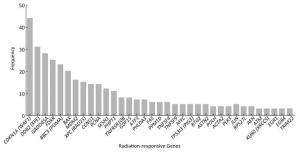
Figure 1. The frequency of top 35 radiation-responsive genes that were previously reported in 79 articles included.
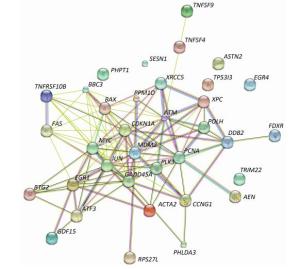
Functional Enrichment Analysis of Radiation-responsive Genes To explore the overall biological functions of significant differential expressed genes after exposure to ionizing radiation, a panel of 35 radiation-responsive genes was categorized using GO enrichment. The top 10 significant biological processes with index of Bayes factor values above 10 are shown in Table 1. Most of differentially expressed radiation-responsive genes are involved in response to DNA damage stimulus, cell proliferation, in response to endogenous stimulus, regulation of cell cycle, DNA repair, regulation of cellular physiological process, cell cycle, and regulation of biological process. KEGG pathway analysis revealed that 12 of 35 differentially expressed radiation-responsive genes were enriched in p53 signaling pathway with the highest network-based association score (XD-score) (XD = 0.744, q < 0.0001) (Table 2). Additionally, the protein-protein interaction network of the panel of 35 radiation-responsive genes was constructed by STRING (Figure 2). Results suggested that most of the proteins interact closely and are directly or indirectly connected with the p53 signal pathway.
Table 1. Gene Ontology Analysis for the 35 Radiation-responsive Genes
Gene Ontology Number of Genes Bayes Factor Genes DNA damage stimulus 9 17 ATM, BTG2, DDB2, GADD45A, PCNA, POLH, SESN1, XPC, XRCC5 Cell proliferation 16 16 ATM, BAX, BTG2, CCNG1, CDKN1A, EGR4, GADD45A, MDM2, MYC, PCNA, PLK3, POLH, PPM1D, SESN1, TNFSF4, TNFSF9 Endogenous stimulus 9 16 ATM, BTG2, DDB2, GADD45A, PCNA, POLH, SESN1, XPC, XRCC5 Regulation of cell cycle 11 16 ATM, BAX, CCNG1, CDKN1A, GADD45A, MDM2, MYC, PCNA, PLK3, PPM1D, SESN1 DNA repair 8 14 ATM, BTG2, DDB2, GADD45A, PCNA, POLH, XPC, XRCC5 Table 2. KEGG Pathway Analysis for the 35 Radiation-responsive Genes
Pathway or Process XD-score* q-value# Overlap/Size p53 signaling pathway 0.74 0.000 12/62 Riboflavin metabolism 0.33 0.841 1/11 Bladder cancer 0.29 0.057 3/38 Nucleotide excision repair 0.26 0.057 3/42 Prion diseases 0.20 0.364 2/35 Cell cycle 0.17 0.002 6/120 Chronic myeloid leukemia 0.14 0.190 3/69 Mismatch repair 0.14 1.000 1/23 Note. *XD-score: absolute Pearson correlation; XD-score significance threshold: 0.25. #q-value: significance of overlap (Fisher test); Significance was determined at 0.05. Validation of Radiation-responsive Genes The alterations of 35 radiation-responsive genes expression in AHH-1 cells were assessed using qRT-PCR at 4 and 24 h after exposure to 0, 1, 3, and 5 Gy of γ-rays irradiation. The expression levels of 18 genes [CDKN1A, DDB2, GADD45A, BAX, MDM2, FDXR, XPC, tumor protein p53 inducible protein3 (PIG3), growth differentiation factor 15 (GDF15), sestrin 1 (SESN1), phosphohistidine phosphatase 1 (PHPT1), pleckstrin homology like domain family A member 3 (PHLDA3), DNA polymerase eta (POLH), astrotactin 2 (ASTN2), ribosomal protein S27 like (RPS27L), tripartite motif containing 22 (TRIM22), polo like kinase 3 (PLK3), and BBC3] were significantly upregulated in a dose-dependent manner at 24 h post-irradiation. In addition, the expression levels of CDKN1A, GADD45A, BBC3, and PLK3 were significantly upregulated in a dose-dependent manner at 4 h after exposures (Figure 3). No dose-response relationship was found at the expression level of other genes.
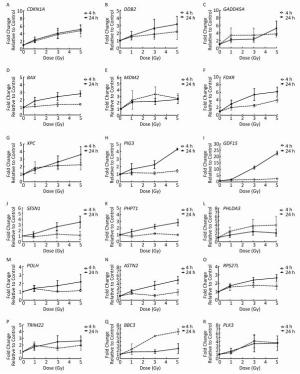
Figure 3. qRT-PCR gene expression alteration in CDKN1A (A), DDB2 (B), GADD45A (C), BAX (D), MDM2 (E), FDXR (F), XPC (G), PIG3 (H), GDF15 (I), SESN1 (J), PHPT1 (K), PHLDA3 (L), POLH (M), ASTN2 (N), RPS27L (O), TRIM22 (P), BBC3 (Q), PLK3 (R) in AHH-1 cell line after in vitro irradiation. Fold changes in expression compared with the control group (0 Gy) are plotted after 4 h and 24 h in response to 1, 3, 5 Gy γ-irradiation. The data represent the mean ± standard deviation (SD) of three independent biological replicates.
In this study, the 35 radiation-responsive genes with the high reproducibility were identified by bibliometric analysis of data retrievals for ionizing radiation-induced responsive genes from 2000 to 2017. Functional enrichment analysis revealed that the p53 signal transduction pathway played a crucial role in response to ionizing radiation. This study was the first to screen the radiation-responsive genes based on a large-scale literature's research. The validation test conducted in this study identified the 18 radiation-responsive genes as the best candidate biomarkers for establishing the ideal gene expression panel. A systematic study will be performed in the future to validate the applicability of the gene panel for dose estimation in human peripheral blood. Further research is needed to establish the gene expression model using a certain number radiation-responsive genes and to determine the impact of potential confounding factors on the accuracy of biological dosimetry based on gene expression in a large sample of human peripheral blood.
Acknowlegements All authors would like to thank Dr. CHEN De Qing for help on review and discussion of the original study protocols.
Conflicts of Interest We declare that we have no conflicts of interest.
Authors' Contributions LIU Qing Jie, LI Shuang and TIAN Mei conceived and designed the experiments; LI Shuang and LU Xue performed the experiments and analyzed the data; FENG Jiang-Bin contributed reagents/materials/analysis tools; LI Shuang wrote the paper. All authors read and approved the final manuscript.
doi: 10.3967/bes2017.112
Identification and Validation of Candidate Radiation-responsive Genes for Human Biodosimetry
-
Abstract: The aim of the present study is to analyze the global research trend of radiation-responsive genes and identify the highly reproducible radiation-responsive genes. Bibliometric methods were applied to analyze the global research trend of radiation-responsive genes. We found 79 publications on radiation-responsive genes from 2000 to 2017. A total of 35 highly reproducible radiation-responsive genes were identified. Most genes are involved in response to DNA damage, cell proliferation, cell cycle regulation, and DNA repair. The p53 signal pathway was the top enriched pathway. The expression levels of 18 genes in human B lymphoblastoid cell line (AHH-1) cells were significantly up-regulated in a dose-dependent manner at 24 h after exposure to 0-5 Gy 60Co γ-ray irradiation. Our results indicate that developing a gene expression panel with the 35 high reproducibility radiation-responsive genes may be necessary for qualitative and quantitative assessment after exposure.
-
Key words:
- Gene expression /
- Biodosimetry /
- Bibliometric /
- Ionizing radiation /
- Dose estimation
-
Figure 3. qRT-PCR gene expression alteration in CDKN1A (A), DDB2 (B), GADD45A (C), BAX (D), MDM2 (E), FDXR (F), XPC (G), PIG3 (H), GDF15 (I), SESN1 (J), PHPT1 (K), PHLDA3 (L), POLH (M), ASTN2 (N), RPS27L (O), TRIM22 (P), BBC3 (Q), PLK3 (R) in AHH-1 cell line after in vitro irradiation. Fold changes in expression compared with the control group (0 Gy) are plotted after 4 h and 24 h in response to 1, 3, 5 Gy γ-irradiation. The data represent the mean ± standard deviation (SD) of three independent biological replicates.
Table 1. Gene Ontology Analysis for the 35 Radiation-responsive Genes
Gene Ontology Number of Genes Bayes Factor Genes DNA damage stimulus 9 17 ATM, BTG2, DDB2, GADD45A, PCNA, POLH, SESN1, XPC, XRCC5 Cell proliferation 16 16 ATM, BAX, BTG2, CCNG1, CDKN1A, EGR4, GADD45A, MDM2, MYC, PCNA, PLK3, POLH, PPM1D, SESN1, TNFSF4, TNFSF9 Endogenous stimulus 9 16 ATM, BTG2, DDB2, GADD45A, PCNA, POLH, SESN1, XPC, XRCC5 Regulation of cell cycle 11 16 ATM, BAX, CCNG1, CDKN1A, GADD45A, MDM2, MYC, PCNA, PLK3, PPM1D, SESN1 DNA repair 8 14 ATM, BTG2, DDB2, GADD45A, PCNA, POLH, XPC, XRCC5 Table 2. KEGG Pathway Analysis for the 35 Radiation-responsive Genes
Pathway or Process XD-score* q-value# Overlap/Size p53 signaling pathway 0.74 0.000 12/62 Riboflavin metabolism 0.33 0.841 1/11 Bladder cancer 0.29 0.057 3/38 Nucleotide excision repair 0.26 0.057 3/42 Prion diseases 0.20 0.364 2/35 Cell cycle 0.17 0.002 6/120 Chronic myeloid leukemia 0.14 0.190 3/69 Mismatch repair 0.14 1.000 1/23 Note. *XD-score: absolute Pearson correlation; XD-score significance threshold: 0.25. #q-value: significance of overlap (Fisher test); Significance was determined at 0.05. -
[1] Blakely WF, Salter CA, Prasanna PG. Early-response biological dosimetry-recommended countermeasure enhancements for mass-casualty radiological incidents and terrorism. Health Phys, 2005; 89, 494-504. doi: 10.1097/01.HP.0000175913.36594.a4 [2] Chaudhry MA. Biomarkers for human radiation exposure. J Biomed Sci, 2008; 15, 557-63. doi: 10.1007/s11373-008-9253-z [3] Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys, 2008; 71, 1236-44. doi: 10.1016/j.ijrobp.2008.03.043 [4] Boldt S, Knops K, Kriehuber R, et al. A frequency-based gene selection method to identify robust biomarkers for radiation dose prediction. Int J Radiat Biol, 2012; 88, 267-76. doi: 10.3109/09553002.2012.638358 [5] Brengues M, Paap B, Bittner M, et al. Biodosimetry on small blood volume using gene expression assay. Health Phys, 2010; 98, 179-85. doi: 10.1097/01.HP.0000346706.44253.5c [6] Knops K, Boldt S, Wolkenhauer O, et al. Gene expression in low-and high-dose-irradiated human peripheral blood lymphocytes:possible applications for biodosimetry. Radiat Res, 2012; 178, 304-12. doi: 10.1667/RR2913.1 [7] Amundson SA, Grace MB, McLeland CB, et al. Human in vivo radiation-induced biomarkers:gene expression changes in radiotherapy patients. Cancer Res, 2004; 64, 6368-71. doi: 10.1158/0008-5472.CAN-04-1883 [8] Dressman HK, Muramoto GG, Chao NJ, et al. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med, 2007; 4, e106. doi: 10.1371/journal.pmed.0040106 [9] Mori M, Benotmane MA, Tirone I, et al. Transcriptional response to ionizing radiation in lymphocyte subsets. Cell Mol Life Sci, 2005; 62, 1489-501. doi: 10.1007/s00018-005-5086-3 [10] Chen C. CiteSpace Ⅱ:Detecting and visualizing emerging trends and transient patterns in scientific literature. Journal of the American Society for Information Science and Technology, 2006; 57, 359-77. doi: 10.1002/(ISSN)1532-2890 -




 下载:
下载:



 Quick Links
Quick Links