-
The genus Vagococcus was first described by Collins et al. and initially consisted of a single species, V. fluvialis. This species was isolated from chicken feces and river water and first described by Hashimoto et al.[1,2]. Teixeira et al. isolated V. fluvialis from human blood and peritoneal fluid, suggesting that it poses a potential threat to human health[3]. Although previous studies have described the isolation and biological characteristics of isolated strains, few have studied their antibiotic resistance and pathogenicity, which have great significance in clinical diagnoses and therapy[4]. Bats are notorious reservoir hosts for some highly pathogenic viruses, including those responsible for causing the severe acute respiratory syndrome.
Herein we report the phenotypic and genotypic characteristics of five V. fluvialis strains. We constructed a 16S phylogenetic tree using other known strains to establish the phylogenetic relationship. Furthermore, using whole genome sequencing (WGS), we studied the virulence and antibiotic resistance of isolated strains. Comparative genomic analysis of the five isolated strains was performed against V. fluvialis BH819 (NZ_FWFD00000000.1) to annotate single nucleotide polymorphisms (SNPs), insertion and deletion (InDel) events, and structural variations (SVs).
Sixty-four alive bats were captured and dissected in the Chinese Center for Disease Control and Prevention. Then, 100 μL slurry of the lung, liver, and spleen was inoculated onto blood agar. Five strains of V. fluvialis were collected and maintained on BHI agar. Total DNA was extracted using a DNA Mini Kit (Qiagen, Germany), according to manufacturer instructions. An approximately 1500-bp sequence of the 16S rRNA gene was amplified by PCR using the following primers: F27, 5′-AGAGTTTGATCMTGGCTCAG-3′ and R1492, 5′-ACGGYTACCTTYTTACGACTT-3′[5]. The positive control was Vagococcus fluvialis. The negative control was water. Phenotypic analysis of the five isolates was performed using conventional biochemical tests[6]. Hemolysis was assessed on Columbia agar containing 5% sheep blood. Motility was determined at 37 °C in a semi-solid medium containing 0.3% noble agar, 1% tryptose, and 0.5% NaCl. Growth at various concentrations of NaCl (1%–8%) and different temperatures (37 °C, 40 °C, 42 °C, and 45 °C) was determined. Oxidation and assimilation of the isolates were detected using API 50CH. Genomes of the five isolates were sequenced on the Illumina PE150 platform. Virulence genes and antibiotic resistance genes were detected using the Virulence Factors of Pathogenic Bacteria (VFDB) and Antibiotic Resistance Genes Database (ARDB), respectively[7,8]. Comparative genomic analysis of the five isolates against V. fluvialis BH819, recommended by NCBI, to annotate SNPs, InDel events, and SVs. Evolutionary analyses were conducted in MEGA7[9].
In total, 192 samples (the lung, liver, and spleen samples of the 64 bats) were detected, and five V. fluvialis strains were isolated. Two strains were isolated from the spleen, three from the liver, and none from the lung (Supplementary Table S1, available in www.besjournal.com). PCR was used to amplify sequences of the 16S rRNA gene from the BF33.1, BF33.2, BF38, BF43, and BF45 isolates. BLASTn indicated that the sequences were 100% similar to those from the V. fluvialis strain TRG15 (MH329632.1). The isolates on blood agar were surrounded by zones of greenish hemolysis. Regarding growth at various concentrations of NaCl (1%–8%), the five isolates could grow at NaCl concentrations of up to 6% only. Moreover, they could grow at 37 °C, 40 °C, and 42 °C, but not at 45 °C. The isolates showed motility in the semi-solid medium. Details about the five isolates producing acid from carbohydrates and general characteristics of the genomes of the five isolates are shown in Supplementary Table S2 and Supplementary Table S3 (available in www.besjournal.com). Then, the annotated genomes were analyzed by the ARDB. All isolates showed potential resistance to bacitracin since they showed the presence of the baca gene. In addition, BF43 showed potential resistance to macrolide, considering the presence of mefA. We identified 86 virulence factors in the five isolates (Supplementary Table S4, available in www.besjournal.com). Of these, 52 were common among the isolates, and most were enzymes and transporter proteins.
Table S1. Number of strains isolated from different organs of bats and the positive rate
Items Lung Liver Spleen Total Number of strains 0 3 2 5 Positive rate (%) 0 4.69 3.13 2.60 Table S2. Details pertaining to the five isolates producing acid from carbohydrates
Details BF33.1 BF33.2 BF38 BF43 BF45 Glycerol + + + + + L-arabinose − + − − + D-ribose + + + + + D-glucose + + + + + D-fructose + + + + + D-mannose + + + + + Mannitol + + + − + Sorbitol + + + + + Methyl-αD-glucopyranoside + + + + + N-acetylglucosamine + + + + + Amygdalin + + + + + Arbutin + + + + + Salicin + + + + + D-cellobiose + + + + + D-maltose + + + + + D-trehalose + + + + + Starch + + + + + D-gentiobiose + + + + + Table S3. General characteristics of the genomes of the five isolates
Characteristics BF33.1 BF33.2 BF38 BF43 BF45 Genome size (bp) 2,964,643 3,044,263 3,036,912 2,687,751 2,745,235 GC content (%) 32.54 32.65 32.66 32.88 32.75 Gene number 2,356 2,925 2,928 2,685 2,693 Genes of genome (%) 66.48 90.71 90.55 90.12 90.34 Table S4. Virulence factors observed in the five strains isolated from bats
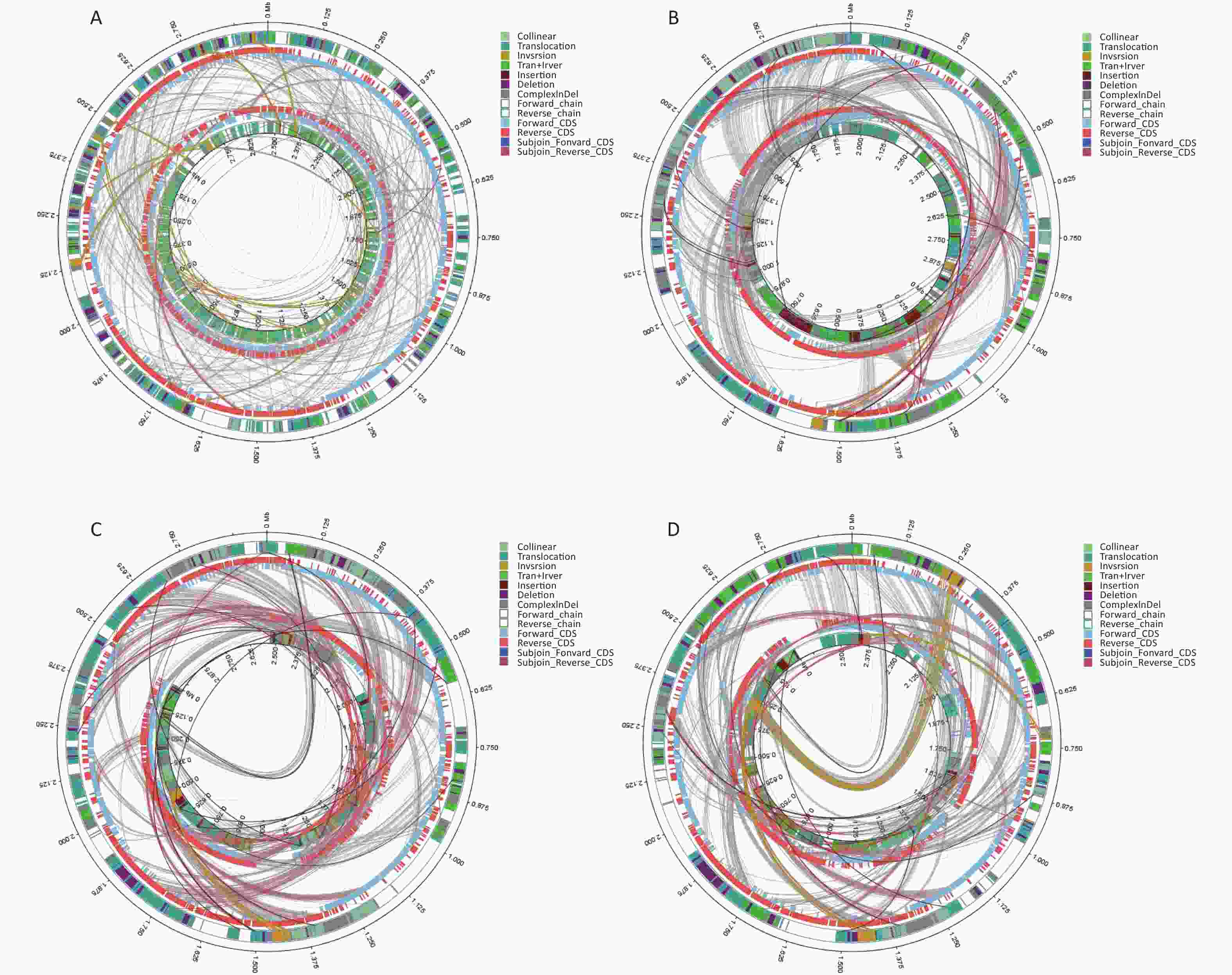
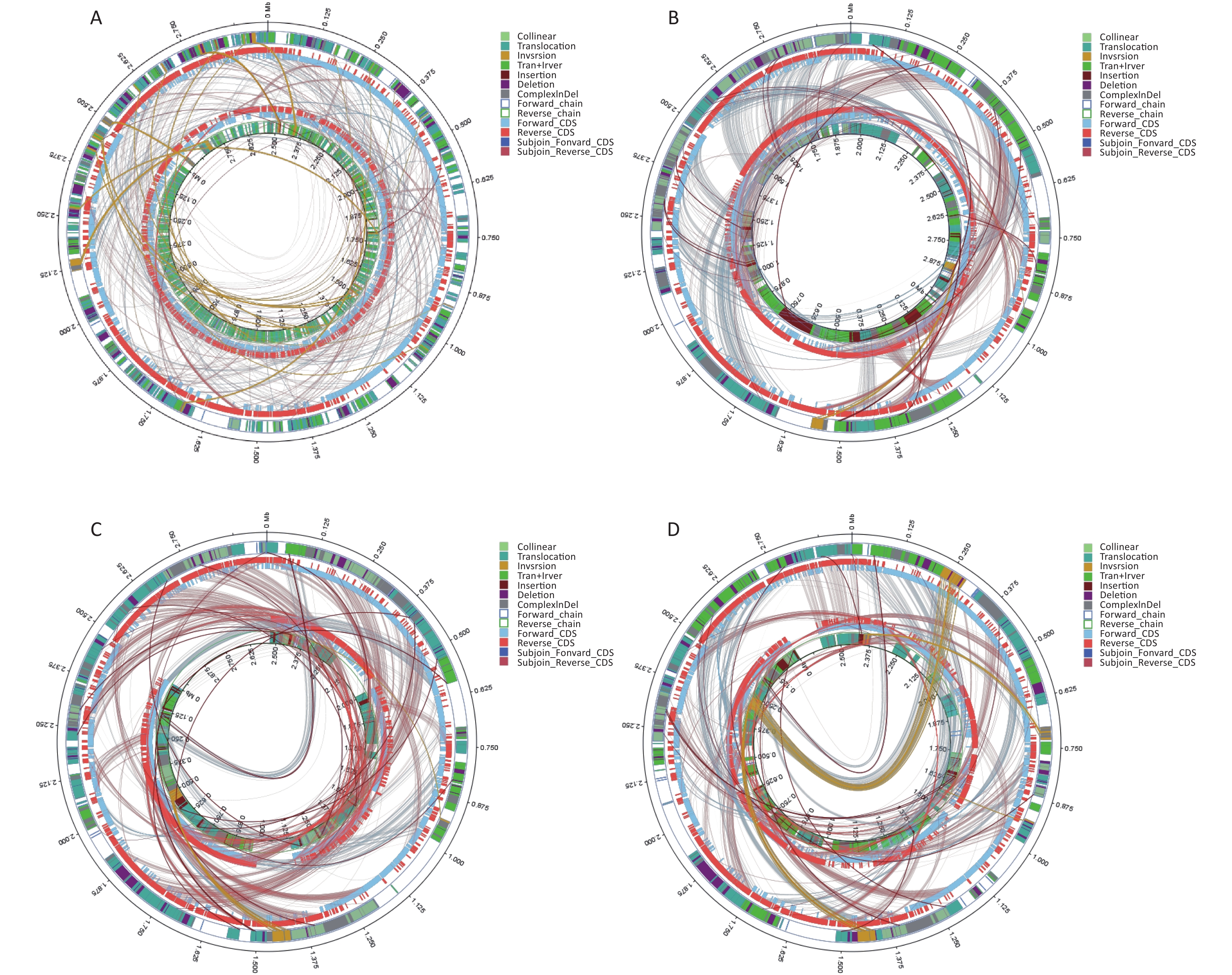
Virulence Factors Gene Product Function BF33.1 BF33.2 BF38 BF43 BF45 (p)ppGpp synthesis and hydrolysis relA GTP pyrophosphokinase Regulation + + + + + ABC transporter fbpC iron-uptake permease ATP-binding protein Iron uptake + + + + + ABC transporter for dispersin aatC ABC transporter ATP-binding protein AatC Adherence + + + + + Accessory secretion factor secA2 preprotein translocase subunit SecA Secretion system + + + + + Ace ace collagen adhesin protein Adherence − + + − − Achromobactin cbrD ABC transporter Iron uptake − + + − − Achromobactin biosynthesis and transport cbrD achromobactin transport ATP-binding protein CbrD Iron uptake − + + − − AdsA adaA Adenosine synthase A Protease − + + − − AgrA/AgrC agrA hypothetical protein Adherence − + + + + Anguibactin fatD ferric anguibactin transport protein Iron uptake − + + + + Auto aut autolysin Invasion + + + + + Autoinducer-2 luxS S-ribosylhomocysteinase Regulation − + + − − Autolysin (GW protein) aut N-acetylmuramoyl-L-alanine amidase family protein Invasion − + + − + Bcp pili srtD Sortase Adherence − + − − − Bee (biofilm enhancer in enterococci) srt1 Srt1 Adherence + + + + + BopD BopD LacI family transcriptional regulator Biofilm formation + + + + + Capsular polysaccharide wbfV/wcvB Predicted UDP-glucose 6-dehydrogenase Regulation + − − − + Capsule Cap, oppF Transporter biosynthesis protein; oligopeptide ABC transporter, permease component Adherence + + + + + ClpC clpC Endopeptidase Clp ATP-binding chain C Stress protein + + + + + ClpE clpE ATP-dependent protease Stress protein − + + + + ClpP clpP ATP-dependent Clp protease proteolytic subunit Stress protein + + + + + Colibactin clbD Putative 3-hydroxyacyl-CoA dehydrogenase Toxin + + + + + Copper exporter ctpV Cation-transporting ATPase V Iron uptake − + + + + Cytolysin cylR2 Cytolysin regulator R2 Toxin + + + − − D-alanine-polyphosphoribitol ligase dltA Putative D-alanine-activating enzyme Toxin + + + + + Ebp pili srtC Sortase Adherence + + + − + EfaA efaA Endocarditis specific antigen Adherence + + + + + EF-Tu tuf Elongation factor Tu Adherence − + + + + Exopolysaccharide mrsA/glmM Phosphoglucosamine mutase + + + + + FbpABC fbpC Iron III ABC transporter, ATP-binding protein Adherence + + + + + Ferrous iron transport feoA Ferrous iron transporter A Iron uptake − + + + − Fibronectin-binding proteins pavA Adherence and virulence protein A Adherence + + + + + Flagella flgG; fliP Flagellar biosynthesis protein Adherence, Invasion + + + − + Glutamine synthesis glnA1 Glutamine synthetase Toxin + + + + + GroEL groEL Chaperonin GroEL + + + + + Hcp secretion island-1 encoded type VI secretion system (H-T6SS) clpV1 Putative ClpA/B-type chaperone Stress protein − + + + + Heme biosynthesis hemG Protoporphyrinogen oxidase Toxin − − − + + Hemolysin hlyA Hemolysin A Toxin + + + + + Hemolysin III hlyIII Hemolysin III Toxin + + + + + HexNAc flg; flh; Fli Flagellar protein Adherence; Invasion + + + + + Histone-like protein (Hlp)/ laminin-binding protein (LBP) hlp Histone-like protein Adherence + + + + + Hyaluronic acid capsule hasC UDP-glucose pyrophosphorylase Antiphagocytosis − + − + − IlpA IlpA Immunogenic lipoprotein A Adherence + + + + + Laminin-binding protein lmb Metal binding lipoprotein Adherence − + + + + Lipoate protein ligase A1 lplA1 Putative lipoate protein ligase A Intracellular growth + + + + + Lipoprotein diacylglyceryl transferase lgt Prolipoprotein diacylglyceryl transferase Adherence + + + + + Lipoprotein-specific signal peptidase II lspA Putative signal peptidase II Adherence + + + + + LisR/LisK lisR Two-component response regulator Regulation + + + + + Listeria adhesion protein lap Hypothetical protein Adherence + + + + + LOS orfM Putative deoxyribonucleotide triphosphate pyrophosphatase Adherence + + + + + LPS gtrB; Fbphi; fabZ Bactoprenol glucosyl transferase Adherence + + + + + Lysine synthesis lysA Diaminopimelate decarboxylase − + + + + Mg2+ transport mgtB Hypothetical protein Magnesium uptake − + + + + MOMP CT396 Molecular chaperone DnaK Adherence + + + + + MprA/B mprA DNA-binding response regulator Regulation + + + + + ND fleQ/flrC FleQ protein Adherence − + + − − Nucleoside diphosphate kinase ndk Nucleoside diphosphate kinase Protease − + + + + Oligopeptide-binding protein oppA Hypothetical protein Adherence + + + + + PblA pblA PblA − − − + − PdgA pdgA Polysaccharide deacetylase Immune evasion + + + + + PDH-B pdhB Pyruvate dehydrogenase E1 component subunit beta + + + + + Periplasmic binding protein-dependent ABC transport systems vctC ABC-type enterochelin transport system, ATPase component + + + + + Peritrichous flagella Che; motA; fliQ Chemotaxis response regulator; flagellar motor protein MotA; flagellar biosynthesis protein Regulation + + + + + PilB-type pili (PGS3) ACI49664 Putative pilus-dedicated sortase Adherence + + + − + Pneumococcal iron uptake piuA Iron-compound ABC transporter, iron-compound-binding protein Iron uptake + + + + + Polar flagella flmH 3-Oxoacyl-ACP reductase − + + + + Polysaccharide capsule lytR Membrane-bound transcriptional regulator LytR Regulation + + + + + Pse5Ac7Ac cheA Chemotaxis histidine kinase Invasion + + + Pse5Ac7Ac, Pse5Ac7Am, Pse8OAc, Pse5Am7AcGlnAc pseB UDP-GlcNAc-specific C4,6 dehydratase/C5 epimerase Motility − + − − + Pyrimidine biosynthesis carA Carbamoyl-phosphate synthase small chain Metabolic adaptation + + + + + RegX3 regX3 DNA-binding response regulator RegX3 Regulation + + + + + Serine protease htrA/degP Serine protease HtrA Adherence + + + + + Serine-threonine phosphatase stp Putative phosphoprotein phosphatase Adherence + + + + + Sigma A sigA/rpoV RNA polymerase sigma factor Protease − + + + + SodB sodB Superoxide dismutase Stress protein + + + + + Sortase A srtA Sortase, putative Adherence + + + + + Streptococcal enolase eno Phosphopyruvate hydratase Secretion system + + + + + Streptococcal lipoprotein rotamase A slrA Peptidyl-prolyl cis-trans isomerase, cyclophilin-type Secretion system + + + + + Streptococcal plasmin receptor/GAPDH plr/gapA Glyceraldehyde-3-phosphate dehydrogenase, type I Adherence + + + + + T3SS mlr6326 Putative DNA invertase Secretion system + + + + + T4SS effectors CBU_1566 Coxiella Dot/Icm type IVB secretion system translocated effector Secretion system + + + + + Trehalose-recycling ABC transporter sugC Probable sugar ABC transporter, ATP-binding protein SugC Iron uptake + + + + + Trigger factor tig/ropA Trigger factor Adherence + + + + + Type IV pili pil Twitching motility protein Adherence − + + + + Type IV pili biosynthesis pilR Type 4 fimbriae expression regulatory protein pilR Adherence + − − − − VirR/VirS virR Hypothetical protein Secretion system + + + + + We conducted comparative genomic analyses of the five isolates against V. fluvialis BH819 recommended by NCBI. SNPs, InDel events, and SVs were annotated (Supplementary Table S5 and Supplementary Table S6, available in www.besjournal.com). Figure 1 shows the SVs observed in the isolated strains aligned against those in the reference strain BH819. Figure 1A, 1B, 1C, 1D, 1E, respectively, show the SVs of the isolated strains BF33.1, BF33.2, BF38, BF43, and BF45 aligned against those of the reference strain BH819. We downloaded data about three whole genomic sequences of V. fluvialis from NCBI and analyzed them. Figure 2 shows the evolutionary relationship among the relevant strains.
Table S5. Comparative genomic analyses of single nucleotide polymorphisms of the five isolates against BH819
Genomic BF33.1 BF33.2 BF38 BF43 BF45 Synonymous 29,934 29,909 29,910 29,975 29981 Non-synonymous 11,630 11,589 11,605 11,599 11,613 Total CDS SNPs 41,812 41,742 41,759 41,820 41,845 Total SNPs 44,924 44,838 44,859 44,935 44,959 Note. CDS, coding sequence; SNPs, single nucleotide polymorphisms. Table S6. Comparative genomic analyses of the insertion and deletion (InDel) events of the five isolates against BH819
Items BF33.1 BF33.2 BF38 BF43 BF45 Frame-shifted 11 7 9 7 8 Start codon 0 0 0 0 1 Stop codon 0 0 0 0 0 Premature stop 0 0 0 0 0 CDS with InDel 24 20 21 20 21 CDS of the reference strain 2,822 2,822 2,822 2,822 2,822 Note. CDS, coding sequence. 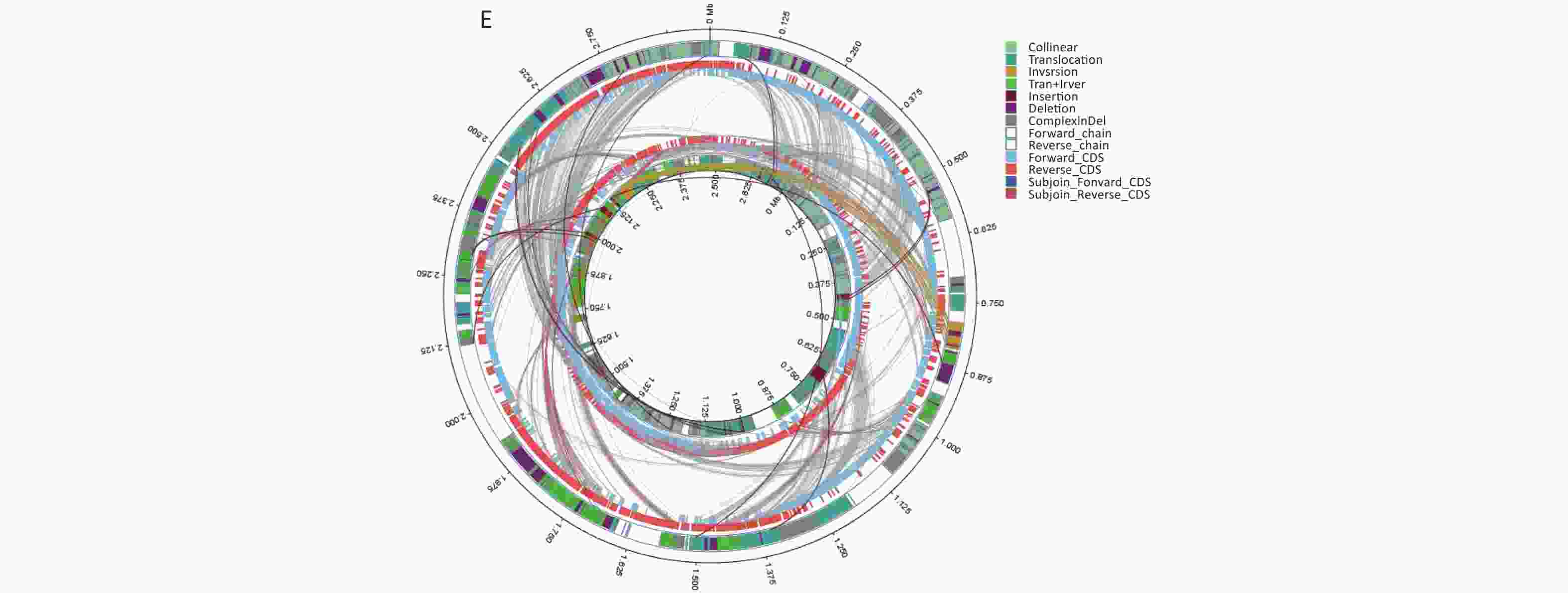
Figure 1. Structural variations (SVs) of the isolated strains aligned against those of the reference strain BH819. (A) BF33.1, (B) BF33.2, (C) BF38, (D) BF43, and (E) BF45. The inner circle represents the genome of the isolated strain, and the outer circle represents that of the reference strain. Collinear: the same linear region; Translocation: the translocation region; Inversion: the inverted region; Tran + Inver: translocation and inverted regions; Insertion: insertion region with a length of ≥ 50 bp; Deletion: a missing region with a length of ≥ 50 bp; ComplexInDel: A complex insertion and deletion (complex indel) is a rare category of genomic SVs. A complex indel presents one or multiple DNA fragments inserted into the genomic location where a deletion occurs; Forward_chain: the forward chain of the genomic sequence at which point the gene coordinates an increase in a clockwise direction; Reverse_chain: the reverse chain of the genomic sequence at which point the gene coordinates an increase in a counterclockwise direction; Forward_CDS: coding sequence (CDS) for translation on the forward strand; Reverse_CDS: CDS for translation on the reverse strand; Subjoin_Forward_CDS: CDS for translation of the complement of the genomic sequence in the forward chain; Subjoin_Reverse_CDS: CDS for translation of the complement of the genomic sequence in the reverse chain.
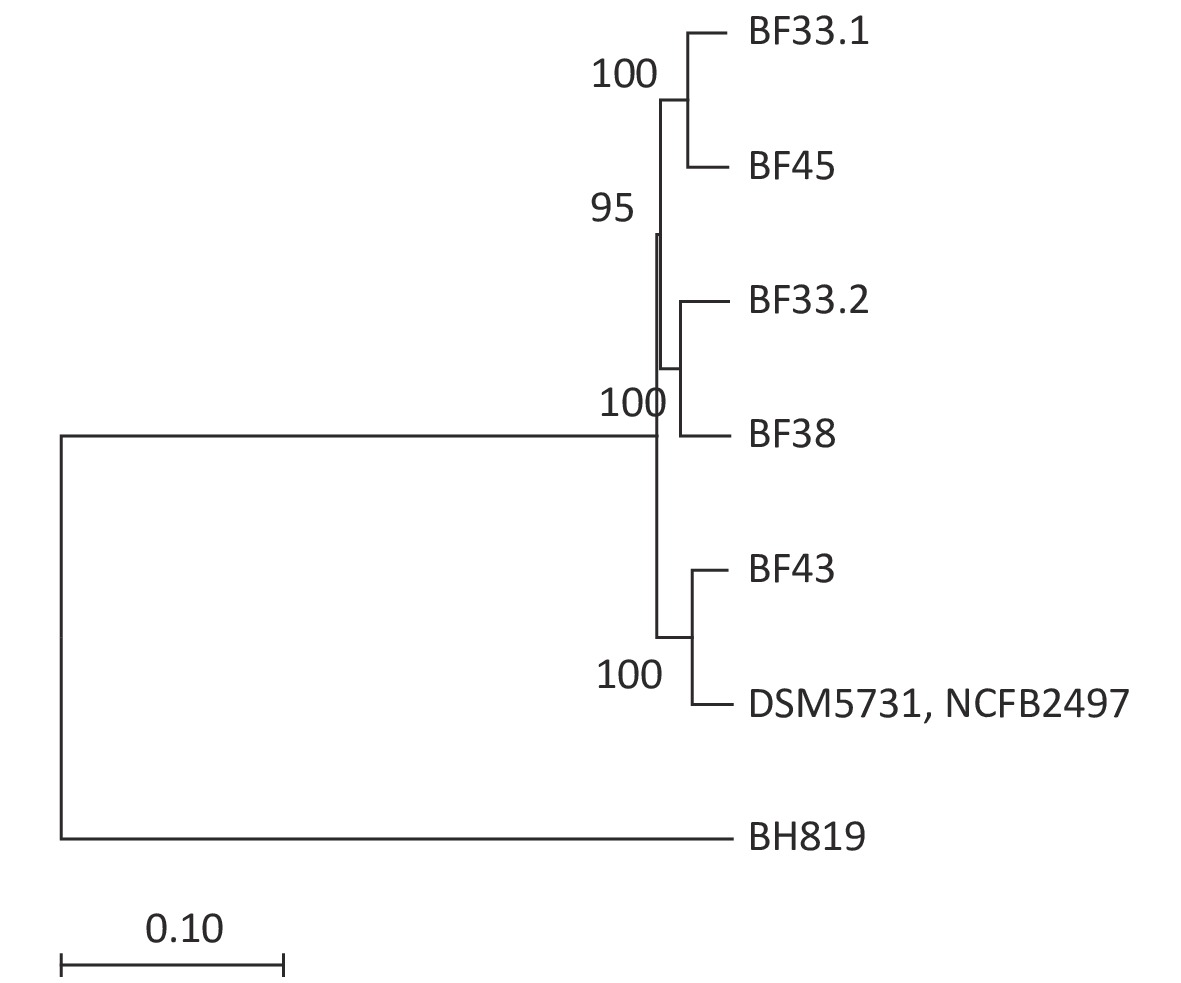
Figure 2. Phylogenetic tree of eight V. fluvialis strains. Data about BH819, DSM5731, and NCFB2497 strains were downloaded from NCBI and then analyzed. The tree was drawn to scale using branch lengths in the same units as those of the evolutionary distances to infer the phylogenetic tree.
This study identified and characterized five novel strains of V. fluvialis using a polyphasic approach, including their phenotypic characterization, the sequencing of the 16S rRNA gene, and WGS. This investigation is the first study to report the isolation of this bacterial species from bats. In a previous study[10], the most common bacteria isolated from individual bats were Escherichia coli, Klebsiella oxytoca, and Serratia marcescens[10], different from the isolated strains that are listed in Supplementary Table S7, available at www.besjournal.com. This discrepancy might be due to the different types of samples collected in the two studies. The previous study swabbed the oral and rectal cavities of bats. In contrast, our study tested a slurry of the lung, liver, and spleen. Also, the differences in etiology between the two studies might be related to differences in the growth environment, species, and diet among bats.
Table S7. Information about bats
The number
of the batBat species Location of samples collection Habitat types from
where bats capturedSamples type Isolated strain type Culture medium
used for strainsNO.3 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.7 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.7 Brown Bat He Nan cave dwelling liver Enterococcus Chocolate Medium NO.7 Brown Bat He Nan cave dwelling liver Staphylococcus epidermidis Blood plate NO.9 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.13 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.14 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.18 Brown Bat He Nan cave dwelling liver Staphylococcus epidermidis Blood plate NO.18 Brown Bat He Nan cave dwelling liver Actinomycetes Agar medium NO.22 Brown Bat He Nan cave dwelling liver Aeromonas veronii Blood plate NO.22 Brown Bat He Nan cave dwelling spleen Enterococcus Chocolate Medium NO.22 Brown Bat He Nan cave dwelling spleen Enterococcus faecalis Chocolate Medium NO.25 Brown Bat He Nan cave dwelling spleen Enterococcus faecalis Chocolate Medium NO.26 Brown Bat He Nan cave dwelling liver Bacillus subtilis Blood plate NO.27 Brown Bat He Nan cave dwelling liver Corynebacterium Blood plate NO.28 Brown Bat He Nan cave dwelling liver Bacillus brevis Blood plate NO.28 Brown Bat He Nan cave dwelling liver Corynebacterium Blood plate NO.28 Brown Bat He Nan cave dwelling spleen Enterococcus Chocolate Medium NO.31 Brown Bat He Nan cave dwelling spleen Aeromonas veronii Chocolate Medium NO.32 Brown Bat He Nan cave dwelling spleen Enterococcus faecalis Chocolate Medium NO.33 Brown Bat He Nan cave dwelling spleen Enterococcus Chocolate Medium NO.36 Brown Bat He Nan cave dwelling spleen staphylococcus Chocolate Medium NO.38 Brown Bat He Nan cave dwelling spleen Klebsiella pneumoniae Chocolate Medium NO.38 Brown Bat He Nan cave dwelling liver Staphylococcus haemolyticus Chocolate Medium NO.40 Brown Bat He Nan cave dwelling liver Staphylococcus haemolyticus Chocolate Medium NO.40 Brown Bat He Nan cave dwelling liver kocuria sp Chocolate Medium NO.40 Brown Bat He Nan cave dwelling liver Enterococcus Chocolate Medium NO.41 Brown Bat He Nan cave dwelling liver Staphylococcus coriolis Chocolate Medium NO.42 Brown Bat He Nan cave dwelling liver Staphylococcus coriolis Chocolate Medium NO.44 Brown Bat He Nan cave dwelling liver Proteus Chocolate Medium NO.44 Brown Bat He Nan cave dwelling lung Bacillus sphaericus Chocolate Medium NO.45 Brown Bat He Nan cave dwelling lung Micrococcus lylae Chocolate Medium NO.45 Brown Bat He Nan cave dwelling lung Enterococcus Chocolate Medium NO.46 Brown Bat He Nan cave dwelling lung Shigella faecalis Chocolate Medium NO.47 Brown Bat He Nan cave dwelling lung Bacillus subtilis Chocolate Medium NO.47 Brown Bat He Nan cave dwelling lung Enterococcus faecalis Chocolate Medium Motility is an essential phenotypic characteristic of V. fluvialis[2]. In this study, all five isolated strains were motile. Other phenotypic traits were similar to those of previously reported strains. This study used the Illumina PE150 platform to sequence the genomes of the five isolates. Virulence genes and antibiotic resistance genes were subsequently detected using the VFDB and ARDB, respectively.
Numerous bacteria have developed antibiotic resistance because of the careless use of antibiotics. The ARDB unifies most publicly available information on antibiotic resistance. This information can be used as a compendium of antibiotic resistance factors and identify resistance genes in newly sequenced genomes. Herein, we used the ARDB to predict drug resistance genes, understand drug resistance mechanisms in the five isolates tested, and discuss the clinically accurate, reasonable, and effective use of drugs. Our results indicated that the five isolates showed potential resistance to bacitracin, which can be attributed to the presence of the baca gene. Bacitracin is mainly used to treat staphylococcal and external skin infections. It damages the bacterial cell wall and protoplast, affecting permeability. Many strains of V. fluvialis have reportedly been isolated from various lesions. It can guide clinical treatment by detecting whether these strains have potential resistance to bacitracin. In addition, BF43 showed potential resistance to macrolides considering the presence of mefA. At a specific concentration, macrolide antibiotics inhibit bacterial protein synthesis by blocking peptidyl transferase activity in 50S ribosomes. The mef efflux pump clears macrolide antibiotics from cells, preventing them from inhibiting bacterial growth.
The VFDB, a database for bacterial virulence factors, was constructed for bacterial pathogens of medical importance. Herein, we identified 32 virulence factors involved in adherence and five virulence factors involved in the invasion. Many genes encoding virulence traits, such as secretion systems, siderophores, catalases, and regulators, are indirectly involved in pathogenesis, and these are equally important for bacteria to establish an infection. Our results indicated that the identified virulence factors involved various functions, such as iron uptake and stress regulation.
In summary, based on our phenotypic and genotypic analyses, we identified five novel strains of V. fluvialis from bats. All five strains were capable of hemolysis, were motile, and could grow at 40 °C but not 45 °C and at 6% NaCl but not 7% NaCl. Moreover, all isolates could produce acid from glycerol, D-ribose, D-glucose, D-fructose, D-mannose, N-acetylglucosamine, amygdalin, arbutin, salicin, D-cellobiose, D-maltose, D-trehalose, starch, and D-gentiobiose. Also, BF33.2 and BF45 could produce acid from L-arabinose. All strains except BF43 could produce acid from mannitol. At present, little genomic information is available for V. fluvialis, which has hindered investigations about antibiotic resistance and pathogenicity mechanisms. In this study, WGS of the five isolates was conducted. All isolates showed potential resistance to bacitracin, and BF43 also showed potential resistance to macrolide. In total, 86 virulence factors were annotated, a large number of which were involved in adherence. We believe that such information should advance our understanding of V. fluvialis. Also, our data should support further studies on drug resistance and pathogenesis. Besides, bats can isolate many other bacteria in addition to Vagococcus fluvialis. The bat hosts of these pathogenic bacteria live near humans, potentially transmitting bacteria to humans and livestock. Chinese food culture maintains that live slaughtered animals provide the best nutrition. This belief may enhance bacterial transmission. Therefore, we should avoid hunting and eating bats.
doi: 10.3967/bes2021.114
-
&These authors contributed equally to this work.
注释: -
Figure 1. Structural variations (SVs) of the isolated strains aligned against those of the reference strain BH819. (A) BF33.1, (B) BF33.2, (C) BF38, (D) BF43, and (E) BF45. The inner circle represents the genome of the isolated strain, and the outer circle represents that of the reference strain. Collinear: the same linear region; Translocation: the translocation region; Inversion: the inverted region; Tran + Inver: translocation and inverted regions; Insertion: insertion region with a length of ≥ 50 bp; Deletion: a missing region with a length of ≥ 50 bp; ComplexInDel: A complex insertion and deletion (complex indel) is a rare category of genomic SVs. A complex indel presents one or multiple DNA fragments inserted into the genomic location where a deletion occurs; Forward_chain: the forward chain of the genomic sequence at which point the gene coordinates an increase in a clockwise direction; Reverse_chain: the reverse chain of the genomic sequence at which point the gene coordinates an increase in a counterclockwise direction; Forward_CDS: coding sequence (CDS) for translation on the forward strand; Reverse_CDS: CDS for translation on the reverse strand; Subjoin_Forward_CDS: CDS for translation of the complement of the genomic sequence in the forward chain; Subjoin_Reverse_CDS: CDS for translation of the complement of the genomic sequence in the reverse chain.
S1. Number of strains isolated from different organs of bats and the positive rate
Items Lung Liver Spleen Total Number of strains 0 3 2 5 Positive rate (%) 0 4.69 3.13 2.60 S2. Details pertaining to the five isolates producing acid from carbohydrates
Details BF33.1 BF33.2 BF38 BF43 BF45 Glycerol + + + + + L-arabinose − + − − + D-ribose + + + + + D-glucose + + + + + D-fructose + + + + + D-mannose + + + + + Mannitol + + + − + Sorbitol + + + + + Methyl-αD-glucopyranoside + + + + + N-acetylglucosamine + + + + + Amygdalin + + + + + Arbutin + + + + + Salicin + + + + + D-cellobiose + + + + + D-maltose + + + + + D-trehalose + + + + + Starch + + + + + D-gentiobiose + + + + + S3. General characteristics of the genomes of the five isolates
Characteristics BF33.1 BF33.2 BF38 BF43 BF45 Genome size (bp) 2,964,643 3,044,263 3,036,912 2,687,751 2,745,235 GC content (%) 32.54 32.65 32.66 32.88 32.75 Gene number 2,356 2,925 2,928 2,685 2,693 Genes of genome (%) 66.48 90.71 90.55 90.12 90.34 S4. Virulence factors observed in the five strains isolated from bats
Virulence Factors Gene Product Function BF33.1 BF33.2 BF38 BF43 BF45 (p)ppGpp synthesis and hydrolysis relA GTP pyrophosphokinase Regulation + + + + + ABC transporter fbpC iron-uptake permease ATP-binding protein Iron uptake + + + + + ABC transporter for dispersin aatC ABC transporter ATP-binding protein AatC Adherence + + + + + Accessory secretion factor secA2 preprotein translocase subunit SecA Secretion system + + + + + Ace ace collagen adhesin protein Adherence − + + − − Achromobactin cbrD ABC transporter Iron uptake − + + − − Achromobactin biosynthesis and transport cbrD achromobactin transport ATP-binding protein CbrD Iron uptake − + + − − AdsA adaA Adenosine synthase A Protease − + + − − AgrA/AgrC agrA hypothetical protein Adherence − + + + + Anguibactin fatD ferric anguibactin transport protein Iron uptake − + + + + Auto aut autolysin Invasion + + + + + Autoinducer-2 luxS S-ribosylhomocysteinase Regulation − + + − − Autolysin (GW protein) aut N-acetylmuramoyl-L-alanine amidase family protein Invasion − + + − + Bcp pili srtD Sortase Adherence − + − − − Bee (biofilm enhancer in enterococci) srt1 Srt1 Adherence + + + + + BopD BopD LacI family transcriptional regulator Biofilm formation + + + + + Capsular polysaccharide wbfV/wcvB Predicted UDP-glucose 6-dehydrogenase Regulation + − − − + Capsule Cap, oppF Transporter biosynthesis protein; oligopeptide ABC transporter, permease component Adherence + + + + + ClpC clpC Endopeptidase Clp ATP-binding chain C Stress protein + + + + + ClpE clpE ATP-dependent protease Stress protein − + + + + ClpP clpP ATP-dependent Clp protease proteolytic subunit Stress protein + + + + + Colibactin clbD Putative 3-hydroxyacyl-CoA dehydrogenase Toxin + + + + + Copper exporter ctpV Cation-transporting ATPase V Iron uptake − + + + + Cytolysin cylR2 Cytolysin regulator R2 Toxin + + + − − D-alanine-polyphosphoribitol ligase dltA Putative D-alanine-activating enzyme Toxin + + + + + Ebp pili srtC Sortase Adherence + + + − + EfaA efaA Endocarditis specific antigen Adherence + + + + + EF-Tu tuf Elongation factor Tu Adherence − + + + + Exopolysaccharide mrsA/glmM Phosphoglucosamine mutase + + + + + FbpABC fbpC Iron III ABC transporter, ATP-binding protein Adherence + + + + + Ferrous iron transport feoA Ferrous iron transporter A Iron uptake − + + + − Fibronectin-binding proteins pavA Adherence and virulence protein A Adherence + + + + + Flagella flgG; fliP Flagellar biosynthesis protein Adherence, Invasion + + + − + Glutamine synthesis glnA1 Glutamine synthetase Toxin + + + + + GroEL groEL Chaperonin GroEL + + + + + Hcp secretion island-1 encoded type VI secretion system (H-T6SS) clpV1 Putative ClpA/B-type chaperone Stress protein − + + + + Heme biosynthesis hemG Protoporphyrinogen oxidase Toxin − − − + + Hemolysin hlyA Hemolysin A Toxin + + + + + Hemolysin III hlyIII Hemolysin III Toxin + + + + + HexNAc flg; flh; Fli Flagellar protein Adherence; Invasion + + + + + Histone-like protein (Hlp)/ laminin-binding protein (LBP) hlp Histone-like protein Adherence + + + + + Hyaluronic acid capsule hasC UDP-glucose pyrophosphorylase Antiphagocytosis − + − + − IlpA IlpA Immunogenic lipoprotein A Adherence + + + + + Laminin-binding protein lmb Metal binding lipoprotein Adherence − + + + + Lipoate protein ligase A1 lplA1 Putative lipoate protein ligase A Intracellular growth + + + + + Lipoprotein diacylglyceryl transferase lgt Prolipoprotein diacylglyceryl transferase Adherence + + + + + Lipoprotein-specific signal peptidase II lspA Putative signal peptidase II Adherence + + + + + LisR/LisK lisR Two-component response regulator Regulation + + + + + Listeria adhesion protein lap Hypothetical protein Adherence + + + + + LOS orfM Putative deoxyribonucleotide triphosphate pyrophosphatase Adherence + + + + + LPS gtrB; Fbphi; fabZ Bactoprenol glucosyl transferase Adherence + + + + + Lysine synthesis lysA Diaminopimelate decarboxylase − + + + + Mg2+ transport mgtB Hypothetical protein Magnesium uptake − + + + + MOMP CT396 Molecular chaperone DnaK Adherence + + + + + MprA/B mprA DNA-binding response regulator Regulation + + + + + ND fleQ/flrC FleQ protein Adherence − + + − − Nucleoside diphosphate kinase ndk Nucleoside diphosphate kinase Protease − + + + + Oligopeptide-binding protein oppA Hypothetical protein Adherence + + + + + PblA pblA PblA − − − + − PdgA pdgA Polysaccharide deacetylase Immune evasion + + + + + PDH-B pdhB Pyruvate dehydrogenase E1 component subunit beta + + + + + Periplasmic binding protein-dependent ABC transport systems vctC ABC-type enterochelin transport system, ATPase component + + + + + Peritrichous flagella Che; motA; fliQ Chemotaxis response regulator; flagellar motor protein MotA; flagellar biosynthesis protein Regulation + + + + + PilB-type pili (PGS3) ACI49664 Putative pilus-dedicated sortase Adherence + + + − + Pneumococcal iron uptake piuA Iron-compound ABC transporter, iron-compound-binding protein Iron uptake + + + + + Polar flagella flmH 3-Oxoacyl-ACP reductase − + + + + Polysaccharide capsule lytR Membrane-bound transcriptional regulator LytR Regulation + + + + + Pse5Ac7Ac cheA Chemotaxis histidine kinase Invasion + + + Pse5Ac7Ac, Pse5Ac7Am, Pse8OAc, Pse5Am7AcGlnAc pseB UDP-GlcNAc-specific C4,6 dehydratase/C5 epimerase Motility − + − − + Pyrimidine biosynthesis carA Carbamoyl-phosphate synthase small chain Metabolic adaptation + + + + + RegX3 regX3 DNA-binding response regulator RegX3 Regulation + + + + + Serine protease htrA/degP Serine protease HtrA Adherence + + + + + Serine-threonine phosphatase stp Putative phosphoprotein phosphatase Adherence + + + + + Sigma A sigA/rpoV RNA polymerase sigma factor Protease − + + + + SodB sodB Superoxide dismutase Stress protein + + + + + Sortase A srtA Sortase, putative Adherence + + + + + Streptococcal enolase eno Phosphopyruvate hydratase Secretion system + + + + + Streptococcal lipoprotein rotamase A slrA Peptidyl-prolyl cis-trans isomerase, cyclophilin-type Secretion system + + + + + Streptococcal plasmin receptor/GAPDH plr/gapA Glyceraldehyde-3-phosphate dehydrogenase, type I Adherence + + + + + T3SS mlr6326 Putative DNA invertase Secretion system + + + + + T4SS effectors CBU_1566 Coxiella Dot/Icm type IVB secretion system translocated effector Secretion system + + + + + Trehalose-recycling ABC transporter sugC Probable sugar ABC transporter, ATP-binding protein SugC Iron uptake + + + + + Trigger factor tig/ropA Trigger factor Adherence + + + + + Type IV pili pil Twitching motility protein Adherence − + + + + Type IV pili biosynthesis pilR Type 4 fimbriae expression regulatory protein pilR Adherence + − − − − VirR/VirS virR Hypothetical protein Secretion system + + + + + S5. Comparative genomic analyses of single nucleotide polymorphisms of the five isolates against BH819
Genomic BF33.1 BF33.2 BF38 BF43 BF45 Synonymous 29,934 29,909 29,910 29,975 29981 Non-synonymous 11,630 11,589 11,605 11,599 11,613 Total CDS SNPs 41,812 41,742 41,759 41,820 41,845 Total SNPs 44,924 44,838 44,859 44,935 44,959 Note. CDS, coding sequence; SNPs, single nucleotide polymorphisms. S6. Comparative genomic analyses of the insertion and deletion (InDel) events of the five isolates against BH819
Items BF33.1 BF33.2 BF38 BF43 BF45 Frame-shifted 11 7 9 7 8 Start codon 0 0 0 0 1 Stop codon 0 0 0 0 0 Premature stop 0 0 0 0 0 CDS with InDel 24 20 21 20 21 CDS of the reference strain 2,822 2,822 2,822 2,822 2,822 Note. CDS, coding sequence. S7. Information about bats
The number
of the batBat species Location of samples collection Habitat types from
where bats capturedSamples type Isolated strain type Culture medium
used for strainsNO.3 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.7 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.7 Brown Bat He Nan cave dwelling liver Enterococcus Chocolate Medium NO.7 Brown Bat He Nan cave dwelling liver Staphylococcus epidermidis Blood plate NO.9 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.13 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.14 Brown Bat He Nan cave dwelling liver Vagococcus fluvialis Chocolate Medium NO.18 Brown Bat He Nan cave dwelling liver Staphylococcus epidermidis Blood plate NO.18 Brown Bat He Nan cave dwelling liver Actinomycetes Agar medium NO.22 Brown Bat He Nan cave dwelling liver Aeromonas veronii Blood plate NO.22 Brown Bat He Nan cave dwelling spleen Enterococcus Chocolate Medium NO.22 Brown Bat He Nan cave dwelling spleen Enterococcus faecalis Chocolate Medium NO.25 Brown Bat He Nan cave dwelling spleen Enterococcus faecalis Chocolate Medium NO.26 Brown Bat He Nan cave dwelling liver Bacillus subtilis Blood plate NO.27 Brown Bat He Nan cave dwelling liver Corynebacterium Blood plate NO.28 Brown Bat He Nan cave dwelling liver Bacillus brevis Blood plate NO.28 Brown Bat He Nan cave dwelling liver Corynebacterium Blood plate NO.28 Brown Bat He Nan cave dwelling spleen Enterococcus Chocolate Medium NO.31 Brown Bat He Nan cave dwelling spleen Aeromonas veronii Chocolate Medium NO.32 Brown Bat He Nan cave dwelling spleen Enterococcus faecalis Chocolate Medium NO.33 Brown Bat He Nan cave dwelling spleen Enterococcus Chocolate Medium NO.36 Brown Bat He Nan cave dwelling spleen staphylococcus Chocolate Medium NO.38 Brown Bat He Nan cave dwelling spleen Klebsiella pneumoniae Chocolate Medium NO.38 Brown Bat He Nan cave dwelling liver Staphylococcus haemolyticus Chocolate Medium NO.40 Brown Bat He Nan cave dwelling liver Staphylococcus haemolyticus Chocolate Medium NO.40 Brown Bat He Nan cave dwelling liver kocuria sp Chocolate Medium NO.40 Brown Bat He Nan cave dwelling liver Enterococcus Chocolate Medium NO.41 Brown Bat He Nan cave dwelling liver Staphylococcus coriolis Chocolate Medium NO.42 Brown Bat He Nan cave dwelling liver Staphylococcus coriolis Chocolate Medium NO.44 Brown Bat He Nan cave dwelling liver Proteus Chocolate Medium NO.44 Brown Bat He Nan cave dwelling lung Bacillus sphaericus Chocolate Medium NO.45 Brown Bat He Nan cave dwelling lung Micrococcus lylae Chocolate Medium NO.45 Brown Bat He Nan cave dwelling lung Enterococcus Chocolate Medium NO.46 Brown Bat He Nan cave dwelling lung Shigella faecalis Chocolate Medium NO.47 Brown Bat He Nan cave dwelling lung Bacillus subtilis Chocolate Medium NO.47 Brown Bat He Nan cave dwelling lung Enterococcus faecalis Chocolate Medium -
[1] Collins MD, Ash C, Farrow JAE, et al. 16S ribosomal ribonucleic acid sequence analyses of lactococci and related taxa. Description of Vagococcus fluvialis gen. nov., sp. nov. J Appl Bacteriol, 1989; 67, 453−60. doi: 10.1111/j.1365-2672.1989.tb02516.x [2] Schleifer KH, Kraus J, Dvorak C, et al. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst Appl Microbiol, 1985; 6, 183−95. doi: 10.1016/S0723-2020(85)80052-7 [3] Teixeira LM, Carvalho MDGS, Merquior VLC, et al. Phenotypic and genotypic characterization of Vagococcus fluvialis, including strains isolated from human sources. J Clin Microbiol, 1997; 35, 2778−81. doi: 10.1128/jcm.35.11.2778-2781.1997 [4] Hamm PS, Dunlap CA, Mullowney MW, et al. Streptomyces buecherae sp. nov., an actinomycete isolated from multiple bat species. Anton Leeuw, 2020; 113, 2213−21. doi: 10.1007/s10482-020-01493-4 [5] Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M. Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons. 1991, 115-75. [6] Facklam R, Elliott JA. Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin Microbiol Rev, 1995; 8, 479−95. doi: 10.1128/CMR.8.4.479 [7] Chen LH, Yang J, Yu J, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res, 2005; 33, D325−8. doi: 10.1093/nar/gki177 [8] Liu B, Pop M. ARDB--antibiotic resistance genes database. Nucleic Acids Res, 2009; 37, D443−7. doi: 10.1093/nar/gkn656 [9] Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol, 2016; 33, 1870−4. doi: 10.1093/molbev/msw054 [10] Cláudio VC, Gonzalez I, Barbosa G, et al. Bacteria richness and antibiotic-resistance in bats from a protected area in the Atlantic Forest of Southeastern Brazil. PLoS One, 2018; 13, e0203411. doi: 10.1371/journal.pone.0203411 -
 20509Supplementary Materials.pdf
20509Supplementary Materials.pdf

-




 下载:
下载:





 Quick Links
Quick Links