-
Methicillin-resistant Staphylococcus aureus (MRSA) is a pathogen that has been identified in the community (community-associated MRSA, CA-MRSA), hospital (hospital-associated MRSA, HA-MRSA), and livestock (livestock-associated MRSA, LA-MRSA), particularly in pigs, and is among the most frequent community-and nosocomial-infections in the world[1-2]. The increasing antimicrobial resistance rates of MRSA have accelerated in recent years, and have posed a serious threat to public health[3-4].In 2011, the total costs due to CA-MRSA was estimated 1.4 billion to 13.8 billion dollars in the United States[5].The control and prevention of MRSA is considered as a serious public health challenge[6-7]. In recent years, MRSA has been isolated from retail meat and livestock, including pig, poultry, and cattle meat[8-10]. Additionally, pig serves as an important reservoir for the spread of LA-MRSA[11].
Epidemiologically, the relative prevalence of MRSA lineages and their subtypes appears to vary geographically[12-13]. ST398 was the first identified LA-MRSA sequence type (ST) and is the predominant ST identified in Europe. ST9 is the most predominant MRSA clone among pigs in the Asian countries, and both ST398 and ST5 are relatively more frequent in pigs in North America[14-20].Specifically, significant geographic variation in the ST398 lineage distribution has been reported in Europe, with spa types t108 and t011 is common in Netherlands and spa type t034 is predominant in Denmark[13, 21-22]. In China, t899 and ST9 are the most prevalent spa types in pigs[16].
Different molecular typing methods of MRSA have been used for epidemiological studies because different methods may provide diverse discriminatory powers. For this reason, the use of more than one subtyping method, such as pulsed-field gel electrophoresis (PFGE), multi-locus sequencing typing (MLST), spa typing, multiple-locus variable number of tandem repeat analysis (MLVA), is essential to more comprehensively assess the genetic diversity of MRSA and to provide the genetic basis for evolutionary and epidemiological studies of MRSA. This study aimed to explore the phylogenetic distribution and population characteristics of MRSA isolated from pigs and retail foods and evaluate the potential risk presented by MRSA to food safety and human public health.
-
In 2011, a total of 56 MRSA strains were isolated from 961 pig nasal swabs obtained from 49 pig farms located in six provinces, and 15 MRSA strains were isolated from 50, 316 retail market food samples located in 32 provinces in China. In detail, two isolates were obtained from ready-to-eat (RTE) vegetable salad, five from cooked meat, two from cooked noodles, three from raw pork meat, two from raw mutton and one from raw beef. The prevalence of S. aureus in the samples was determined using the qualitative detection method according to National Food Safety Standards of China document GB 4789.10-2010. Briefly, an aliquot 25 g from each sample was transferred to a sterile glass flask containing 225 mL of 10% saline solution (Land Bridge, Beijing, China). Following homogenization, the solutions were incubated at 37 ℃ for 24 h; while, the nasal swabs were incubated in a 5 mL 10% saline solution (Land Bridge). Loopfuls of the resulting cultures were streaked onto Baird-Parker Agar plate and Blood Agar plate (Land Bridge), respectively, then incubated at 37 ℃ for 24-48 h. Putative S. aureus isolates were tested for coagulase activity, and were further confirmed using API STAPH test strips (bioMerieux, Marcyl'Etoile, France). Finally, all MRSA isolates were then confirmed by detection of 16S rRNA, nuc, and mecA by PCR[23]. All confirmed MRSA isolates were stored in brain heart infusion broth with 40% glycerol in a -80 ℃ freezer before further genotypic characterization.
-
Antimicrobial susceptibility of all MRSA isolates against 13 antimicrobials was determined by the broth dilution method and interpreted according to the Clinical and Laboratory Standards Institute guidelines (CLSI)[24]. These antimicrobials were benzyl penicillin (BEN), oxacillin (OXA), gentamicin (GEN), ciprofloxacin (CIP), levofloxacin (LEV), moxifloxacin (MXF), erythromycin (ERY), clindamycin (CLI), vancomycin (VAN), nitrofurantoin (NIT), rifampicin (RIF), tetracycline (TET), and trimethoprim-sulfamethoxazole (SXT). S. aureus ATCC 29213 was used as the control for the antimicrobial susceptibility testing.
-
The frozen strains were cultured in brain heart infusion broth and incubated for overnight at 37 ℃. A TIANamp Bacterial DNA extraction kit (DNA Kit DP302, Beijing, China) was used to extract the genomic DNA from the isolates according to the manufacturer's instructions, which were adapted for Gram-positive bacteria through pretreatment with lysostaphin (0.1 g/L). A NanoDrop-2000 spectro-photometer (Thermo Fisher Scientific, NH, USA) was used to evaluate the quality and quantity of DNA. Samples diluted in sterile deionized water at a concentration of 50 ng/μL were used as DNA templates for PCR amplification.
-
Seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) were used in the MLST analysis, which was performed according to the protocol described by Enright et al.[25].The alleles and the ST were assigned according to the MLST database (http://www.mlst.net/). eBURST software (http://eburst.mlst.net) was used to cluster STs into clonal complexes (CC). Strains that diverged at no more than one of the seven MLST loci were considered as a single CC. Double-locus variant were included in the CC if the linking single-locus variant was present in the MLST database.
-
The x region of the spa gene was amplified according to the protocol previously described by Applied Biosystems (Applied Biosystems, Foster City, USA)[26]. The DNA was then directly sequenced using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction (Applied Biosystems, Foster City, USA) on an ABI 3730 sequencer. The sequences were analyzed and spa types were assigned using the spa Type Finder (http://fortinbras.us/cgi-bin/spaTyper/spaTyper.pl) and the Ridom Spa Server database (http://spa.ridom.de).
-
MLVA was performed as previously described[27]. Eight VNTR loci (VNTR09_01, VNTR21_01, VNTR81_01, VNTR67_01, VNTR63_01, VNTR24_01, VNTR61_01, and VNTR61_02) were amplified in three multiplex PCRs using an Applied Biosystems 9700 PCR machine (Applied Biosystems, Foster City, USA). The PCR products were subjected to capillary electrophoresis on a 3730xl genetic analyzer (Applied Biosystems, Foster City, USA). The number of repeats of each VNTR locus was analyzed by the Gene-Marker software (Softgenetics, State College, USA). The calculated numbers of repeats of the eight VNTR loci were combined into a string of eight integers. This string representing the MLVA profile was then submitted to the MLVA database (www.mlva.net) to obtain the corresponding MLVA complex. The MLVA patterns were analyzed using Bionumerics software (Applied Math, Belgium) for clustering.
-
Total genomic DNA for PFGE analysis was extracted according to the protocol described by Murchan et al.[28]. Briefly, the target DNA of the tested strains was digested with SmaⅠ restriction enzyme (20 units/μL, New England Biolabs) in accordance with the supplier's instructions and separated by electrophoresis on a 1.0 agarose SeaKem Gold gel using the CHEF DR Ⅲ system (Bio-Rad, Hercules, California). The PFGE patterns were interpreted with bionumerics software (Applied Math, Belgium) using Unweighted Pair Group Method with Arithmetic Mean (UPGMA). Bands that were smaller than 20.5 kb were not included in the analysis. Patterns that were indistinguishable computationally and by visual inspection were assigned the same pattern designation. Salmonella Braenderup H9812 was used as a positive control. The reference standard, plug preparation, and restriction digestion of this strain was performed according to the protocol described by Ribot et al.[29].
-
The genetic diversity and discriminatory ability of typing were calculated by the Simpson's index of diversity (diversity index, DI). The formula is as follows:
$$DI = 1 - \frac{1}{{\left[ {N\left( {N - 1} \right)} \right]}}\sum\limits_{j - 1}^s {{n_j}} \left( {{n_j} - 1} \right)$$ (1) In this formula, nj is the number of strains belonging to the jth type, and N is the total number of strains in the sample population.
-
MRSA were isolated from 56 of the pig nasal swabs (5.80%, 56/961) and from 15 of the 50, 316 retail food samples.
-
MLST was successfully performed for all (n = 71) isolates and the results are summarized in Table 1 and Figure 1. The MLST analysis revealed eight STs (ST9, ST10, ST59, ST338, ST398, ST630, ST903, and ST1376). The evolutionary relationships were determined by the algorithm eBURST and the results revealed that these STs belonged to five CCs (CC8, CC9, CC10, CC59, and CC398) (Figure 2 and Table 2). In this study, ST9 was as the most frequent ST type (CC9) among both pig-and food-associated isolates. Among the 56 isolates from pigs, ST9 (54/56) was the prevalent strain; ST1376 (1/56) and ST398 (1/56) were also identified. Among 15 isolates from the retail food samples, ST9 was the prevalent strain followed in decline order by ST630, ST338, ST59, ST398, ST10, and ST903 (Table 2).
Table 1. Diversity Indices (DI) of the genotyping methods for MRSA isolates
Samples Method No. of isolates No. of types Average No. of per type DI All tested MRSA MLST 71 8 8.9 0.262 spa 71 8 8.9 0.371 MLVA 71 16 4.4 0.503 PFGE 68 51 1.3 0.989 Pig MRSA MLST 56 3 18.7 0.071 spa 56 1 56 0.000 MLVA 56 6 9.3 0.236 PFGE 55 40 1.4 0.983 Food MRSA MLST 15 7 1.9 0.867 spa 15 8 1.9 0.791 MLVA 15 10 1.5 0.924 PFGE 13 11 1.2 0.981 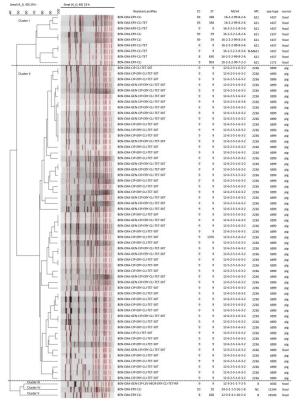
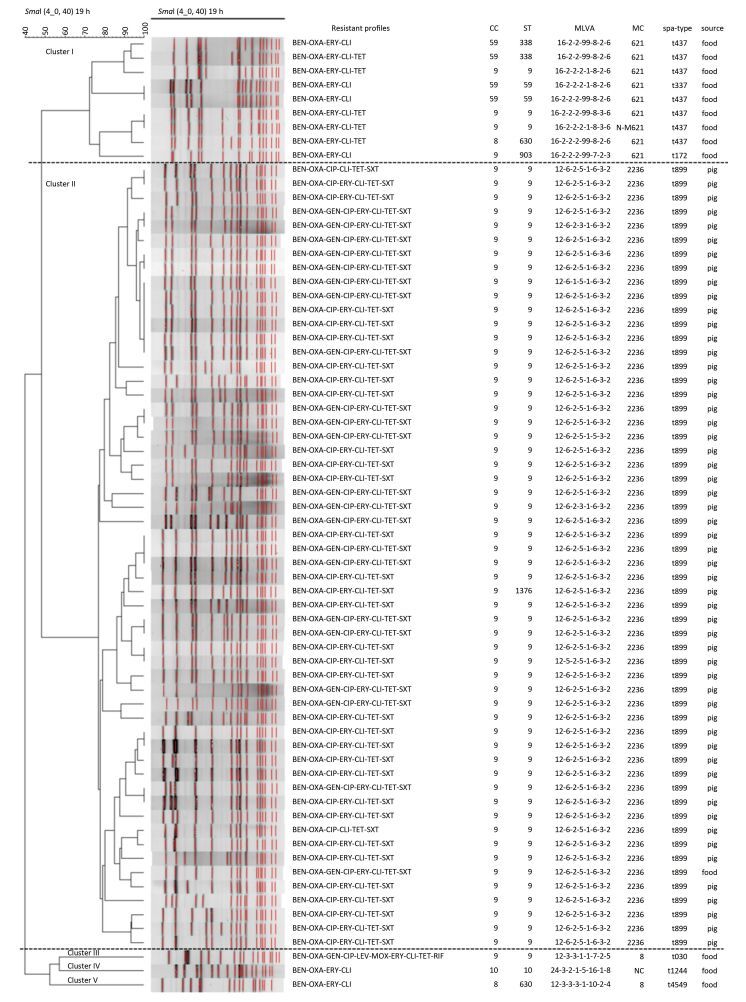
Figure 1. Resistant profiles, clone relationships, and the sample sources of 68 MRSA isolates, established with SmaI PFGE analysis. CC = clone complexes, ST = sequence types, MLVA = MLVA profiles, MC = MLVA complexes.
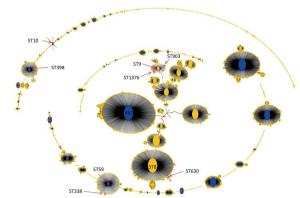
Figure 2. Population snapshot analyses by eBURST on 5782 S.aureus strains that belonged to 2766 STs in S.aureus MLST database. Pink circles labeled with red arrows and the ST names in black font are used for sequence types identified in this study. ST9, ST903, and ST1376 belonged to CC9. ST59 and ST338 belonged to CC59. ST630 belonged to CC8. ST398 belonged to CC398. ST10 belonged to CC10.
Table 2. Molecular typing and antimicrobial profile of MRSA Isolates
Samples CC-MLST spa t ype MLVA c omplex PFGE c luster Resistant p rofile No. of Isolates Pig CC9-ST9 t899 MC2236 Ⅱ BEN-OXA-ERY-CLI-TET-CIP-SXT, BEN-OXA-ERY-CLI-TET-GEN-CIP-SXT 54 CC9-ST1376 t899 MC2236 Ⅱ BEN-OXA-ERY-CLI-TET-CIP-SXT 1 CC398-ST398 t034 MC2236 - BEN-OXA-ERY-CLI-TET 1 Food CC8-ST630 t437 MC621 Ⅰ BEN-OXA-ERY-CLI-TET 1 (1 raw beef) CC8-ST630 t4549 MC8 Ⅴ BEN-OXA-ERY-CLI 1 (1 RTE vegetable salad) CC9-ST9 t437 MC621/N-MC621 Ⅰ BEN-OXA-ERY-CLI 3 (1 raw mutton and 2 cooked meat) CC9-ST9 t030 MC8 Ⅲ BEN-OXA-ERY-CLI-TET-GEN-CIP-LEV-MOX-RIF 1 (1 cooked noodles) CC9-ST9 t899 MC2236 Ⅱ BEN-OXA-ERY-CLI-TET-GEN-CIP-SXT 1 (1 raw pork meat) CC9-ST903 t172 MC621 Ⅰ BEN-OXA-ERY-CLI 1 (1 cooked meat) CC10-ST10 t1244 NMC Ⅳ BEN-OXA-ERY-CLI 1 (1 cooked meat) CC59-ST59 t437 MC621 Ⅰ BEN-OXA-ERY-CLI 1 (1 RTE vegetable salad) CC59-ST59 t337 MC621 Ⅰ BEN-OXA-ERY-CLI 1 (1 cooked meat) CC59-ST338 t437 MC621 Ⅰ BEN-OXA-ERY-CLI, BEN-OXA-ERY-CLI-TET 2 (1 raw mutton and 1 cooked noodles) CC398-ST398 t034 MC398 -* BEN-OXA-ERY-CLI 2 (2 raw pork meat) Note.*No PFGE cluster generated. -
The spa typing revealed eight spa types of among the 71 isolates (Figure 1 and Tables 1-2). The t899 type (55/71, 78.87%) was the prevalent spa type followed by one t034 among MRSA isolates from pigs (Table 2). The t437 type was the prevalent spa type of the MRSA isolates from retail food samples, followed in order of prevalence by t034, t030, t172, t337, and t4549.
-
In total, the 71 MRSA isolates were assigned into 16 MLVA types (Figure 1 and Tables 1-2), which were clustered into six MLVA complexes (MC8, MC398, MC621, MC2236, N-MC398, and N-MC621) with the exception of one isolate, which had a profile of 24-3-2-1-5-16-1-8 and could not be assigned into any MLVA complex (no complex, Figure 3). The predominant MLVA type was 12-6-2-5-1-6-3-2, which belonged to MC2236, constituting 70.42% (50/71) of all isolates and existing both in pig-and food-associated isolates. Fifty-five out of 56 (98.21%) pig-associated isolates were MC2236, including six MLVA types, and MLVA type 12-6-2-5-1-6-3-2 was the predominant MLVA type including 49 isolates. The remaining pig-associated isolate was N-MC398, with an MLVA type of 14-0-2-4-1-6-1-2. Eight out of 15 (53.33%) food-associated isolates were MC621, including four MLVA types, and 16-2-2-2-99-8-2-6 was the predominant MLVA type including four isolates. The other MLVA complexes were MC8, MC398, MC2236, N-MC621, and one that did not match to any complex.

Figure 3. Minimum spanning tree of the MRSA isolates as typed by MLVA. Clustering of MLVA profiles was done using a categorical coefficient. In the minimum spanning tree the MLVA types are displayed as circles. The size of each circle indicates the number of isolates for this particular type. Thick solid lines connect types that differ in a single VNTR locus and a thin solid line connects types that differ in two VNTR loci. The color of the halo surrounding the MLVA types denotes that types belonged to the same complex. VNTR, variable number of tandem repeat.
-
Among 71 MRSA isolates subjected to PFGE typing, three isolates were SmaI non-type-able (belonging to ST398). The remaining 68 isolates were differentiated into 51 PFGE types, which were further grouped into five clusters by PFGE pattern and all had more than 72% similarity (Figure 1 and Tables 1-2). The predominant PFGE cluster was cluster Ⅱ and included 56 isolates (55 pig-associated isolates and one food-associated isolate originating from raw pork meat), which included 41 PFGE types. All MRSA isolates in cluster Ⅱ were characterized as CC9 (ST9/ST1376)-t899-MC2236. In this cluster, one pig-associated isolate was MLST type ST1376 and the others were ST9. cluster Ⅰ included nine food-associated isolates of seven PFGE types. The vast majority of MRSA isolates in cluster Ⅰ were CC9-t437-MC621/N-MC621 and CC59-t437-MC621. Two isolates were identical by PFGE but had different MLVA and spa types (Figure 1). Each of the other three clusters included only one food-associated isolate. The DI of PFGE, MLVA, MLST, and spa typing for all tested isolates were 0.989, 0.503, 0.371, and 0.262, respectively.
-
The association between antimicrobial resistance phenotypes and genotypes is shown in Table 2 and Figure 2. In this study, all MRSA isolates showed resistance to BEN, OXA, ERY, and CLI, and sensitivity to VAN and NIT. The resistance rate to CIP, TET, and SXT was almost 80%; 32.40% of isolates were resistant to GEN and 1.40% of isolates were resistant to LEV, MOX, and RIF (Table 3). All MRSA isolates showed MDR sharing common resistance profile in each clone type. Isolates identified as CC9-ST9 clones with t899-MC2236-PFGE cluster Ⅱ and t030-MC8-PFGE cluster Ⅲ exhibited broader antimicrobial resistance profiles compared to other clones studied here (Figure 1 and Table 2).
Table 3. Antimicrobial resistance patterns of MRSA isolates from pigs and foods
Antimicrobial agent Resistant (%) Pigs (n = 56) Food (n = 15) Total Benzyl penicillin 100.00% 100.00% 100.00% Oxacillin 100.00% 100.00% 100.00% Gentamicin 13.33% 37.50% 32.39% Ciprofloxacin 13.33% 98.21% 80.28% Levofloxacin 6.67% 0.00% 1.41% Moxifloxacin 6.67% 0.00% 1.41% Erythromycin 100.00% 100.00% 100.00% Clindamycin 100.00% 100.00% 100.00% Vancomycin 0.00% 0.00% 0.00% Tetracycline 46.67% 100.00% 88.73% Nitrofurantoin 0.00% 0.00% 0.00% Rifampicin 6.67% 0.00% 1.41% Trimethoprim-sulfamethoxazole 6.67% 100.00% 80.28% -
This study investigated the genetic diversity of MRSA isolated from pigs and retail food samples. The relationship between antimicrobial resistance phenotype and genotype was also determined. The level of antimicrobial resistance among food animals has steadily increased and has become a major health problem in the world. Strategies to eliminate or decrease the prevalence of MRSA in pigs and generally in livestock are a public health priority.
Comparing to similar studies in China, the prevalence rate (5.80%, 56/961) of MRSA among pig samples in this study is lower than previously reported in four provinces (11.4%, 58/509) and Hong Kong (16.0%, 16/100) in China[14, 16], but was higher than those reported from other countries, such as Malaysia (1.40%, 5/360) and Japan (0.90%, 1/115), respectively[30-31]. Over the last few decades, antimicrobial resistance of MRSA has been frequently reported in pigs or retail food samples[32-33]. In addition, pig-associated MRSA strains showed lower resistant to SXT (6.67%), similar to the results from a survey conducted in four Chinese provinces (all showed susceptible to SXT)[16], but lower than reported for Taiwan, China (90%)[32]. A possible explanation of this pattern is that the standards or polices for the application of antimicrobials during pig breeding differ in different regions. For example, the higher rate of resistance to SXT among pig MRSA isolates in Taiwan, China might be attributed to the residual effects of previous low-dose use of this agent as a feed additive in Taiwan, China[32].
The ST9 (CC9) strains were found in both pig and retail food samples. This prevalence of ST9 is consistent with that previously reported for pigs in China and other regions of the world[16]. The prevalence of ST9 has also been documented in retail pork, beef, turkey, and chicken samples in the United States, Germany, and England[34-36]. ST9 isolates have also been reported as a dominant clone among pig farmers in China, and other countries[16, 37]. Additionally, a recent Chinese study has confirmed that three ST9 MRSA were isolated from three patients with no contact with livestock, suggesting that LA-MRSA strains can cause humans infections[38]. MRSA is the leading cause of infections, ranging from mild skin and soft tissue infections to life threatening diseases such as septicemia, necrotizing fasciitis, endocarditis, and necrotizing pneumonia[39]. Therefore, it is necessary to urgently take control measures to delay the development of LA-MRSA isolates, especially ST9 clone. In our study, three MRSA isolates (1 isolate from pig and 2 isolated from raw pork meat) were characterized as ST398 (CC398). This sequence type is the most prevalent type in pigs in Europe and North America[40-41], and is also detected in cow milk and poultry[42-43]. Interestingly, interspecies transmission of MRSA ST398 (from livestock to human) has been reported in many countries[44-45].Although ST398 MRSA isolates occupied a small proportion in this study, they still have the potential risk of dissemination between different geographic regions and hosts. Besides, one MRSA characterized as ST630 (CC8) was obtained in this study. ST630 was the most common sequence type in milk of cows with mastitis in the mid-east of China[46]. Other sequence types, such as ST903/ST1376 (CC9), ST59/ST338 (CC59), and ST10 (CC10), were also found in this study. It has already been reported that CA-MRSA infection can be disseminated through a community or hospital[47-48]. Pigs or food, especially retail meat, could become important reservoirs for MRSA and increase the potential risk of infections in humans.
In this study, t899 was found as the most frequently distributed genotype among pig samples, followed by ST1376. The genotype ST9-t899 was previously identified as the predominant clone among pigs in China[49], which is consistent with our findings. Variant t437 was the most common isolate from retail food samples (46.70%, 7/15) in this study, including raw pork meat, RTE vegetable salad, cooked meat and cooked noodles, which is in agreement with previous reports from Asia, Europe, and particularly China[50-51]. The data of this study may suggest that the CC59-t437 clone might have been transmitted from Asia to Europe via trade and travelling. Furthermore, we also found three t034 with ST398 strains in both pig and food samples. These strains are typically associated with LA-MRSA[40-41]. Patients with this spa type usually work in close contact with a major livestock reservoir of MRSA[52]. The other spa types, t030, t172, t1244, and t4549, were also reported as CA-MRSA or HA-MRSA causing infections in China[53-55].
In this study, 68 MRSA isolates were typed by PFGE and five pulse clusters of isolates were identified. MLVA yielded 16 types that were clustered into 11 distinct MLVA complexes. These results appeared to coincide with MLST clonal complexes. The MLVA approach was at least as discriminatory as PFGE and twice as discriminatory as spa typing. The current observations of this study are in complete agreement with those reported by Schouls et al.[27].
This study revealed the molecular characteristics of pig-and food-associated MRSA in China. Consistent with a previous report, CC9-ST9-t899-MC2236 was the predominant clone isolated from pigs. The genotyping results of MRSA among retail food samples showed more diversity, and CC9-ST9-t437-MC621 was the predominant clone. Additionally, some CA-MRSA and HA-MRSA were also detected among retail food samples (t030, t172, t1244, and t4549)[53-55].
To the best of our knowledge, this is the first study to investigate the genotypic diversity of MRSA isolated from different geographical regions of China. The results of the present study are important giving a deep insight in the epidemiology of MRSA because covered different regions of China, examined a great number of MRSA isolates from pigs and retail foods, and used several different typing methods. However, there were few limitations in this study. First, the samples were collected retrospectively and selection bias may have been introduced. Secondly, the study subjects may not represent the general population. MRSA may be disseminated circulate among livestock, food, community, and hospital, making its control and prevention difficult. As all MRSA isolates in this study were MDR, a future large-scale, multi-population and food item-based surveillance of MRSA should be conducted to obtain comprehensive information on the prevalence of MRSA in various animal populations and food items.
-
The authors declare no competing financial interests.
-
WANG Wei: Experimental design, Data analysis, Manuscript editing; LIU Feng: Experimental design, Data analysis; Zulqarnain Baloch: Experimental design, Data analysis, Manuscript writing; ZHANG Cun Shan: Antimicrobial susceptibility test; MA Ke: MLST analysis; PENG Zi Xin: spa typing analysis; YAN Shao Fei: MLVA analysis; HU Yu Jie: PFGE analysis; GAN Xin: PFGE analysis; DONG Yin Ping: Data management; BAI Yao: Data analysis; LI Feng Qin: Data analysis; YAN Xiao Mei: Data analysis; MA Ai Guo: Experimental design, Manuscript editing; XU Jin: Experimental design, Manuscript editing.
-
The data supporting the findings contained within this manuscript will be shared upon request submitted to the corresponding author.
doi: 10.3967/bes2017.076
Genotypic Characterization of Methicillin-resistant Staphylococcus aureus Isolated from Pigs and Retail Foods in China
-
Abstract:
Objective To investigate the genotypic diversity of Methicillin-resistant Staphylococcus aureus (MRSA) isolated from pigs and retail foods from different geographical areas in China and further to study the routes and rates of transmission of this pathogen from animals to food. Methods Seventy-one MRSA isolates were obtained from pigs and retail foods and then characterized by multi-locus sequencing typing (MLST), spa typing, multiple-locus variable number of tandem repeat analysis (MLVA), pulsed-field gel electrophoresis (PFGE), and antimicrobial susceptibility testing. Results All isolated MRSA exhibited multi-drug resistance (MDR). Greater diversity was found in food-associated MRSA (7 STs, 8 spa types, and 10 MLVA patterns) compared to pig-associated MRSA (3 STs, 1 spa type, and 6 MLVA patterns). PFGE patterns were more diverse for pig-associated MRSA than those of food-associated isolates (40 vs. 11 pulse types). Among the pig-associated isolates, CC9-ST9-t899-MC2236 was the most prevalent clone (96.4%), and CC9-ST9-t437-MC621 (20.0%) was the predominant clone among the food-associated isolates. The CC9-ST9 isolates showed significantly higher antimicrobial resistance than other clones. Interestingly, CC398-ST398-t034 clone was identified from both pig-and food-associated isolates. Of note, some community-and hospital-associated MRSA strains (t030, t172, t1244, and t4549) were also identified as food-associated isolates. Conclusion CC9-ST9-t899-MC2236-MDR was the most predominant clone in pigs, but significant genetic diversity was observed in food-associated MRSA. Our results demonstrate the great need for improved surveillance of MRSA in livestock and food and effective prevention strategies to limit MDR-MRSA infections in China. -
Figure 2. Population snapshot analyses by eBURST on 5782 S.aureus strains that belonged to 2766 STs in S.aureus MLST database. Pink circles labeled with red arrows and the ST names in black font are used for sequence types identified in this study. ST9, ST903, and ST1376 belonged to CC9. ST59 and ST338 belonged to CC59. ST630 belonged to CC8. ST398 belonged to CC398. ST10 belonged to CC10.
Figure 3. Minimum spanning tree of the MRSA isolates as typed by MLVA. Clustering of MLVA profiles was done using a categorical coefficient. In the minimum spanning tree the MLVA types are displayed as circles. The size of each circle indicates the number of isolates for this particular type. Thick solid lines connect types that differ in a single VNTR locus and a thin solid line connects types that differ in two VNTR loci. The color of the halo surrounding the MLVA types denotes that types belonged to the same complex. VNTR, variable number of tandem repeat.
Table 1. Diversity Indices (DI) of the genotyping methods for MRSA isolates
Samples Method No. of isolates No. of types Average No. of per type DI All tested MRSA MLST 71 8 8.9 0.262 spa 71 8 8.9 0.371 MLVA 71 16 4.4 0.503 PFGE 68 51 1.3 0.989 Pig MRSA MLST 56 3 18.7 0.071 spa 56 1 56 0.000 MLVA 56 6 9.3 0.236 PFGE 55 40 1.4 0.983 Food MRSA MLST 15 7 1.9 0.867 spa 15 8 1.9 0.791 MLVA 15 10 1.5 0.924 PFGE 13 11 1.2 0.981 Table 2. Molecular typing and antimicrobial profile of MRSA Isolates
Samples CC-MLST spa t ype MLVA c omplex PFGE c luster Resistant p rofile No. of Isolates Pig CC9-ST9 t899 MC2236 Ⅱ BEN-OXA-ERY-CLI-TET-CIP-SXT, BEN-OXA-ERY-CLI-TET-GEN-CIP-SXT 54 CC9-ST1376 t899 MC2236 Ⅱ BEN-OXA-ERY-CLI-TET-CIP-SXT 1 CC398-ST398 t034 MC2236 - BEN-OXA-ERY-CLI-TET 1 Food CC8-ST630 t437 MC621 Ⅰ BEN-OXA-ERY-CLI-TET 1 (1 raw beef) CC8-ST630 t4549 MC8 Ⅴ BEN-OXA-ERY-CLI 1 (1 RTE vegetable salad) CC9-ST9 t437 MC621/N-MC621 Ⅰ BEN-OXA-ERY-CLI 3 (1 raw mutton and 2 cooked meat) CC9-ST9 t030 MC8 Ⅲ BEN-OXA-ERY-CLI-TET-GEN-CIP-LEV-MOX-RIF 1 (1 cooked noodles) CC9-ST9 t899 MC2236 Ⅱ BEN-OXA-ERY-CLI-TET-GEN-CIP-SXT 1 (1 raw pork meat) CC9-ST903 t172 MC621 Ⅰ BEN-OXA-ERY-CLI 1 (1 cooked meat) CC10-ST10 t1244 NMC Ⅳ BEN-OXA-ERY-CLI 1 (1 cooked meat) CC59-ST59 t437 MC621 Ⅰ BEN-OXA-ERY-CLI 1 (1 RTE vegetable salad) CC59-ST59 t337 MC621 Ⅰ BEN-OXA-ERY-CLI 1 (1 cooked meat) CC59-ST338 t437 MC621 Ⅰ BEN-OXA-ERY-CLI, BEN-OXA-ERY-CLI-TET 2 (1 raw mutton and 1 cooked noodles) CC398-ST398 t034 MC398 -* BEN-OXA-ERY-CLI 2 (2 raw pork meat) Note.*No PFGE cluster generated. Table 3. Antimicrobial resistance patterns of MRSA isolates from pigs and foods
Antimicrobial agent Resistant (%) Pigs (n = 56) Food (n = 15) Total Benzyl penicillin 100.00% 100.00% 100.00% Oxacillin 100.00% 100.00% 100.00% Gentamicin 13.33% 37.50% 32.39% Ciprofloxacin 13.33% 98.21% 80.28% Levofloxacin 6.67% 0.00% 1.41% Moxifloxacin 6.67% 0.00% 1.41% Erythromycin 100.00% 100.00% 100.00% Clindamycin 100.00% 100.00% 100.00% Vancomycin 0.00% 0.00% 0.00% Tetracycline 46.67% 100.00% 88.73% Nitrofurantoin 0.00% 0.00% 0.00% Rifampicin 6.67% 0.00% 1.41% Trimethoprim-sulfamethoxazole 6.67% 100.00% 80.28% -
[1] Karkaba A, Benschop J, Hill KE, et al. Characterisation of methicillin-resistant Staphylococcus aureus clinical isolates from animals in New Zealand, 2012-2013, and subclinical colonisation in dogs and cats in Auckland. New Zeal Vet J, 2016; 7, 1-6. http://explore.tandfonline.com/cfp/est/tnzv-1q2017-vsi [2] Wang D, Wang Z, Yan Z, et al. Bovine mastitis Staphylococcus aureus:antibiotic susceptibility profile, resistance genes and molecular typing of methicillin-resistant and methicillin-sensitive strains in China. Infect Genet Evol, 2015; 31, 9-16. doi: 10.1016/j.meegid.2014.12.039 [3] Lu C, Guo Y, Wang S, et al. Decreased vancomycin MICs among methicillin-Resistant Staphylococcus aureus clinical isolates at a Chinese tertiary hospital over a 12-year period. Front Microbiol, 2016; 7, 1714. doi: 10.3389/fmicb.2016.01714/pdf [4] Earls MR, Kinnevey PM, Brennan GI, et al. The recent emergence in hospitals of multidrug-resistant community-associated sequence type 1 and spa type t127 methicillin-resistant Staphylococcus aureus investigated by whole-genome sequencing:Implications for screening. PLoS One, 2017; 12, e0175542. doi: 10.1371/journal.pone.0175542 [5] Centers for Disease Control and Prevention. Antibiotic resistance Threats in the United States, 508. Available online at: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf. [2013-05-11] [6] Zhao C, Sun H, Wang H, et al. Antimicrobial resistance trends among 5608 clinical Gram-positive isolates in China:Results from the Gram-Positive Cocci Resistance Surveillance program (2005-2010). Diagn Microbiol Infect Dis, 2012; 73, 174-81. doi: 10.1016/j.diagmicrobio.2012.03.003 [7] Wang H, Liu YD, Sun HL, et al. In vitro activity of ceftobiprole, kinezolid, tigecycline, and 23 other antimicrobial agents against Staphylococcus aureus isolates in China. Diagn Microbiol Infect Dis, 2008; 62, 226-9. doi: 10.1016/j.diagmicrobio.2008.06.003 [8] Kluymans JA. Methicillin-resistant Staphylococcus aureus in food products:cause for concern or case for complacency? Clin Microbiol Infect, 2010; 16, 11-5. doi: 10.1111/j.1469-0691.2009.03110.x [9] Harper AL, Ferguson DD, Leedom Larson KR, et al. An overview of livestock-associated MRSA in agriculture. J Agromedicine, 2010; 15, 101-4. doi: 10.1080/10599241003627110 [10] De Boer E, Zwartkruis-Nahuis JT, Wit B, et al. Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int J Food Microbiol, 2009; 134, 52-6. doi: 10.1016/j.ijfoodmicro.2008.12.007 [11] Morgan M. Methicillin-resistant Staphylococcus aureus and animals:zoonosis or humanosis? J Antimicrob Chemot, 2008; 62, 1181-7. doi: 10.1093/jac/dkn405 [12] Battisti A, Franco A, Merialdi G, et al. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet Microbiol, 2010; 142, 361-6. doi: 10.1016/j.vetmic.2009.10.008 [13] European Food Safety Authority. Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008. EFSA Journal, 2009; 7, 1376. doi: 10.2903/j.efsa.2009.1376 [14] Guardabassi L, O'Donoghue M, Moodley A, et al. M Novel lineage of methicillin-resistant Staphylococcus aureus, Hong Kong. Emerg Infect Dis, 2009, 15, 1998-2000. doi: 10.3201/eid1512.090378 [15] Frana TS, Beahm AR, Hanson BM, et al. Isolation and characterization of methicillin-resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS One, 2013; 8, e53738. doi: 10.1371/journal.pone.0053738 [16] Cui S, Li J, Hu C, et al. Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J Antimicrob Chemother, 2009; 64, 680-3. doi: 10.1093/jac/dkp275 [17] Smith TC, Gebreyes WA, Abley MJ, et al. Methicillin-resistant Staphylococcus aureus in pigs and farm workers on conventional and antibiotic-free swine farms in the USA. PLoS One, 2013; 8, e63704. doi: 10.1371/journal.pone.0063704 [18] Smith TC, Male MJ, Harper AL, et al. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One, 2009; 4, e4258. doi: 10.1371/journal.pone.0004258 [19] Khanna T, Friendship R, Dewey C, et al. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol, 2008; 128, 298-303. doi: 10.1016/j.vetmic.2007.10.006 [20] Molla B, Byrne M, Abley M, et al. Epidemiology and genotypic characteristics of methicillin-resistant Staphylococcus aureus strains of porcine origin. J Clin Microbiol, 2012; 50, 3687-93. doi: 10.1128/JCM.01971-12 [21] Agerso Y, Hasman H, Cavaco LM, et al. Study of methicillin resistant Staphylococcus aureus (MRSA) in Danish pigs at slaughter and in imported retailed meat reveals a novel MRSA type in slaughter pigs. Vet Microbiol, 2012; 157, 246-50. doi: 10.1016/j.vetmic.2011.12.023 [22] de Neeling AJ, van den Broek MJ, Spalburg EC, et al. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol, 2007; 122, 366-72. doi: 10.1016/j.vetmic.2007.01.027 [23] Louie L, Goodfellow J, Mathieu P, et al. Rapid Detection of Methicillin-Resistant Staphylococci from Blood Culture Bottles by Using a Multiplex PCR Assay. J Clin Microbiol, 2002; 2786-90. [24] Clinical Laboratory Standards Institute. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline. Wayne Pennsylvania: Clinical Laboratory Standards Institute, 2015. [25] Enright MC, Day NPJ, Davies CE, et al. Multi-locus Sequence Typing for Characterization of Methicillin-Resistant and Methicillin-Susceptible Clones of Staphylococcus aureus. J Clin Microbiol, 2000; 38, 1008-15. [26] Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol, 2013; 41, 5442-8. [27] Schouls LM, Spalburg EC, van Luit M, et al. Multiple-locus Variable Number Tandem Repeat Analysis of Staphylococcus Aureus:Comparison with Pulsed-Field Gel Electrophoresis and spa-Typing. PLoS One, 2009; 4, e5082. doi: 10.1371/journal.pone.0005082 [28] Murchan S, Kaufmann ME, Deplano A, et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus:a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol, 2003; 41, 1574-85. doi: 10.1128/JCM.41.4.1574-1585.2003 [29] Ribot EM, Fair M, Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis, 2006; 3, 59-67. doi: 10.1089/fpd.2006.3.59 [30] Neela V, Mohd Zafrul A, Mariana NS, et al. Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J Clin Microbio, 2009; 47, 4138-40. doi: 10.1128/JCM.01363-09 [31] Baba K, Ishihara K, Ozawa M, et al. Isolation of methicillin resistant Staphylococcus aureus (MRSA) from pig in Japan. Int J Antimicrob Ag, 2010; 36, 352-4. doi: 10.1016/j.ijantimicag.2010.06.040 [32] Lo YP, Wan MT, Chen MM, et al. Molecular characterization and clonal genetic diversity of methicillin-resistant Staphylococcus aureus of pig-origin in Taiwan. Comp Immunol Microb, 2012; 35, 513-21. doi: 10.1016/j.cimid.2012.05.001 [33] Charlene RJ, Johnnie AD, John BB. Prevalence and characterization of methicillin-resistant Staphylococcus aureus isolates from retail Meat and humans in Georgia. J Clin Microb, 2013; 51, 1199-207. doi: 10.1128/JCM.03166-12 [34] Hanson BM, Dressler AE, Harper AL, et al. Prevelence of Staphylococcus aureus and methicillin-susceptible Staphylococcus aureus (MRSA) on retail meat in Iowa. J Infect Public Heath, 2011; 4, 169-74. doi: 10.1016/j.jiph.2011.06.001 [35] Meemken D, Blaha T, Tegeler R, et al. Livestock associated methicillin-resistant Staphylococcus aureus (LaMRSA) isolated from lesions of pigs at necropsy in northwest Germany between 2004 and 2007. Zoonoses Public Heath, 2010; 57, e143-8. doi: 10.1111/j.1863-2378.2009.01313.x [36] Dhup V, Kearns AM, Pichon B, et al. First report of identification of livestock-associated MRSA ST9 in retail meat in England. Epidemiol Infect, 2015; 143, 2989-92. doi: 10.1017/S0950268815000126 [37] Armand Lefevre L, Ruimy R, Andremont A. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg Infect Dis, 2005; 11, 711-4. doi: 10.3201/eid1105.040866 [38] Yu F, Lu C, Liu Y, et al. Emergence of quinupristin/dalfopristin resistance among livestock-associated Staphylococcus aureus ST9 clinical isolates. Int J Antimicrob Agents, 2014; 44, 416-9. doi: 10.1016/j.ijantimicag.2014.06.020 [39] Rodríguez-Lázaro D, Ariza-Miguel J, Diez-Valcarce M, et al. Foods confiscated from non-EU flights as a neglected route of potential methicillin-resistant Staphylococcus aureus transmission. Int J FoodMicrobiol, 2015; 209, 29-33. [40] Bouiller K, Gbaguidi-Haore H, Hocquet D, et al. Clonal complex 398 methicillin-susceptible Staphylococcus aureus bloodstream infections are associated with high mortality. Clin Microbiol Infect, 2016; 22, 451-5. doi: 10.1016/j.cmi.2016.01.018 [41] Graveland H, Wagenaar JA, Heesterbeek H, et al. Methicillin resistant Staphylococcus aureus ST398 in veal calf farming:human MRSA carriage related with animal antimicrobial usage and farm hygiene. PLoS One, 2010; 5, e10990. doi: 10.1371/journal.pone.0010990 [42] Bardiau M, Yamazaki K, Duprez JN, et al. Genotypic and phenotypic characterization of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk of bovine mastitis. Lett Appl Microbiol, 2013; 57, 181-6. doi: 10.1111/lam.2013.57.issue-3 [43] Nemati M, Hermans K, Lipinska U, et al. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry:first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob Agents Chemother, 2008; 52, 3817-9. doi: 10.1128/AAC.00613-08 [44] Golding GR, Bryden L, Levett PN, et al. Livestock-associated methicillin-resistant Staphylococcus aureus sequence type 398 in humans, Canada. Emerg Infect Dis, 2010; 16, 587-94. doi: 10.3201/eid1604.091435 [45] Van Cleef BAGL, Monnet DL, Voss A, et al. Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerg Infect Dis, 2011; 17, 502-5. doi: 10.3201/eid1703.101036 [46] Zhang L, Li Y, Bao H, et al. Population structure and antimicrobial profile of Staphylococcus aureus strains associated with bovine mastitis in China. Microb Pathog, 2016; 97, 103-9. doi: 10.1016/j.micpath.2016.06.005 [47] Graveland H, Wagenaar JA, Bergs K, et al. Persistence of livestock associated MRSA CC398 in humans is dependent on intensity of animal contact. PLoS One, 2011; 6, e16830. doi: 10.1371/journal.pone.0016830 [48] Van den Broek IVF, Van Cleef B a. GL, Haenen A, et al. Methicillin-resistant Staphylococcus aureus in people living and working in pig farms. Epidemiol Infect, 2009; 137, 700-8. doi: 10.1017/S0950268808001507 [49] Song JH, Hsueh PR, Chung DR, et al. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries:an ANSORP study. J Antimicrob Chemother, 2011; 66, 1061-9. doi: 10.1093/jac/dkr024 [50] Glasner C, Pluister G, Westh H, et al. Staphylococcus aureus spa type t437:identification of the most dominant community-associated clone from Asia across Europe. Clin Microbiol Infec, 2015; 21, 163. e1-8. https://www.narcis.nl/publication/RecordID/oai%3Apure.rug.nl%3Apublications%2F5d2782d3-34df-4de5-aafc-b7da0b24c380 [51] Wang X, Li GH, Xia XD, et al. Antimicrobial Susceptibility and Molecular Typing of Methicillin-Resistant Staphylococcus aureus in Retail Foods in Shaanxi, China. Foodborne Pathog Dis, 2014; 11, 281-6. doi: 10.1089/fpd.2013.1643 [52] Kock R, Brakensiek L, Mellmann A, et al. Cross-border comparison of the admission prevalence and clonal structure of methicillin-resistant Staphylococcus aureus. J Hosp Infect, 2009; 71, 320-6. doi: 10.1016/j.jhin.2008.12.001 [53] Guo XC, Guang YB, Yong ZC, et al. Prevalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int J Food Microbiol, 2015; 211, 142-7. doi: 10.1016/j.ijfoodmicro.2015.07.018 [54] Ho PL, Chiu SS, Chan MY, et al. Molecular epidemiology and nasal carriage of Staphylococcus aureus and methicillin-resistant S. aureus among young children attending day care centers and kindergartens in Hong Kong. J Infection, 2012; 64, 500-6. doi: 10.1016/j.jinf.2012.02.018 [55] He WQ, Chen HB, Zhao CJ, et al. Prevalence and molecular typing of oxacillin-susceptible mecA-positive Staphylococcus aureus from multiple hospitals in China. Diagn Mic Infec Dis, 2013; 77, 267-9. doi: 10.1016/j.diagmicrobio.2013.07.001 -




 下载:
下载:



 Quick Links
Quick Links