-
West Nile virus (WNV), an arthropod-borne virus, is a member of the Japanese encephalitis virus (JEV) serocomplex of the Flaviviridae family[1]. WNV infections occur mainly through the bite of Culex spp. mosquitoes, and wild birds are recognized as the main reservoirs and amplifying hosts[2]. WNV infections can result in West Nile fever and neurological diseases in humans, and can present as an asymptomatic infection or a mild acute febrile illness, or progress to meningitis, encephalitis, and acute paralysis[3]. Although there are nine proposed lineages of WNV, most human outbreaks of WN encephalitis have been attributed to lineage 1 and 2[1].
Recombinase-aided amplification (RAA) is a new isothermal amplification technology that does not require a classical thermostable enzyme[4]. It has already been successfully applied to detect bacterial and viral pathogens[4-5]. In this study, we developed an internally controlled RT-RAA assay for WNV, using either a real-time fluorescence device or a lateral flow dipstick (LFD) for the visual observation. The analytical specificity and sensitivity of the assays were evaluated and the test results were compared with the data from real-time reverse transcription-polymerase chain reaction (RT-qPCR) assays using archived mosquito samples.
The virus was isolated from Culex pipiens specimens collected in the Xinjiang Uygur Autonomous Region in 2011, with the work being conducted in the laboratory of the Department of Viral Encephalitis, Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention (IVDC, China CDC)[6]. Vero cells were cultivated in minimal essential medium containing 2% fetal bovine serum and penicillin-streptomycin and then infected with the WNV (XJ11129 strain; GenBank: JX442279). Total RNA was extracted from the mosquitoes and from the WNV-infected Vero cells using a genomic DNA/RNA commercial kit (QIAamp Viral RNA Mini Kit; QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions.
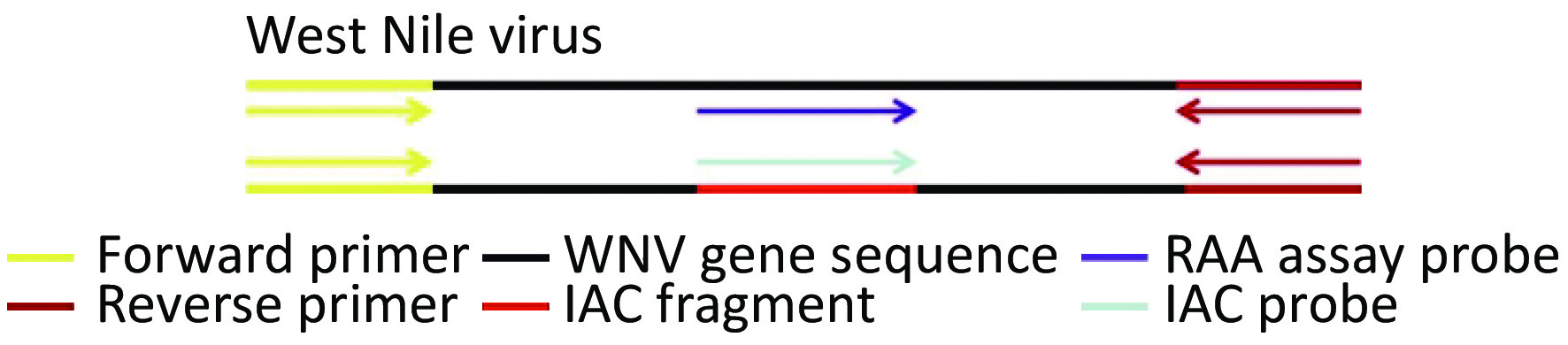
The WNV primers and probes were designed according to the principles of RAA primer and probe design. An internal amplification control (IAC) probe was designed as a short fragment complementary to the template region of the internally controlled plasmid (Figure 1). These primer and probe sequences are presented in Supplementary Table S1 available in www.besjournal.com. The targeted sequence (nt232-nt420; GenBank accession no.: KC407666.1), which was 189 base pairs in length, was cloned by TsingKe Biotech Corp. (Beijing, China). The IAC probe was inserted into the recombinant plasmid of WNV, replacing the corresponding probe sequence of the virus. Accordingly, the IAC template was a recombinant plasmid consisting of the IAC probe sequence, a short gene sequence of the rose rosette virus.
Primer/probe Sequence (5′-3′) Primer length (bp) Product size (bp) References RT-RAA WNV-F ATTTGTGTTGGCTCTCTTGGCGTTCTTCAGGT 32 189 This paper WNV-R CTTTTGTTTTGAGCTCCGCCGATTGATAGCAC 32 WNV-Pa TGTGAACAAACAAACAGCGATGAAACACC/iFAMdT/T/idSp//iBHQdT/GAGTTTTAAGAAGGA 50 WNV-Pa(IAC) GTAAGGTGCTAGACTAAAATTGTTGGGACTT/iHEXdT/G/idSp/A//iBHQ1dT/CTCTGAAGTAAAAGG 51 RT-RAA-LFD WNV-LFD-F ATTTGTGTTGGCTCTCTTGGCGTTCTTCAGGT 32 189 This paper WNV-LFD-R Biotin-CTTTTGTTTTGAGCTCCGCCGATTGATAGCAC 32 WNV-LFD-Pa TGTGAACAAACAAACAGCGATGAAACACCTT/idSp/TGAGTTTTAAGAAGGA[C3-spacer] 48 RT-qPCR qPCR-F GCACTGAGAGGACTGCCCAT 20 101 [22] qPCR-R TGGGTGAGGGTAGCATGACA 20 qPCR-P TACCAGACATCCGCAGTGCCCAGA 24 Note. aProbe modifications: FAM, 6-carboxyfuorescein; HEX, 5-hexachlorofuorescein; THF, tetrahydrofuran; BHQ, black hole quencher; C3-spacer, 3′ phosphate blocker. Table S1. The oligonucleotides of RT-RAA and RT-qPCR
We developed an internally controlled duplex RT-RAA assay for WNV RNA through the incorporation of an IAC template and a corresponding IAC exo probe. The real-time RT-RAA reaction was performed in a 50 μL volume using an exo probe kit (commercial RT-RAA kit from Qitian Bio-Tech Co. Ltd., Jiangsu, China), which contained 25 μL of rehydration buffer, 0.42 μmol/L of the forward and reverse primers, 0.12 μmol/L of the target-specific exo probe and IAC exo probe, 1 μL of the IAC template, 1 μL of the viral RNA or DNase-free water (blank control), and 14 mmol/L of magnesium acetate. The tubes were placed into the fluorescence detection device (QT-1620, Qitian Bio-Tech Co. Ltd, Jiangsu, China) with a setting of 39.0 °C for 30 min. The blank control was included in each run. For the RT-RAA-LFD method, the reaction system was the same as that for the fluorescence method except that there was no internal reference probe and internal reference template. The tubes were placed into the detection device (QT-RAA-B6100, Qitian Bio-Tech Co. Ltd., Jiangsu, China) with a setting of 39.0 °C for 40 min. The amplification product was then diluted 10 times with phosphate-buffered saline and tested using the LFD. The LFD detection result was considered positive if both the test line and control line were simultaneously positive; was doubtful and needed to be retested if only the test line was positive; and was negative if the control line was positive while the test line was negative.
The WNV recombinant plasmid at a dilution range between 105 and 101 copies and the live virus RNA at a dilution range between 1.6 × 105 and 1.6 × 100 pfu/μL were prepared to test the sensitivity of the duplex RT-RAA assays for WNV in eight reaction replicates, respectively. The specificity of the duplex RT-RAA assays for WNV was examined by cross-reaction assays, using a virus control panel that comprised the Zika virus, Kadipiro virus, JEV, Getah virus, yellow fever virus, dengue virus, Tahyna virus, Sindbis virus, and tick-borne encephalitis virus. The optimum concentration of the IAC template was examined by testing different copies of the IAC plasmid (1 × 10, 1 × 102, 1 × 103) in the presence of various WNV recombinant plasmid concentrations (between 101 and 105 copies). The practical performance of the duplex RT-RAA assay with the IAC was evaluated by testing 32 mosquito samples. The RT-qPCR assay was carried out simultaneously as a parallel test[6].
The amplification of the IAC template (102 copies) did not significantly inhibit the amplification of low copies of the target gene (Figure 2; where 10 copies of the WNV plasmid still had an obvious amplification curve in the presence of 100 copies of IAC). As shown in Figure 2, the higher the concentration of the virus target plasmid was, the earlier was the peak and the later was the peak of the corresponding 102 copies of the internal control. The results indicated that the duplex RAA assays were capable of detecting 10 copies of WNV plasmid or 1.6 pfu of cultured WNV RNA per reaction (Table 1). The duplex RT-RAA assays also demonstrated excellent analytical specificity, as the test with the virus control panel turned out to be negative, indicating that no cross-reaction had occurred. The analytical sensitivity of the RT-RAA-LFD assay for the virus RNA was approximately 1,000 plasmid copies per reaction.
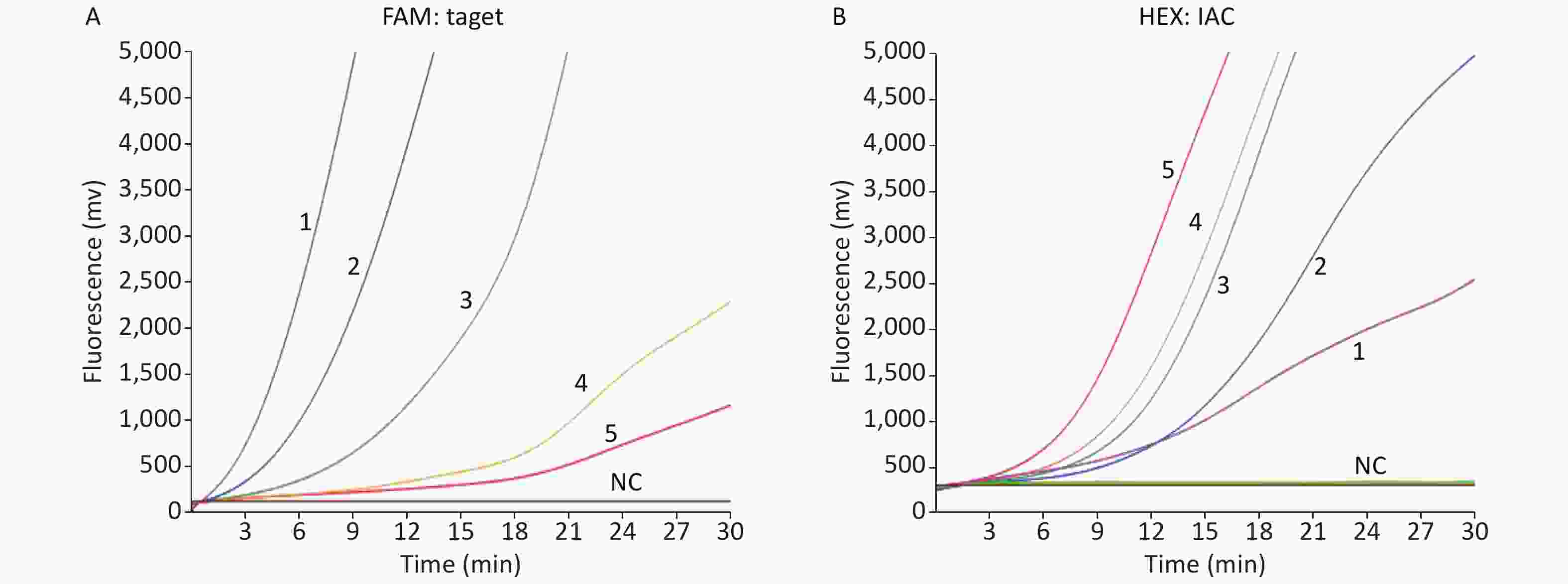
Figure 2. Demonstration of WNV internally controlled duplex RT-RAA assays reaction. (A) Amplification curves 1 to 5 indicate a dilution range from 105 to 101 copies/reaction of WNV recombinant plasmid respectively in the presence of 100 copies of the IAC in the FAM (6-carboxyfuorescein) detection channel. (B) Amplification curves for 100 copies of the IAC in the presence of 105 to 101 copies of WNV recombinant plasmid (curves 1 to 5), respectively, in the HEX (5-hexachlorofuorescein) detection channel.
Serially diluted WNV culture medium (copies/reaction) Serially diluted WNV plasmid No. replicates tested No. detection Detection rate (%) 1.6 × 105 1 × 105 8 8 100 1.6 × 104 1 × 104 8 8 100 1.6 × 103 1 × 103 8 8 100 1.6 × 102 1 × 102 8 8 100 1.6 × 101 1 × 101 8 8 100 1.6 × 100 − 8 8 100 Table 1. Sensitivity of duplex RT-RAA assays using cultured WNV RNA and plasmid
In total, 32 mosquito samples were tested with the duplex RT-RAA, RT-RAA-LFD, and RT-qPCR assays. The RT-qPCR results showed that five samples were positive for WNV, with their CT values ranging from 19.41 to 25.45. The results of the RT-RAA assays, whether by the real-time fluorescence method or LFD method, were in complete agreement with those obtained by RT-qPCR (Supplementary Table S2 available in www.besjournal.com).
Methods RT-qPCR RT-RAA Compare with RT-qPCR Kappa value Positive Negative Total Positive (%) Negative (%) RT-RAA Positive 5 0 5 Negative 0 27 27 100 100 1 Total 5 27 32 RT-RAA-LFD Positive 5 0 5 Negative 0 27 27 100 100 1 Total 5 27 32 Table S2. The results of the duplex RT-RAA assays and RT-RAA-LFD assays compared with the RT-qPCR results
WNV is a serious threat to human health worldwide. Even though there has been no WNV epidemic in China to date, the virus has been isolated from mosquito specimens collected in Xinjiang in western China[7]. Many nucleic acid-based detection assays for WNV have been reported. The detection limit of the SYBR Green I-based one-step real-time RT-PCR assay for WNV is 20 copies and 100 pfu[8]. The reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for WNV reportedly has an exceptionally high sensitivity (0.1 pfu/reaction), and the RT-LAMP-LFD assay has demonstrated a detection limit of 102 copies/μL of synthetic RNA and 102 pfu/μL[9]. However, despite the high sensitivity of most molecular detection techniques, the requirement of a high-precision thermal cycler prevents their wide use as routine diagnostic tools in places such as private clinics or budget-limited laboratories. Moreover, these methods consume more than 2−3 h of time. Although RT-LAMP is more sensitive and time saving than RT-PCR for WNV detection, the design of 4−6 LAMP primers is a relatively hard task.
We have developed a simple and rapid isothermal amplification assay for WNV based on a one-step real-time RT-RAA with IAC. The detection sensitivity was 10 copies/reaction for the plasmid and 1.6 pfu/reaction for the virus RNA, which were similar to the sensitivity results reported with RT-PCR and RT-qPCR. Our developed assays also showed good specificity for WNV. Additionally, the RT-RAA assay has the distinct advantages of simplicity and a shorter turn-around time due to the portable fluorescence device and 30 min reaction time at 39.0 °C, compared with the expensive thermal cycler equipment and 3−4 h needed in the case of RT-qPCR. Although the RT-LAMP assay has higher sensitivity than the RT-RAA assay, the former requires 60 min of reaction time at a constant temperature of 63.0 °C, and each test costs more than the RT-RAA assay with its IAC[5, 10]. Hence, the RT-RAA assay for WNV is of great value to the resource-limited setting, such as in areas in Africa where WNV infections cause a serious burden to the healthcare system. In addition, the proposed RT-RAA-LFD assay for WNV offers another valuable tool for device-free visual detection of the virus, which is well suited to field use. The results of the RT-RAA assay of the 32 mosquito samples were the same as those obtained by RT-PCR assay.
The introduction of IAC effectively eliminated false-negative or invalid results. Through the introduction of this competitive internal inference strategy, the target sequence and the IAC are amplified with one common set of primers under the same conditions, with only the probes being different, thereby reducing the risk of interactions among multiple primers. By optimizing the amount of primers, the ratio of target probes to IAC probes, and the amount of IAC plasmids used, the inhibitive impact of the IAC on the detection sensitivity of the target can be minimized.
Although the methods were designed to detect both WNV lineage 1 and 2, only lineage 1 virus and mosquito samples were tested in this study owing to the unavailability of other resources. Additionally, the RT-RAA-LFD assay for WNV had a relatively low sensitivity and thus more optimization of this assay is needed in the future.
All the authors approved the final manuscript and have no conflict of interest to declare.
Development of an Internally Controlled Reverse Transcription Recombinase-aided Amplification Assay for the Rapid and Visual Detection of West Nile Virus
doi: 10.3967/bes2019.116
- Received Date: 2019-06-03
- Accepted Date: 2019-09-23
-
Key words:
- WNV /
- Detection /
- RT-RAA /
- Lateral flow dipstick (LFD)
Abstract: West Nile virus (WNV) causes West Nile fever and West Nile encephalitis. Because infection by WNV creates serious public health problems, its simple, rapid, and visual detection is very important in clinical practice, especially in resource-limited laboratories. We have developed a rapid, specific, and highly sensitive internally controlled reverse transcription recombinase-aided amplification (RT-RAA) assay to detect WNV, using both real-time fluorescence and the lateral flow dipstick (LFD) at 39.0 °C for 30 min. The analytical sensitivity of the RT-RAA assay was 10 plasmid copies and 1.6 pfu per reaction with real-time fluorescence, and 1,000 plasmid copies per reaction with the LFD. No cross-reaction with other control viruses was observed. Compared with the RT-qPCR assay, the RT-RAA assay demonstrated 100% sensitivity and 100% specificity for WNV.
| Citation: | FAN Guo Hao, SHEN Xin Xin, LI Fan, LI Xin Na, BAI Xue Ding, ZHANG Rui Qing, WANG Rui Huan, LEI Wen Wen, WANG Huan Yu, MA Xue Jun, WU Gui Zhen. Development of an Internally Controlled Reverse Transcription Recombinase-aided Amplification Assay for the Rapid and Visual Detection of West Nile Virus[J]. Biomedical and Environmental Sciences, 2019, 32(12): 926-929. doi: 10.3967/bes2019.116 |







 Quick Links
Quick Links
 DownLoad:
DownLoad:
