-
Since the highly pathogenic avian influenza (HPAI) virus A/goose/Guangdong/1/1996 (Gs/GD, H5N1) was first detected in China, it has posed a threat to public health [1,2]. To date, the H5 Gs/GD lineage HPAI viruses have evolved into ten genetically distinct hemagglutinin (HA) clades (0–9) and multiple subclades [3]. Clade 2.3.4.4 viruses with different neuraminidase (NA) subtypes (N2, N6, or N8) have become the most prevalent viruses in recent years [4-6]. Among the H5Nx viruses, influenza A (H5N1) and A (H5N6) viruses have been reported to infect humans [7]. From the World Health Organization (WHO) statistics on human infections with avian influenza viruses, H5N1 cases have been gradually decreasing [8], but confirmed cases of H5N6 infection have been repeatedly detected in China [7] since the first case in Sichuan in 2014 [9]. According to recent data (2020-01-20), 24 confirmed cases of A (H5N6) virus infection, including seven deaths, have been reported in China by the WHO [10]. A considerable genetic diversity exists among clade 2.3.4.4 viruses [11], which have spread to various countries [12-15]. H5N6 viruses pose a potential risk to public health, and thus, the development of candidate vaccine viruses (CVVs) is needed.
In the absence of a universal influenza vaccine, the development of subtype-specific vaccines is essential for pandemic preparedness. The WHO recommends the development of CVVs for zoonotic influenza viruses based on the changes in the genetic and antigenic characteristics of these viruses twice a year; two H5N6 viruses, namely, A/Hubei/29578/2016 (HB29578)-like virus and A/Fujian-sanyuan/21099/2017 (FJ21099)-like virus of clade 2.3.4.4, have been recently proposed [16,17]. The developed CVVs should match those in circulation, as recommended by the WHO; however, mutations occasionally occur after passage in eggs, resulting in dissatisfactory vaccine effectiveness. Our previous work and other studies have reported amino acid substitutions in the HA of serially passaged viruses in eggs during the preparation of CVVs [18-20]. Therefore, the genetic stability of CVVs must be maintained. In accordance with the WHO recommendations and using reverse genetics (RG) technology, we constructed two CVVs with a (6 + 2) gene constitution, namely, RG-HB29578 and RG-FJ21099. To determine the genetic stability of the two CVVs after multiple passages in eggs, we performed complete genome sequencing and assessed their growth ability in MDCK cells, antigenic identity with the original virus, and pathogenicity in vitro and in vivo. The results confirmed that both CVVs retained their original antigenicity and were attenuated in vitro and in vivo. These genetically stable CVVs with high growth characteristics may therefore be valuable for pandemic preparedness.
-
The HA and NA genes were synthesized (GenScript, China) based on the sequences of HB29578 and FJ21099 viruses, modifying LRERRRKRGLF to LRETRGLF for RG-HB29578 and LREKRRKRGLF to LRETRGLF for RG-FJ21099 at the cleavage site for HA (Figure 1). Cleavage site modifications were thought to reduce pathogenicity for avian embryos and poultry [21]. The synthesized genes were cloned into vector pHW2000. (6 + 2) H5N6 reassortant influenza viruses (RG-HB29578 and RG-FJ21099) were generated by a RG approach. RG (6 + 2) virus harbored a mutant HA gene, the NA gene of wild virus, and six internal genes of influenza A/PR/8/1934 (H1N1) virus. The Vero cell cultures were transfected with eight plasmids harboring a mutant HA gene, the NA gene of RG-HB29578 or RG-FJ21099, and six internal genes of influenza A/Puerto Rico/8/34 (PR8; H1N1) virus using TurboFect transfection reagent (Thermo Fisher Scientific, R0531). After a 72 h incubation, the supernatant was passaged to 9-day-old specific-pathogen-free (SPF) eggs to amplify the generated viruses. Then, a first-passage viral stock with positive HA titers were obtained and designated V1E1 (V:Vero, E:egg). V1E2–V1E10 virus stocks were prepared by inoculation into SPF eggs using limiting dilution. At each passage, the embryonated eggs were checked, and dead embryos were recorded. At each passage, allantoic fluids with positive HA titers were collected and stored at 80 °C prior to processing. All viruses were sequenced in our laboratory using next-generation sequencing as previously described[22]. BioEdit (Version 7.1.3.0) was used to analyze the entire genome sequences. The 50% tissue culture infective dose (TCID50) was determined in MDCK cells and calculated by the Reed-Muench method.

Figure 1. Modified sequence in HA cleavage site of reassortant viruses. (A) A/Hubei/29578//2016 (HB29578). (B) A/Fujian-sanyuan/21099//2017 (FJ21099). Multiple basic amino acids RRK were deleted in both RG viruses at the HA cleavage site. ↓: proteolytic cleavage site. Changes in sequences are in bold.
-
The ability of viruses to grow in MDCK cells and eggs was determined by analyzing multiple replication cycles. Reassortant viruses were tested using TCID50 and inoculated at the same multiplicity of infection (0.001) in MDCK cells and inoculated at the same titer of 104 TCID50/mL in eggs. At 24, 48, 72, and 96 h post inoculation (p.i.), supernatants and allantoic fluid were collected and stored at −80 °C. Hemagglutination assay was used to determine the virus titer with 1% turkey erythrocytes. The data were analyzed by two-way analysis of variance (ANOVA) using the GraphPad Prism 5 software package (version 5.0).
-
The viral plaque characteristics were determined in MDCK cells. As previously described [19], cells were infected with diluted virus with or without N-p-tosyl-L-phenylalanine chloromethyl ketone (TPCK)-treated trypsin and incubated at 37 °C for 18–20 h. The cells were then fixed with 4% paraformaldehyde with Triton X-100. Next, the cells were immunolabeled with antinucleoprotein monoclonal antibody (Merck Millipore; MAB8257 and MAB8258) and secondary goat-anti-mouse antibody (KPL, 074–1806). Finally, TrueBlue Peroxidase Substrate (KPL, 5510–0030) was added to visualize the plaques using an ELISpot Reader (AID, version 4.0).
-
As described in the Manual of Diagnostic Tests and Vaccines for Terrestrial Animals [23], virus with HA titer > 16 was used. Ten 6-week-old SPF chickens were injected intravenously with 100 µL 1/10 virus dilution and were examined for 10 days. During daily observation, the chickens were scored 0 if they were normal, 1 if sick, 2 if severely sick, and 3 if deceased, as described in the World Organization for Animal Health (OIE) Guidelines [23]. The intravenous pathogenicity index (IVPI) was also determined.
-
Ferrets were prescreened for the absence of antibodies to seasonal influenza viruses and the tested viruses. Several ferrets were used to prepare antisera, and the others were used for attenuation assessment. Antisera to reassortant viruses were produced in ferrets by inoculating them intranasally (500 µL per nostril) with viruses. Blood samples were collected on 14 or 15 d p.i. (dpi) and were tested by hemagglutination inhibition (HI) assay to determine whether boosting was required. Ferrets were boosted intradermally with purified virus if the preboost HI titers were < 80. Blood sample was collected on 30 dpi.
In pathogenicity attenuation tests, the ferrets were divided into three groups, including RG-HB29578, RG-FJ21099, and PR8. In accordance with the WHO guidelines [24], groups of 6–8-month-old ferrets (n = 4) were intranasally inoculated with 106 TCID50 of each virus in 1 mL sterile phosphate buffered saline (PBS). Nasal washes were collected from the infected ferrets every other day for 7 d. Two ferrets in each group were euthanized on 3 dpi, and samples were collected for the determination of virus replication from the following organs: spleen, intestine, lungs, brain, olfactory bulb of the brain, and nasal turbinate. The remaining two ferrets were observed for body weight and survival for two weeks. Virus titrations of nasal washes and various tissue organs were determined by TCID50 assays.
-
Hemagglutination and HI assays were performed by using 1% turkey erythrocytes as described in the Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza by the WHO [25]. Antigenic characterization of reassortant viruses was performed using ferret antisera in a HI assay. The HI titers were presented as the reciprocal value of the highest serum dilution that inhibited hemagglutination.
-
All animal experiments were conducted in accordance with the Guidelines for Animal Experiments described and approved by the Animal Care Welfare Committee of the National Institute for Viral Disease Control and Prevention, China CDC.
-
Two reassortant viruses (RG-HB29578 and RG-FJ21099) were obtained using the (6 + 2) RG method in vero cells. To confirm the reassortant viruses, we sequenced the first-generation rescued viruses RG-HB29578 V1E1 and RG-FJ21099 V1E1 (initial growth in Vero cells followed by one passage in eggs). Sequence analysis indicated that the cleavage site of the HA sequence was consistent with the desired cleavage motif (Figure 1A and 1B). No other mutations were detected when comparing the HA and NA sequences with those of the parental strains HB29578 and FJ21099. Six internal gene segments were derived from the high-yield PR8 strain. The sequencing results for RG-HB29578 V1E1 and RG-FJ21099 V1E1 viruses confirmed that the gene sequence was identical to that carried by plasmids used in their construction.
-
Rescued V1E1 virus was propagated in embryonated eggs for an additional nine passages, and the resulting passages were designated as V1E2–V1E10 to determine the genetic stability of the HA and NA genes. No mortality among the eggs was observed for any of the virus passages. The HA titers of RG-HB28578 for passages V1E2 to V1E10 were 256–512 HA units, with the exception of V1E1 (HA 128) and RG-FJ21099, between 256–512 for passages V1E1 to V1E10 (Table 1). The two CVVs from each passage were sequenced and compared with their respective V1E1 sequences. The HA sequence of the two CVVs retained the modified motif at the cleavage site, and no mutations had occurred from V1E2 to V1E10. Sequence analysis of the NA gene of the two CVVs from V1E2 to V1E10 revealed no mutations compared with the respective NA gene sequences of V1E1. In the internal gene, sequencing results revealed a mixture of amino acids (A/T) at position 648 in basic protein 1 (PB1) of RG-HB29578 V1E10 compared with a homozygous A at the same position of V1E1 to V1E9. At position 347 in nucleoprotein (NP), a mutation from E to K first appeared in RG-HB29578 V1E7, and at position 384 in NP, a mixture of amino acids (R/K) was found in RG-FJ21099 V1E10 (Table 1).
Viruses Passage historya HA titer LogTCID50/mLb
(mean ± SD)IVPI PB1 648 NP 372 384 RG-HB29578 V1E1 128 − − A E R V1E2 256 7.34 ± 0.13 0 A E R V1E3 256 − − A E R V1E4 256 − − A E R V1E5 256 − − A E R V1E6 256 − − A E R V1E7 256 7.54 ± 0.12 0 A K R V1E8 256 − − A K R V1E9 512 − − A K R V1E10 512 7.81 ± 0.21 0 A/T K R RG-FJ21099 V1E1 256 − − A E R V1E2 256 8.28 ± 0.18 0 A E R V1E3 256 − − A E R V1E4 512 − − A E R V1E5 256 − − A E R V1E6 512 − − A E R V1E7 512 − − A E R V1E8 256 − − A E R V1E9 512 − − A E R V1E10 512 8.44 ± 0.08 0 A E R/K PR8 C1E2 1,024 8.48 ± 0.16 − A E R Note. a: V. Vero cells; E. eggs; C. MDCK cells; bMean ± standard deviations represent three replicates; TCID50 was determined by hemagglutination assay and calculated by the Reed-Muench method. The 1% Turkey red blood cells were used; IVPI: intravenous pathogenicity index; −: excluded from analysis. Changed amino acids are in bold. Table 1. Virus titers and amino acid changes of CVVs during passage in eggs
-
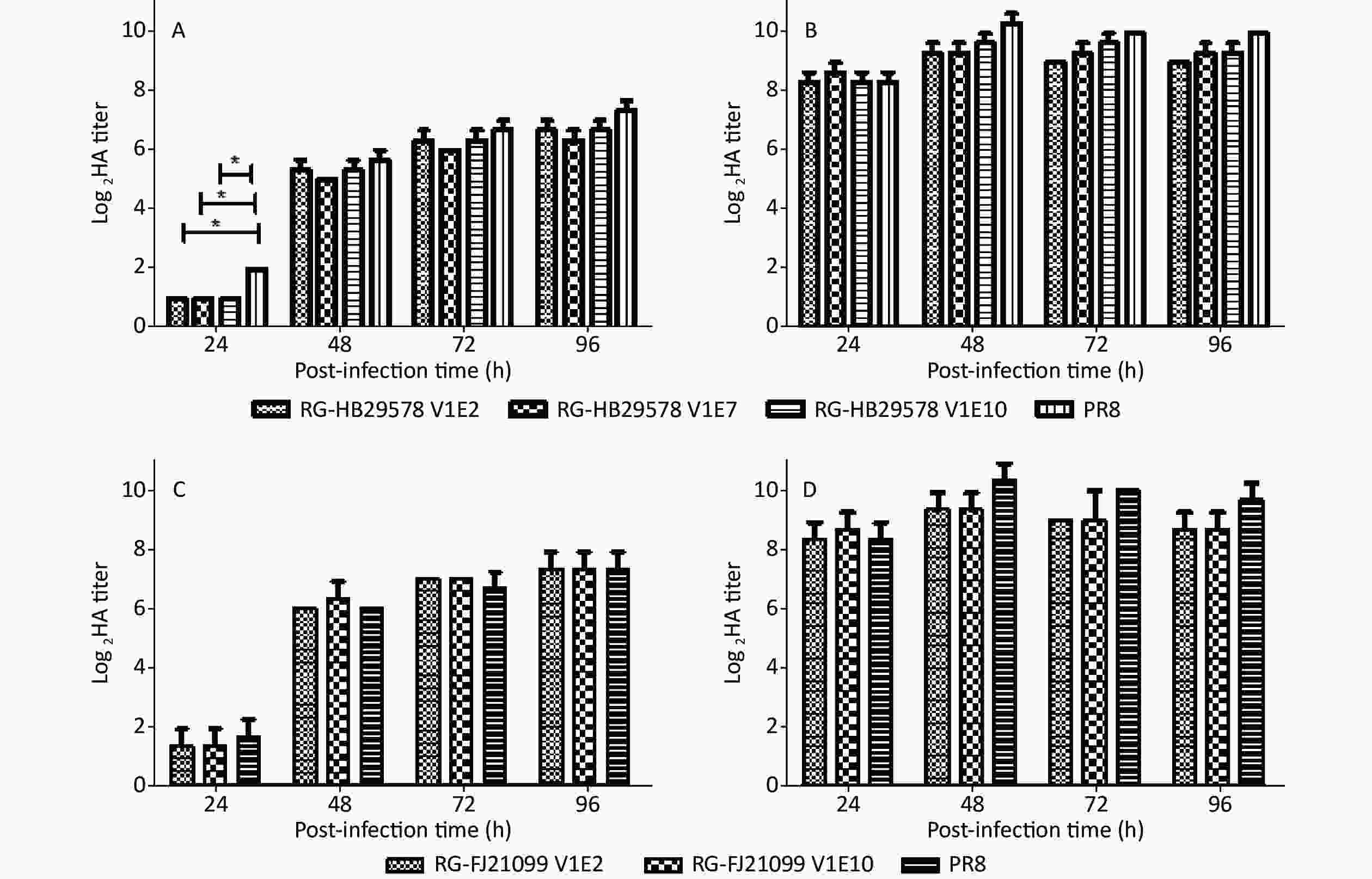
The viral growth characteristics were assessed in MDCK cells and in eggs as per previous reports to analyze the replication efficiency of different passage viruses [26,27]. No significant difference was observed among RG-HB29578 V1E2, V1E7, and V1E10 at any of the indicated time points, although a single amino acid mutation was recorded at position 372 from E to K in NP of V1E7 and one mixture of amino acids A/T at position 648 in PB1 of V1E10 (Table 1, Figure 2A and 2B). No significant differences were detected between RG-HB21099 V1E2 and V1E10 at any of the indicated time points, although one mixture of amino acids R/K was detected at position 384 in NP of V1E10 (Table 1, Figure 2C and 2D). All detected viruses had higher titers in embryonated eggs than in cells at each time point, and these viral titers were all above 28. We observed that the detected viruses had similar growth characteristics to that of PR8 at most p.i. time points in MDCK cells, except for RG-HB29578 at 24 h p.i. (*P < 0.05).
-
For CVVs derived from highly pathogenic H5 strains by RG, the HA was modified from possessing multiple basic amino acids at the HA cleavage site to a single basic amino acid; as a result, such viruses were unable to form plaques in the absence of added trypsin [24]. The CVVs should maintain a HA cleavage site consistent with a low pathogenic phenotype upon multiple passages in embryonated eggs. We selected V1E1 and V1E10 of the two CVVs to determine whether they maintained a low pathogenic phenotype using a TPCK-treated trypsin-dependent plaque formation assay in MDCK cells. The results showed that none of the tested viruses can form plaques in the absence of TPCK-treated trypsin, whereas in the presence of TPCK-treated trypsin, plaques can be identified, indicating the low pathogenicity of these viruses (Figure 3).
Figure 3. Replication of two CVVs in MDCK cells with or without TPCK-treated trypsin. V1Ex: V1Ex with initial growth in Vero cells followed by x passage in eggs; the original viruses were diluted from 10−1 to 10−6, and 10−2 to 10−6 diluted viruses were inoculated in MDCK cells. Mock cells were inoculated with PBS.
-
To confirm that the antigenicity of the two CVVs, which were passaged ten times, was consistent with the homologous wild-type viruses, we tested the reactivity of ferret antisera to RG-HB29578 (V1E2 and V1E10) and RG-FJ21099 (V1E2 and V1E10) viruses (Table 2). RG-HB29578 (V1E2 and V1E10) were immunogenic and induced HI antibody titers (160 and 640, respectively) against homologous viruses. RG-FJ21099 (V1E2 and V1E10) were immunogenic and induced HI antibody titers (160) against homologous viruses. Antisera to RG-HB29578 (V1E2 and V1E10) can inhibit the wild-type HB29578 virus well. Antisera to RG-FJ21099 (V1E2 and V1E10) reacted well with wild-type FJ21099 virus. Antisera to RG-HB29578 inhibited neither wild-type FJ21099 nor reassortant RG-FJ21099, and similarly, antisera to RG-FJ21099 inhibited neither wild-type HB29578 nor reassortant RG-HB29578. The above results indicate that both CVVs maintained consistent antigenicity to wild-type viruses after 10 passages, with no cross-reactivity between them.
Antigens Passage history Antisera RG-HB29578 V1E2 RG-HB29578 V1E10 RG-FJ21099 V1E2 RG-FJ21099 V1E10 HB-29578 wt E2 80 320 < 20 < 20 FJ-21099 wt E2 < 20 < 20 80 80 RG-HB29578 V1E2 160 640 < 20 < 20 RG-HB29578 V1E10 160 640 < 20 < 20 RG-FJ21099 V1E2 < 20 < 20 160 160 RG-FJ21099 V1E10 < 20 < 20 160 160 Note. wt: Wild-type. Boldface denotes titers of ferret sera with homologous antigens. Table 2. Antigenicity analysis of two CVVs
-
To verify the low pathogenicity of the two CVVs, we tested viral pathogenicity in chickens. SPF chickens (n = 10) were inoculated with 1:10 diluted HA titers of RG-HB29578 (V1E2 and V1E10) and RG-FJ21099 (V1E2 and V1E10) via the intravenous route. All chickens remained healthy throughout the 10-day observation period with no mortalities (Table 1). The IVPI was calculated in accordance with the standards of OIE [23]. The IVPI for both viruses was zero. These results demonstrate that both CVVs exhibited similar pathogenic characteristics that are indicative of low pathogenic avian influenza virus after 10 passages. Pathogenicity was unaffected by the substitutions in PB1 and/or NP.
Ferrets have been used extensively as an ideal indicator of influenza virus virulence in humans [28]. The attenuation of CVVs in ferrets must be demonstrated in accordance with WHO guidelines. Thus, two RG viruses were tested in ferrets, with PR8 as the control virus. All ferrets survived the two-week observation period with slight weight loss, and the most weight loss was observed in the PR8-infected group (Figure 4). Nasal discharge was observed in all infected animals on 1–3 dpi. In the nasal wash samples, viral titers were detected at 1, 3, and 5 dpi. in the three groups but were not detected at 7 dpi (Table 3). In the nasal turbinate samples, viral titers were detected in all the infected animals on day 3, and CVVs titers were similar to those of PR8 (Table 3). In the lung samples, viral titers were detected in both PR8-infected animals and one of the animals for CVVs (Table 3). Viruses were not detected in any other organs (i.e., the spleen, intestine, brain, or olfactory bulb of the brain). Viruses were only detected in the respiratory system, including the lungs, nasal turbinate, and nasal wash fluid.
Virus Animal No. Nasal washesa Nasal turbinatesb Lungb Other organsb,c Day 1 Day 3 Day 5 Day 7 Day 3 Day 3 Day 3 PR8 1 4.00 2.23 3.83 / 2 4.17 2.00 2.00 / 3 2.50 2.00 / 4 2.17 1.67 / RG-HB29578 1 2.5 2.25 2.00 / 2 2.83 2.77 2.00 / 3 2.50 / / 4 1.67 1.33 / RG-FJ21099 1 2.00 2.50 1.73 / 2 2.67 2.23 2.00 / 3 2.33 1.50 / 4 1.67 / / Note. aLog10TCID50/mL. bLog10TCID50/g. cOther organs include brain, spleen, intestine, and olfactory bulb of the brain. /, Denotes that no virus titer was detected. Table 3. Virus titer in different organs
Figure 4. Body weight change (%) post-infection. The change in weight is shown as an average with the standard deviation. Ferrets (n = 2 for each group) were lightly anesthetized via inhalation of isoflurane and inoculated intranasally with 1.0 mL containing 106 50% TCID50 RG-HB29578 or RG-FJ21099 or PR8 influenza virus.
-
In China, H5N6 virus first emerged in 2010, and since then, it has circulated extensively in domestic and wild birds[29]. Four years later, the first human infection of H5N6 was confirmed in China [9]. Although transmission of the infection from person to person is rare[30], the H5N6 viruses are the only clade 2.3.4.4 viruses that have been reported in humans and thereby present an increased threat to public health. Currently, vaccination is the most effective measure to prevent influenza infection. Vaccine production requires matching high-yield CVVs. Optimal CVVs can not only be obtained in high-yield but also retain the same antigenicity as wild-type circulating viruses. However, influenza viruses are susceptible to mutation during propagation in eggs. In this study, we evaluated the genetic stability of two H5N6 CVVs after 10 passages in eggs.
The sequencing results confirmed that HA and NA consistently maintained their sequence from V1E1 to V1E10, but several substitutions were observed in PB1 and NP. Amino acid substitutions in viral proteins can often influence viral replication or pathogenicity. To investigate the effect of the substitutions in PB1 and NP, we first assessed the replication of RG-HB29578 (V1E2, V1E7, and V1E10) and RG-FJ21099 (V1E2, and V1E10). The results indicated no significant differences among RG-HB29578 V1E2, V1E7, and V1E10, or between RG-FJ21099 V1E2 and V1E10. The detected viruses reached similar titers to the control virus PR8 at most postinfection time points, which are the usual time of harvesting, and showed better growth characteristics in embryonated eggs than in cells. Both CVVs had similar high growth characteristics to PR8. Next, we assessed the pathogenicity in chickens. The IVPI data in chickens showed no difference among these viruses, and all of them exhibited low pathogenicity. We therefore speculated that the substitutions in PB1 and NP did not affect viral replication or pathogenicity.
The results of the HI assay revealed a four-fold difference between the HI titers of the antisera to RG-HB29578 and RG-FJ21099. However, both antisera inhibited the homologous wild-type viruses and RG viruses after 10 passages. These results confirm that both CVVs retained their original antigenicity. Phenotypic diversity is a characteristic of clade 2.3.4.4 viruses [31]. Although the two studied viruses belong to clade 2.3.4.4 and share 96.27% of the putative consensus protein sequence in the HA gene, they belong to a different subgroup of the clade. HB29578 belongs to clade 2.3.4.4 d, and FJ21099 belongs to clade 2.3.4.4 b [32]. These genetic changes can induce antigenic change, resulting in the poor cross-reactivity between these two viruses when testing with HI test with ferret antisera. Based on our result, the cross-reactivity based on HI test was low (HI titer < 20).
Attenuated vaccine strains of highly pathogenic influenza viruses for vaccine production must be developed. Although previous CVV studies have shown that CVVs constructed by the reported method are safe [33,34], further assurances of safety are needed because influenza viruses exhibit strain-specific characteristics. In vitro and in vivo assays were therefore carried out. In the in vitro assay, the CVVs cannot replicate in the absence of added trypsin. Following the WHO guidelines [23], we tested the pathogenicity in vivo, including in chickens and ferrets. The IVPI of the two CVVs was zero, which is below the OIE standard of less than 1.2 [23]. In previous reports, in addition to the respiratory tract, viruses were detected in the brain and other organs of ferrets infected with HPAI H5N6, where the virus induced severe pneumonia [35,36]. In our study, the viral titers of the two CVVs were similar to those of PR8, and the viruses showed restricted replication in the respiratory tract of ferrets. Altogether, the in vivo results indicate that the viruses showed the characteristics of low pathogenicity in chickens and ferrets, similar to other CVVs in previous studies [37], and controlled virus PR8.
Influenza viruses, which reside as a vast silent reservoir in aquatic birds, are difficult to eradicate. Predicting the subtype that will cause the next influenza epidemic or pandemic is extremely difficult, and implementing positive measures, such as preparing CVVs, is currently the best course of action. Selecting the optimal CVVs is crucial for vaccine manufacture. In this study, we obtained two CVVs of HPAI H5N6 and assessed them in vitro and in vivo. The results confirmed that the CVVs were genetically stable and showed high growth characteristics. We propose that these CVVs may be important in preparing for a potential H5N6 pandemic.
-
We obtained two stable CVVs of HPAI H5N6 with high growth characteristics that may aid our preparedness for a potential H5N6 pandemic.
-
We thank Dr. LU Xuan Cheng, Dr. LIU Xiao Yu, and the technical staff in the Laboratory Animal Center of Chinese Center for Disease Control and Prevention for their support to the animal experiment.
-
LIU Li Oi, LI Zi, JIAO Ming, LU Jian, ZHOU Jian Fang, LI Xi Yan, LIU Jia, GUO Jun Feng, XIAO Ning, and ZHAO Xiang carried out the experiments, collected the data, and participated in drafting the manuscript. LIU Li Qi and WANG Da Yan participated in the design of the experiment and wrote the manuscript. All authors read and approved the final manuscript.
-
All authors declare no conflicts of interest.
Development and Assessment of Two Highly Pathogenic Avian Influenza (HPAI) H5N6 Candidate Vaccine Viruses for Pandemic Preparedness
doi: 10.3967/bes2020.088
- Received Date: 2020-02-05
- Accepted Date: 2020-07-22
-
Key words:
- Highly pathogenic avian influenza H5N6 virus /
- Genetic stability /
- Candidate vaccine virus /
- Reverse genetic technology
Abstract:
| Citation: | LIU Li Qi, LI Zi, JIAO Ming, LU Jian, ZHOU Jian Fang, LI Xi Yan, LIU Jia, GUO Jun Feng, XIAO Ning, ZHAO Xiang, WANG Da Yan. Development and Assessment of Two Highly Pathogenic Avian Influenza (HPAI) H5N6 Candidate Vaccine Viruses for Pandemic Preparedness[J]. Biomedical and Environmental Sciences, 2020, 33(9): 670-679. doi: 10.3967/bes2020.088 |








 Quick Links
Quick Links
 DownLoad:
DownLoad: