-
Over the past several years, the most widely used in vitro tests for mutagenicity have been those performed on bacteria. These include the Ames test, SOS, and SOS/umu chromotest [1,2]. The Ames test is the most commonly used system, but it is not automatable and requires several strains to detect different types of mutagens. Faster alternative tests include the SOS chromotest and the umu test, which are based on SOS promoter-linked Lac Z, which is activated by the SOS response to DNA damage in Escherichia coli and Salmonella typhimurium, respectively. The most important drawback of bacterial tests is that they are not capable of emulating the cell cycle controls or the repair mechanisms found in eukaryotic cells because the gene structure, gene regulation, and metabolism for prokaryotic cells are quite different from those of eukaryotic cells. In this way, the use of prokaryotic cells in mutagenicity screening for chemical substances has certain limitations.
Mammalian cell tests such as the single-cell gel electrophoresis assay, V79/HGPRT gene mutation test, and the test of in vitro mammalian cell transformation have no such drawbacks with respect to gene structure, gene regulation, or metabolism. However, the cells require relatively intense experimental culture conditions and their resistance to chemical toxicity is poor. In these ways, these tests are generally unsuitable for large-scale screening.
The yeast cell (Saccharomyces cerevisiae) is a specific kind of eukaryotic cell. It has the same advantages as bacteria including its simple genetic background, short cell cycle, fast cultivation, modest nutritional requirements, and strong resistance to harmful chemicals. This excellent yeast model has gene structure, gene regulation, metabolism, and protein post-translational modifications and processes that are similar to those of mammalian cells [3,4]. As a result, given proper biotechnological techniques, yeast can be an ideal host for construction of recombinant cells for rapid and high-throughput screening of chemical mutagens.
DNA damage in eukaryotic cells leads to the gene expression involved in DNA repair. The DNA damage checkpoint pathway responsible for activation of DNA-damage-inducible genes in Saccharomyces cerevisiae yeast has been the focus of intensive research during the past decade [5,6]. One of the frequently used tools in the DNA damage checkpoint pathway in Saccharomyces has been the gene RNR3 [7]. RNR3 is part of the ribonucleotide reductase (RNR) system. Under normal growth conditions, the expression of RNR3 was nearly undetectable in cells. However, this gene is expressed 5- to 10-fold, and in some reports more than 100-fold, during DNA breakage and synthesis blockade. These properties of the RNR3 have been used in the identification of a number of important genes involved in the DNA damage caused by chemical products [8]. Some researchers have fused this gene promoter with some reporter genes to construct recombinant yeast cells for screening of chemical mutagens [9, 10]. However, the normalization of reporter genes has routinely been achieved by quantifying total protein of the yeast lysate or on the basis of the optical density (OD600) of the yeast culture, while the optical density measurements can yield highly variable results. The dual luciferase assay is performed by sequentially measuring the firefly and Renilla luciferase activities of the same sample, with the results expressed as the ratio of firefly to Renilla luciferase activity (Fluc/Rluc). Although luciferase has been used as a highly sensitive reporter, especially in animal cells, yeast cells have to be ruptured for determination of luciferase activity, which is a time-consuming procedure. We used green fluorescent protein (yEGFP) as a reporter gene. The codons of yEGFP protein are optimized for yeasts, and its amino acid coding is biased to the preferred usage of yeasts. yEGFP emits green fluorescing light that can be measured directly without cell rupture. The chromophore of yEGFP is formed by intramolecular cyclization and subsequent dehydrogenation without adding any cofactors. The green fluorescent protein itself is rather small, highly soluble, and stable across a broad pH range.
Several variants have been developed using directed mutagenesis, such as DsRed1, yDsRed2, DsRed-Express2, and yDsRed-Express2. yDsRed and yDsRed-Express2 with the S. cerevisiae codon usage were constructed in our laboratory. Comparison to the fluorescence intensities of the three DsReds showed that DsRed-express-2 emission was the strongest, followed by yDsRed-Express2, then DsRed1, and the weakest fluorescence intensity was yDsRed. Thus, the RFP strength has nothing to do with the codon preference in yeast cells. This is different from GFP. Therefore, we selected DsRed-Express2 as an internal control (the second signal) in yeast cells to normalize the cell number and the cell state. DsRed-Express-2 is a noncytotoxic DsRed (Discosoma sp. red fluorescent protein) variant that retains favorable properties, such as brightness, fast maturation, and photostability, of this protein. In this paper, we developed the yEGFP/DsRed-Express-2 reporter assay system in which the quantitation of yEGFP gene expression correlates to the effects of DNA damage, while the second DsRed-Express-2 reporter gene provides an internal control by which experimental values can be normalized to minimize experimental variability. This allows for the sequential quantitative measurement of both yEGFP and DsRed-Express-2 fluorescent density directly in situ with no need to add substrate and cofactors. This system could be easily adapted into a simple, rapid, convenient, high-throughput, automated screening for DNA damaging chemicals.
We used four red fluorescent protein genes, DsRed, DsRed-Express-2, yDsRed, and yDsRed-Express-2. These were selected to determine which was the best for fluorescing in yeast cells. Comparison of the fluorescence intensities of the four DsReds showed that DsRed-express-2 emission was the strongest, followed by yDsRed-Express2, then DsRed, and the weakest fluorescence intensity was yDsRed. This indicated that the RFP strength has nothing to do with the codon preference in yeast cells. This is different from GFP. Therefore, we selected DsRed-Express2 as an internal control (the second signal) in yeast cells to normalize the cell number and the cell state (Table 1).
Items Replicates OD600 Fluoresce W303-1A/DsRed 24 0.10 ± 0.004 547.5 ± 21.64 W303-1A/yDsRed 24 0.12 ± 0.005 259.6 ± 16.41 W303-1A/DsRed-Express-2 24 0.11 ± 0.003 1138.8 ± 99.25* W303-1A/yDsRed-Express-2 24 0.10 ± 0.004 835.5 ± 124.98 Note. *P < 0.05 vs. other three fluorescent protein genes. Table 1. The fluorescence intensities of four red fluorescent protein genes in yeast cells (mean ± S)
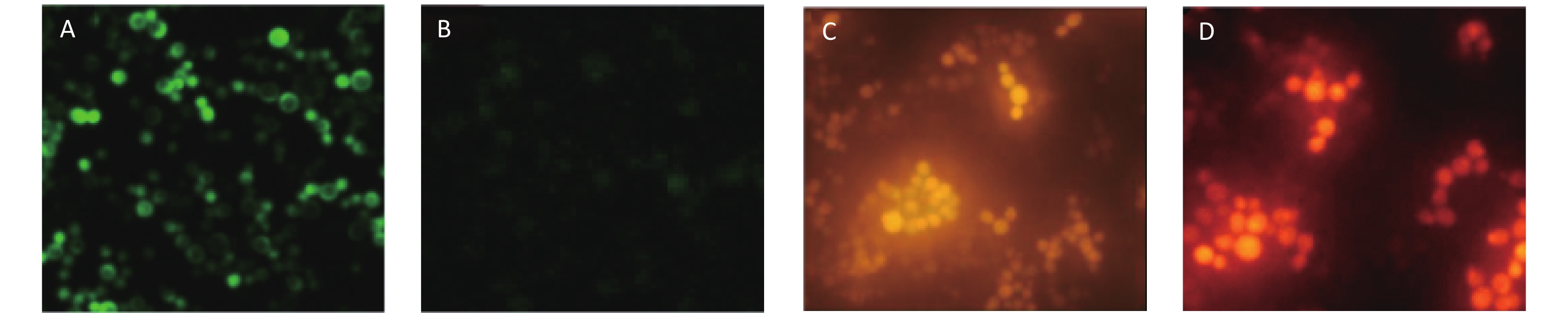
The yeast cells (Yeast W303-1A/RNR3-yEGFP) were transformed with fusion cassettes that contained a functional RNR3 promoter [10] and a gene encoding yEGFP, plus GPD promoter and a gene encoding DsRed-Express-2. The recombinant yeast cells were examined by epifluorescence microscopy after exposure to 200 mg/L methyl methanesulfonate (MMS) for 12 h to ensure the RNR3-yEGFP cells would express yEGFP in response to DNA damage. The cytosensors that exposed MMS emitted intense green fluorescence (Figure 1A), but the sensors that were not exposed to MMS exhibited weak fluorescence (Figure 1B). However, the fluorescent density of DsRed-Express-2 was found to have nothing to do with the dose-dependent MMS (Figure 1C and D). These results confirm that the fluorescence of yEGFP is driven by the DNA-damage-sensitive RNR3 promoter upon exposure to a genotoxin, and the fluorescence of DsRed-Express-2 can be used as a control for normalizing the assay.
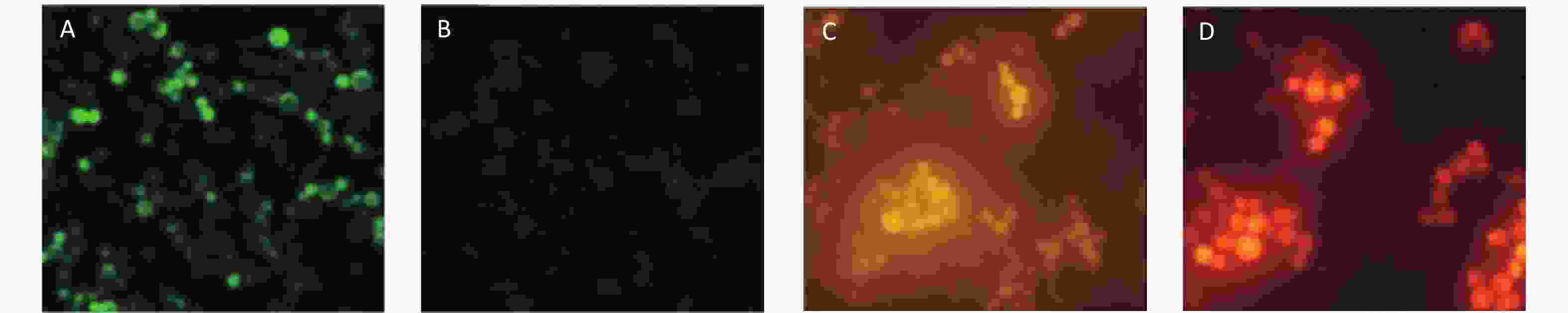
Figure 1. Epifluorescence images of W303-1A/RNR3-yEGFP/DsRed-Express-2 exposed to MMS. (A) Green fluorescence images of the sensor strain exposed to 200 mg/L MMS. (B) Fluorescence images of the sensor strain exposed to 0 mg/L MMS. (C) Red fluorescence images of the sensor strain exposed to 200 mg/L MMS and (D) Red fluorescence images of the sensor strain exposed to 0 mg/L MMS. Cells were grown to log phase in SD/-trp,ura and MMS was added at the indicated concentration. Cells were exposed to MMS for 12 h before imaging.
W303-1A/RNR3-yEGFP/DsRed-Express-2 was treated with different concentrations of test substances of DNA alkylating agents such as MMS and chlorambucil. The fold induction (FI) was determined at 0, 4, 6, 8, 12, 16, and 20 h after the cells were exposed. Fluorescent levels measured are expressed as relative fluorescence units (RFU), i.e., the ratio of yEGFP/DsRed-Express-2 fluorescence intensity. Induction of the yeast sensors by each treatment was calculated as fold induction (FI):
$$ {\rm{FI}} = {\rm{RFUt}}/{\rm{RFUc}} $$ where RFUt is the RFU of the yeast sensor with treatments (measured in at least three replicates) and RFUc is the RFU of the sensor for blank solvent controls (measured in at least three replicates). Each FI value was expressed as the mean and the standard deviation. A tested chemical inducing greater than 1.5 FI was considered to be positive [9].
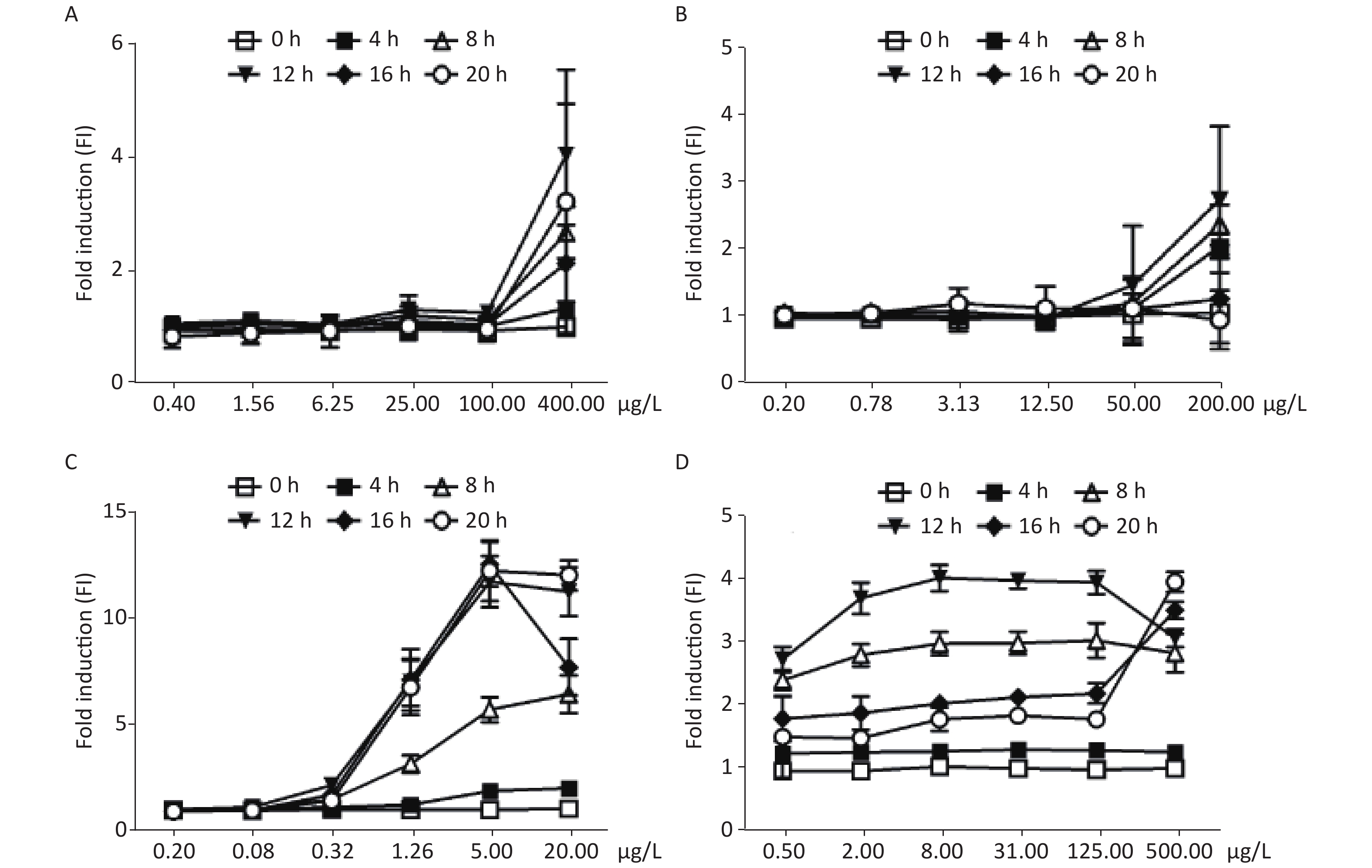
As shown in Figure 2A and B, the FIs of MMS and chlorambucil increased with incubation time in a yeast culture. After 4 h, only a weak increase in fluorescence could be observed. The fluorescence signal peaked after approximately 8–12 h incubation. Distinct FIs with peaks of 4.12 and 2.80 were found for MMS and chlorambucil at 12 h, respectively. However, the time-dose effect relationship decreased at 16 and 20 h of chlorambucil treatment (FI < 1.5), most likely due to the yeast detoxification.
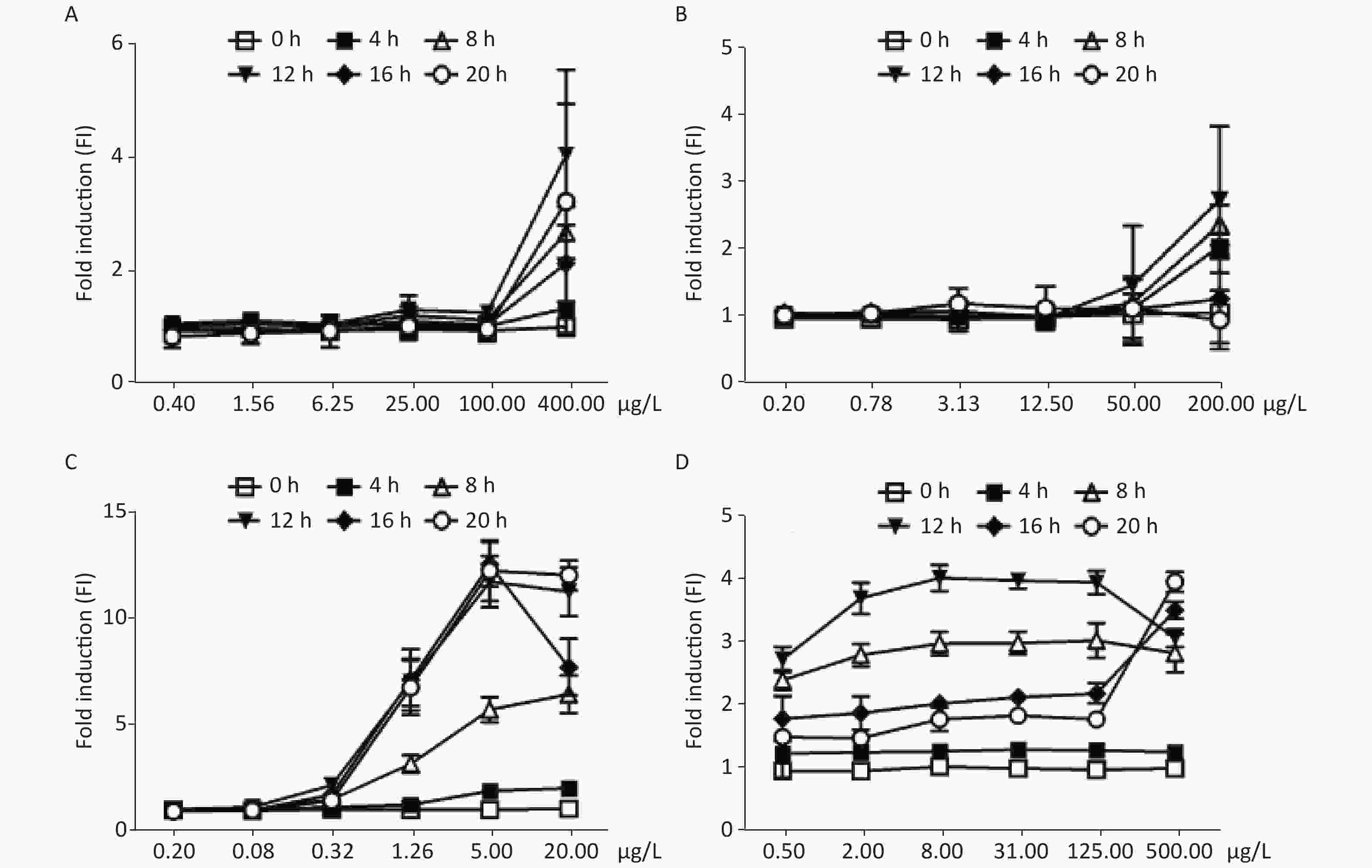
Figure 2. Induction of W303-1A/RNR3-yEGFP/DsRed-Express-2 by various chemical mutagens. (A): MMS; (B): chlorambucil; (C): 4-NQO; (D): 5-FU. Results are the average of at least three independent experiments.
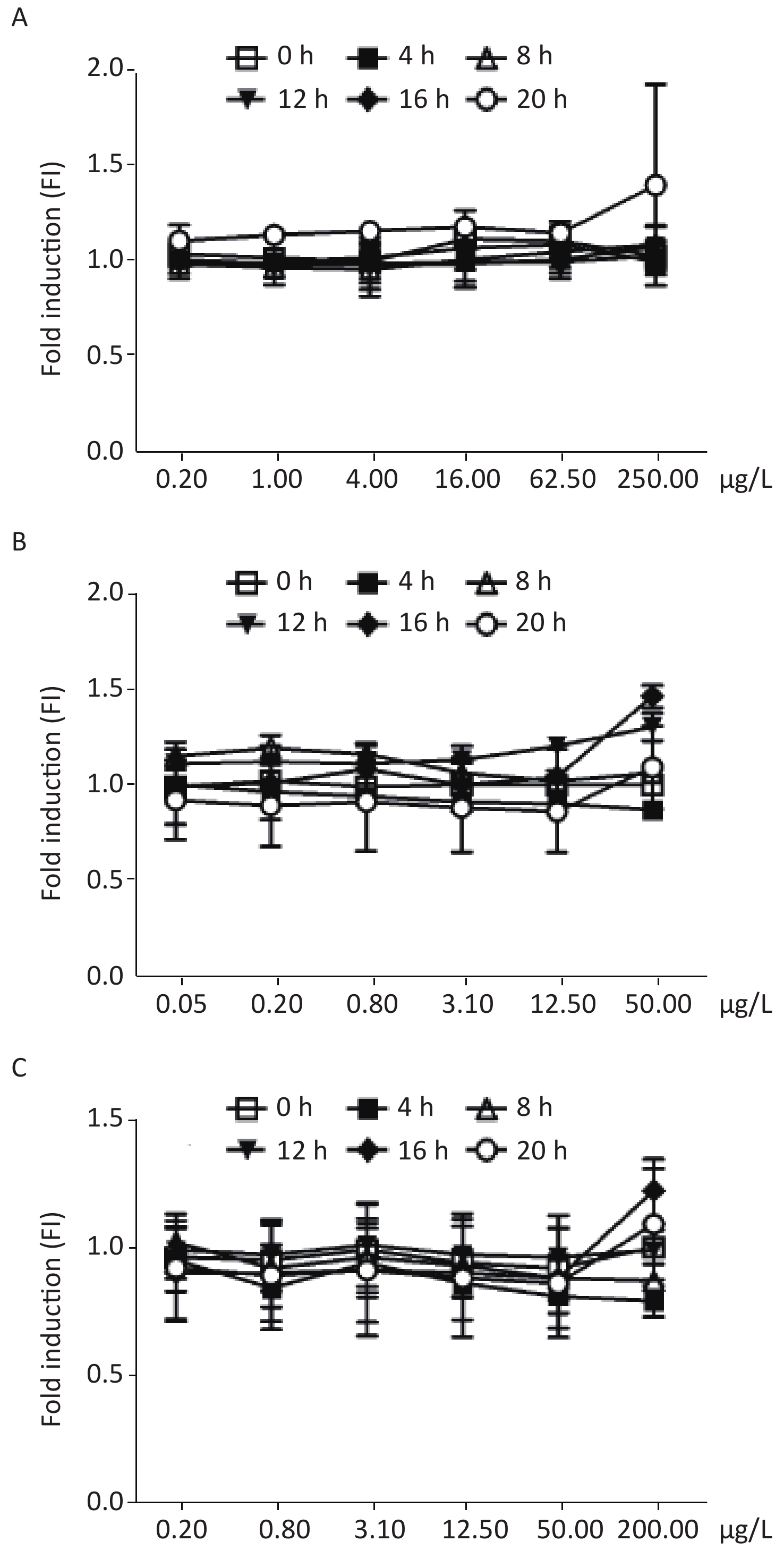
Figure 2C shows the dose-dependent relationships resulting from exposure of the biosensor to 4-nitroquinoline-N-oxide (4-NQO), which is a DNA cleaving agent. The time-dose response curve of 4-NQO genotoxin showed slight increases after 4 h and showed a peak FI of 3.87 at after 12 h of incubation. This change after 12 h of exposure may be due to either the drug’s toxic action on cells or the degradation of the genotoxin. The maximum FIs of the other DNA cleaving agents, such as cis-platinum, bleomycin, and phleomycin, when exposed to different concentrations of the DNA-damaging agents, were below the 1.5-fold established positive result threshold (Supplementary Figure S1 available in www.besjournal.com).
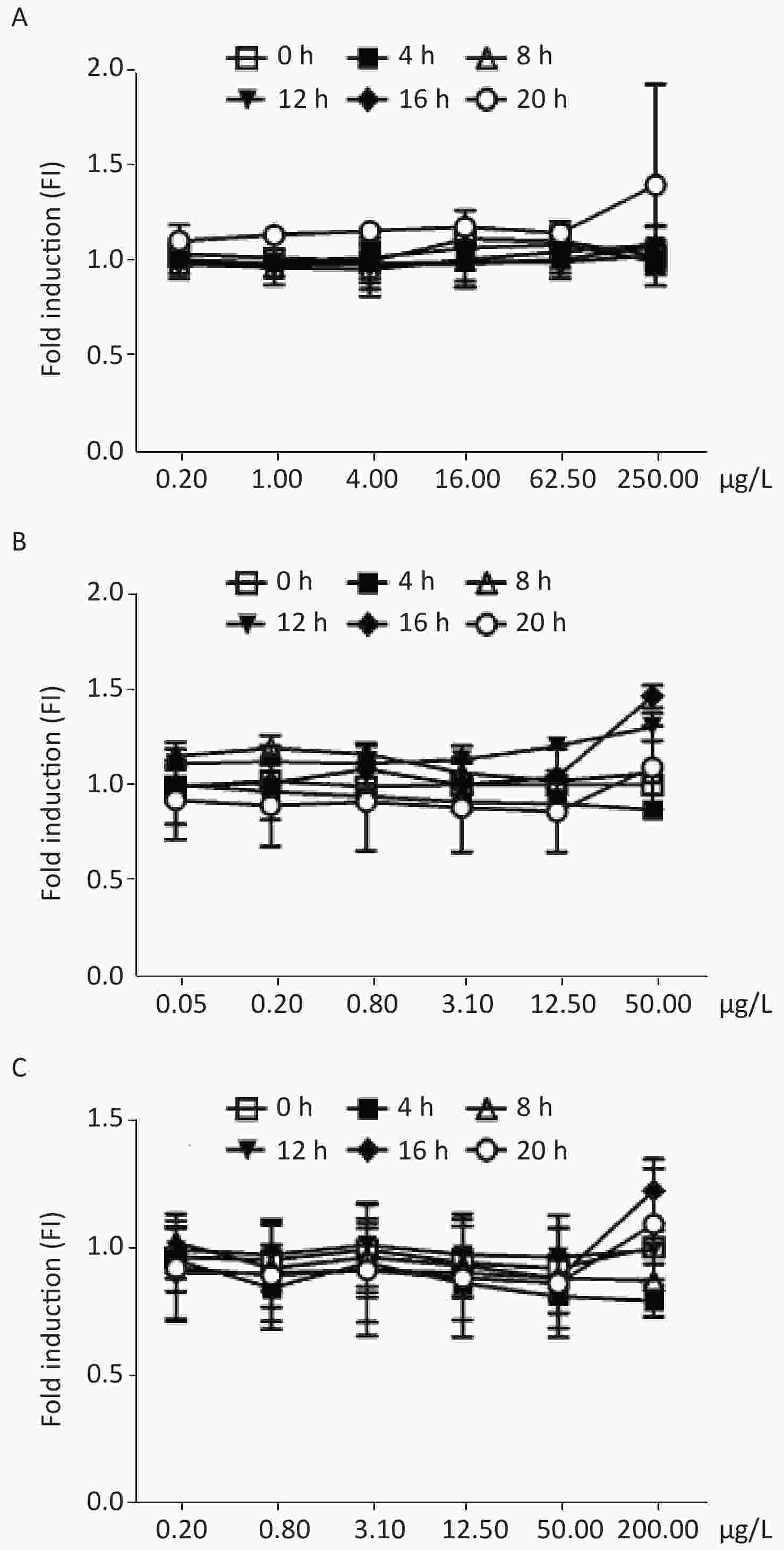
Figure S1. Induction of W303-1A/RNR3-yEGFP/DsRed-Express-2 by various chemical mutagens. (A): cis-platinum; (B): bleomycin; (C): phleomycin. Results are the average of at least three independent experiments.
The yeast censor responded to exposure to different concentrations of 5-fluorouracil (5-FU), an inhibitor of DNA polymerases. It was found that the response curve began to increase after exposure for 4 h, and an FI greater than 2.0 was observed in the group above the concentration of 0.5 μg/mL treated for 8 h, but the response curve increased slowly within the FI range from 2.38 to 3.01. Similar results were observed when cells were exposed to 5-FU for 12 h with a minimum FI of 2.71 and maximum of 4.08 induction (Figure 2D). This was most likely due to yeast detoxification over time.
In our research, some genotoxic compounds, such as MMS, chlorambucil, 4-NQO, and 5-FU were found to trigger a detectable and reproducible level of yEGFP/DsRed-Express-2 for this system. Furthermore, the ratio of yEGFP/DsRed-Express-2 was not induced by other toxic biochemicals, including colchicine, canavanine, and tetracycline. These results suggest that this system will be a valuable supplement to traditional genotoxicity assays.
Dual Fluorescent Protein (yEGFP/DsRed-Express-2) Bioassay System for Rapid Screening for Chemical Mutagens Based on RNR3 Regulation in Saccharomyces Cerevisiae
doi: 10.3967/bes2021.057
- Received Date: 2020-10-07
- Accepted Date: 2021-03-04
| Citation: | LIU Xing, CHEN Geng, LU Gang Yu, YAO Jia, ZHU Fang Yu, XU Jun, LI Xiang Ming. Dual Fluorescent Protein (yEGFP/DsRed-Express-2) Bioassay System for Rapid Screening for Chemical Mutagens Based on RNR3 Regulation in Saccharomyces Cerevisiae[J]. Biomedical and Environmental Sciences, 2021, 34(5): 421-424. doi: 10.3967/bes2021.057 |








 Quick Links
Quick Links
 DownLoad:
DownLoad: