-
Legionella, a genus of pathogenic Gram-negative bacteria, is widely present in natural water sources and artificial water systems. A total of 65 species and > 70 serogroups of Legionella have been characterized[1]. Lipopolysaccharide (LPS), the main component of the outer membrane of Legionella, is not only associated with toxicity, but also provides the basis for classification in serotyping[2]. LPS consists of lipid A, core polysaccharide, and O-antigen. O-antigens are glycopolymers expressed on the cell surface of Gram-negative bacteria. Variability in the O-antigen structure constitutes the basis for the establishment of serotyping. Based on variations in the O-antigen, Legionella can be divided into 15 serogroups[3]. Approximately 84% of Legionella infections are caused by L. pneumophila serogroup 1 (LP1)[4], and Legionnaires’ disease caused by non-L. pneumophila strains of Legionella accounts for approximately 5%–10% of all cases[5].
LPS comprises 0–1.6% of the dry weight of L. pneumophila. LPS has been shown to stimulate morphological changes and alterations in gene expression in almost all host cells, leading to the uncontrolled expression of host cytokines, severe infection, and septic shock[6]. Gene clusters encoding enzymes for the synthesis of Legionella LPS provide model candidates for studying molecular evolution at the DNA level. The Legionella O-antigen contains a number of isomers and derivatives of the monosaccharide pseudaminic acid, such as legionaminic acid and 4-epilegionaminic acid[7]. This monosaccharide is rarely present in the O-antigen of bacteria, thus accounting for the unique O-antigen structure of Legionella.
In the present study, various Legionella serogroups were identified by PCR, and comparisons were made among the O-antigen-specific genes. The PCR method has been tested previously for its specificity and sensitivity[8, 9], and LPS composition may be a determinant of serogroup specificity, as defined by the immunofluorescence-based serotyping schema for L. pneumophila and other Legionella species[2].
Whole genome sequences from 97 strains of Legionella were analyzed in the present study. Complete details of these strains, including the strain name, source, place of isolation, serotype, and time of isolation, are provided in Supplementary Table S1 available in www.besjournal.com. The data generated in this Whole Genome Shotgun project have been deposited in the National Center for Biotechnology Information (NCBI) under the BioProjectID PRJNA281151 with accession numbers LBAW00000000, LBHK00000000, LBAX00000000, LUCB00000000, LCUA00000000, LBAY00000000, LAVP00000000, and LBMS00000000.
Name of strains BioSample number SRA number Bioproject Source of strains Place of isolation Serotype Time of isolation AH104 SAMN05439981 SRS1609486 PRJNA331678 cooling tower water anhui LP1 2006 BJ-20 cooling tower water beijing LP1 2002 BJ-23 SAMN05439998 SRS1609494 PRJNA331678 cooling tower water beijing LP1 2006 BJ-9 SAMN05440000 SRS1609496 PRJNA331678 cooling tower water beijing LP1 2005 FS_4_1103abu SAMN05440002 SRS1609499 PRJNA331678 spring water beijing LP1 2011 Hu6 SAMN05440003 SRS1609501 PRJNA331678 cooling tower water huhehaote LP1 2008 ICDC-LP001 SAMN05440004 SRS1609500 PRJNA331678 sputum specimens beijing LP1 2011 ICDC-LP002 SAMN05440005 SRS1609502 PRJNA331678 sputum specimens beijing LP1 2012 JNLH86 SAMN05440006 SRS1609503 PRJNA331678 cooling tower water shandong LP1 2006 JX1 SAMN05440007 SRS1609504 PRJNA331678 cooling tower water jiangxi LP1 2008 NX0702 SAMN05440008 SRS1609505 PRJNA331678 environmental water ningxia LP1 2008 Qin1 SAMN05440009 SRS1609506 PRJNA331678 cooling tower water qinhuangdao LP1 2008 SH003 SAMN05440011 SRS1609509 PRJNA331678 air conditioning water shanghai LP1 2008 SH013 environmental water shanghai LP1 2008 SH078 environmental water shanghai LP1 2008 SH095 SAMN05440012 SRS1609510 PRJNA331678 air conditioning water shanghai LP1 2008 SH135 SAMN05440013 SRS1609511 PRJNA331678 air conditioning water shanghai LP1 2008 SH202 SAMN05440014 SRS1609512 PRJNA331678 air conditioning water shanghai LP1 2009 SZ026 SAMN05440015 SRS1609513 PRJNA331678 cooling tower water shenzhen LP1 2005 SZ059 SAMN05440016 SRS1609514 PRJNA331678 cooling tower water shenzhen LP1 2005 SZ069 SAMN05440017 SRS1609515 PRJNA331678 cooling tower water shenzhen LP1 2005 SZ099 SAMN05440018 SRS1609516 PRJNA331678 cooling tower water shenzhen LP1 2005 SZ2012006 SAMN05440019 SRS1609517 PRJNA331678 spring water shenzhen LP1 2012 SZ2012007 SAMN05440020 SRS1609518 PRJNA331678 spring water shenzhen LP1 2012 TL-12 SAMN05440021 SRS1609520 PRJNA331678 spring water beijing LP1 2005 WD_4_1102a SAMN05440022 SRS1609521 PRJNA331678 spring water beijing LP1 2011 WD_4_1102b-36 SAMN05440023 SRS1609522 PRJNA331678 spring water beijing LP1 2011 WD_9_1102a SAMN05440024 SRS1609523 PRJNA331678 spring water beijing LP1 2011 WX2011029 SAMN05440025 SRS1609524 PRJNA331678 cooling tower water wuxi LP1 2010 WX2011036 SAMN05440026 SRS1609525 PRJNA331678 cooling tower water wuxi LP1 2010 WX2011046 SAMN05440027 SRS1609526 PRJNA331678 cooling tower water wuxi LP1 2010 Yu237 SAMN05440029 SRS1609528 PRJNA331678 environmental water chongqing LP1 2005 ZJ030014 cooling tower water zhejiang LP1 2006 ZJ050052 SAMN05440030 SRS1609529 PRJNA331678 cooling tower water zhejiang LP1 2006 ATCC33153 SAMN05439982 SRS1609487 PRJNA331678 lung tissue Philadelphia LP1 1976 L.pneumophila_2300_99_Alcoy_uid48801 SAMN02604292 None PRJNA18743 sputum specimens Spain LP1 1999 L.pneumophila_Corby_uid58733 SAMN02603241 None PRJNA17491 Human isolate Oxford LP1 1985 L.pneumophila_Lens_uid13126 SAMEA2240936 ERS353852 PREB4698 Clinical isolate France LP1 2003 L.pneumophila_Lorraine_uid170535 SAMEA3138425 ERS610360 PRJNA67921 Cooling tower water France LP1 1987 L.pneumophila_Paris_uid13127 SAMN06270327 SRS1938716 PRJNA368718 Cooling tower water Paris LP1 1985 L.pneumophila_Philadelphia_1_ uid57609 SAMN05180048 SRS1473718 PRJNA323476 Human lung Philadelphia LP1 1976 BJ46 cooling tower water beijing LP2 2006 SZ098 cooling tower water shenzhen LP2 2005 ATCC33154 SAMN05439983 SRS1609497 PRJNA331678 lung tissue Philadelphia LP2 1976 FS-10-1101a-1 spring water beijing LP3 2011 WD12-1101b spring water beijing LP3 2011 ATCC33155 SAMN05439984 SRS1609508 PRJNA331678 portable water Chicago LP3 1985 SH005 environmental water shanghai LP4 2008 SH106 environmental water shanghai LP4 2008 ATCC33156 SAMN05439985 SRS1609519 PRJNA331678 lung tissue Los Angeles LP4 1988 BJ56 cooling tower water beijing LP5 2006 JX007 cooling tower water jiangxi LP5 2008 ATCC33216 SAMN05439986 SRS1609530 PRJNA331678 lung tissue Dallas LP5 1980 BJ7 SAMN05439999 SRS1609495 PRJNA331678 cooling tower water beijing LP6 2002 BJ-7 cooling tower water beijing LP6 2002 FS_10_1101a-3 SAMN05440001 SRS1609498 PRJNA331678 spring water beijing LP6 2011 NX0701 environmental water ningxia LP6 2008 L.pneumophila_Thunder_Bay_uid206517 SAMN02603729 None PRJNA168333 Clinical isolate Canada LP6 1994 Sctan SAMN05440010 SRS1609507 PRJNA331678 sputum specimens sichuan LP7 2008 ATCC33823 SAMN05439992 SRS1609561 PRJNA331678 Human lung Chicago LP7 1981 L.pneumophila_ uid170534 SAMN05439992 SRS1609561 PRJNA331678 Human lung Chicago LP7 1981 JH1102 spring water beijing LP8 2011 ATCC35096 SAMN05439994 SRS1609490 PRJNA331678 postmortem lung specimen California LP8 1983 ATCC35289 SAMN05439995 SRS1609491 PRJNA331678 open-lung biopsy specimen California LP9 1984 ATCC43283 SAMN05439997 SRS1609493 PRJNA331678 sputum specimens Netherland LP10 1994 WZ1519012 domestic water wenzhou LP11 2015 ATCC43130 SAMN05439996 SRS1609492 PRJNA331678 lung tissue New York LP11 1986 2011-0116 spring water beijing LP12 2011 JN-3-1103 spring water beijing LP12 2011 SH181 environmental water shanghai LP12 2009 WD-10-1105-1 spring water beijing LP12 2010 L.pneumophila_ATCC_43290_uid86885 SAMN02650977 None PRJNA239272 lung tissue Denver LP12 1986 SH122 SAMN05440028 SRS1609527 PRJNA331678 environmental water shanghai LP13 2008 SH167 SAMN05439987 SRS1609531 PRJNA331678 environmental water shanghai LP13 2009 WD-4-1102 SAMN05439988 SRS1609532 PRJNA331678 spring water beijing LP14 2011 WX2012012 SAMN05439989 SRS1609533 PRJNA331678 cooling tower water wuxi 2月14日 2012 ATCC33217 SAMN05439990 SRS1609534 PRJNA331678 lung tissue Atlanta bozemanii 1980 ATCC33218 SAMN05439991 SRS1609488 PRJNA331678 Human blood via yolk sac Atlanta micdadei 1980 ATCC33297 SAMN05439993 SRS1609489 PRJNA331678 soil Atlanta gormanii 1980 ATCC33623 SAMEA3138431 ERS610366 PRJNA174470 Jordan river Bloomington jordanis 1982 ATCC33761 SAMN02743988 SRS1368331 PRJNA222237 cooling tower water Oak Ridge oakridgenisis 1983 ATCC35072 SAMN02471332 None PRJNA39279 Grinding machine coolant fluid Atlanta feeleii 1984 Legionella_anisa_Linanisette_ uid199703 SAMN01916508 None None Tap water Los Angeles anisa 1985 Legionella_cherrii_DSM_19213_ uid222237 SAMN01917904 None None Thermally altered water Minnesota cherrii 1985 Legionella_drancourtii_LLAP12_uid56003 SAMN02743993 SRS1431785 environmental water source UK drancourtii 1983 Legionella_dumoffii_NY_23 PRJNA223037 Cooling tower water New York dumoffii 1980 Legionella_dumoffii _ Tex-KL SAMN02743989 SRS1368330 PRJNA221077 Postmortem lung speciemen Houston dumoffii 1979 Legionella_fairfieldensis_ATCC_49588_uid223037 SAMN02744032 SRS1368371 PRJNA234422 Cooling tower water Australia fairfieldensis 1987 Legionella_geestiana_DSM_21217_ uid221077 SAMN02951933 None Hot water tap London geestiana 1993 Legionella_lansingensis_DSM_19556_ATCC_49751 _uid23442 PRJNA36681 Bronchoscopy washings of patient Lansing lansingensis 1987 Legionella_longbeachae_D_4968_uid42207 SAMEA2272007 ERS379470 PRJEA39579 Human lung California longbeachae 1981 Legionella_longbeachae_NSW150_uid39579 SAMN02441510 None Human lung California longbeachae 1981 Legionella_moravica_DSM_19234_uid185635 PRJNA185635 Cooling tower water Czechoslovakia moravica 1988 Legionella_oakridgensis_OR_10 _uid183980 SAMN02641506 None PRJNA183980 Industrial cooling water Pennsylvan oakridgensis 1983 Legionella_sainthelensi_ATCC_35248 _uid223118 SAMN02744009 SRS1430353 PRJNA223118 Spring water Helens sainthelensi 1984 Legionella_shakespearei_DSM_23087_uid199258 SAMN04274794 None PRJNA285910 Cooling tower water England shakespearei 1992 Legionella_wadsworthii_DSM_21896_ATCC_33877_uid234416 SAMN02744130 SRS710358 PRJNA234416 sputum specimens Wadsworth wadsworthii 1982 Table S1. Details of 97 strains of Legionella
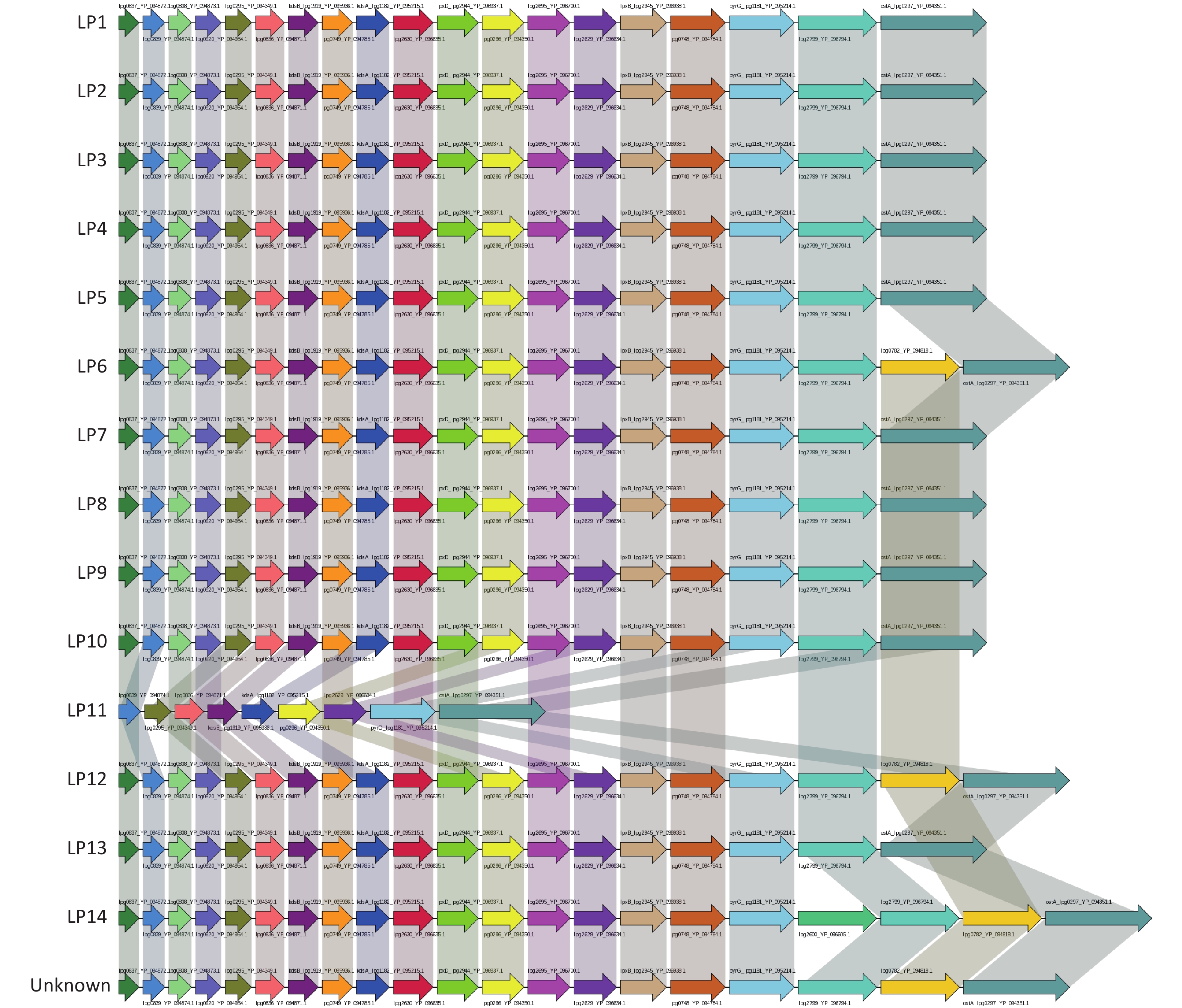
The LPS synthesis gene sequences of all Legionella strains were downloaded in NCBI and multiple annotated databases (including NR, SwissProt, KEGG, COG, TCDB, go, PHI, VFDB, ARDB, Secretory-protein, T3SS, and CAZy). The LPS synthesis gene structure of each serogroup of Legionella was found to be similar. Among the LPS core genes of 15 serotypes (including serogroups LP1–14 and an unknown), the genomes of serotypes LP1, LP2, LP3, LP4, LP5, LP7, LP8, LP9, LP10, and LP13 contained 19 core genes. LP11 contained fewer LPS genes than LP1–LP5, LP7–LP10, and LP13, and lacked the genes lpg0837_YP_094872.1, lpg0838_YP_094873.1, lpg0920_YP_094954.1, lpg0749_YP_094785.1, lpg2630_YP_096635.1, lpxD_lpg2944_YP_096937.1, lpg2695_YP_096700.1, lpxB_lpg2945_YP_096938.1, lpg0748_YP_094784.1, and lpg2799_YP_096794.1 (Supplementary Figure S1, available in www.besjournal.com).
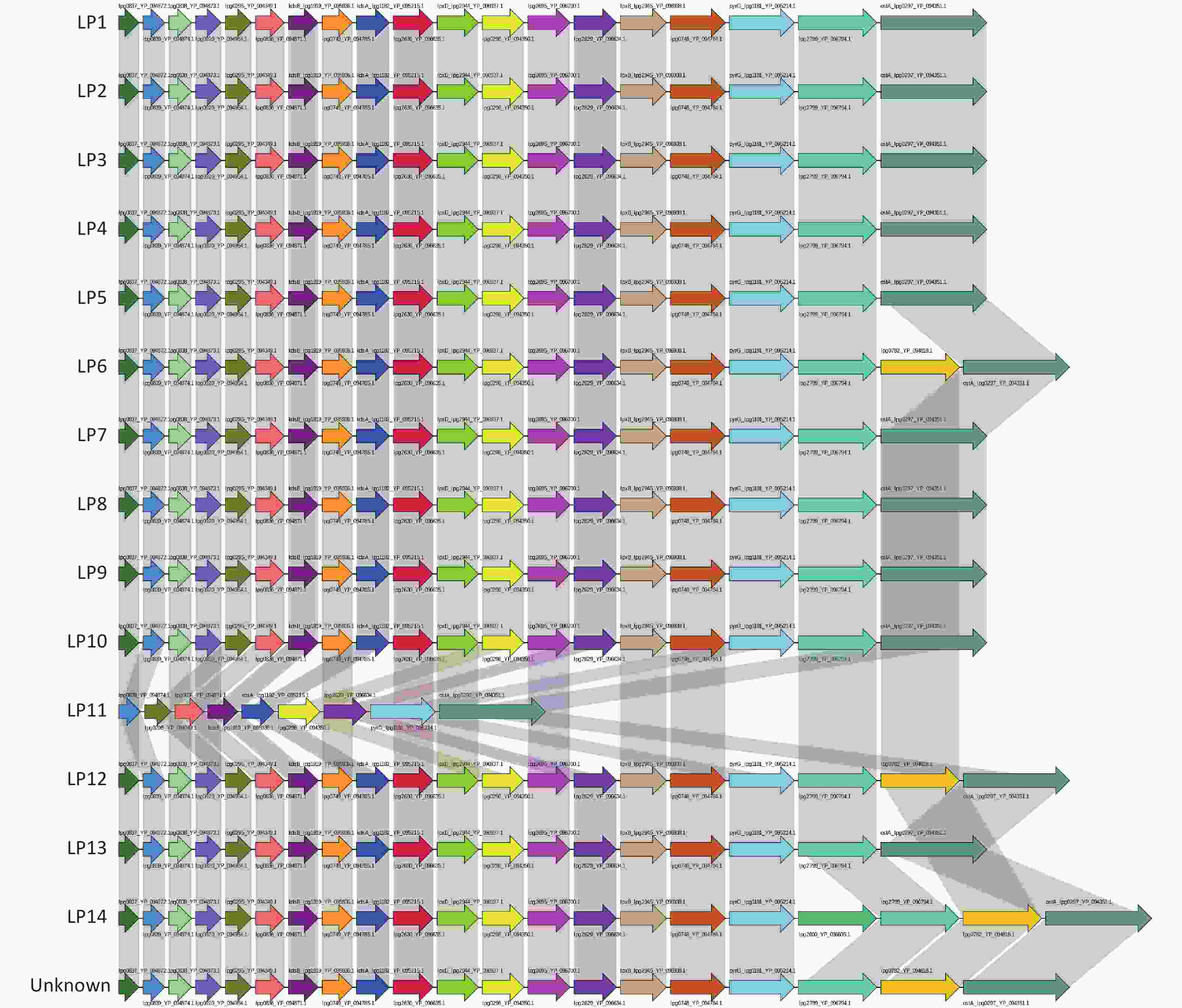
Figure S1. Core gene LPS structure of Legionella pneumophila in each serogroup. The results obtained revealed that there are 15 LPS core genes in each serogroup. Each arrow represents one core gene. Identical genes are indicated using the same color.
Core-pan analysis of all serogroups (LP1–14 and the unknown) was then performed. Numbers of LPS core and pan genes, and the core-pan differences between each serogroup of Legionella, are shown in Supplementary Table S2 available in www.besjournal.com. For the pan genes of LPS, the following differences were observed: Among the 15 serotypes, LP1 and the unknown contained the lpg0777_YP_094813.1 gene (an O-acetyltransferase), and unlike the other Legionella types, LP4, LP5, LP8, LP9, LP11, and LP13 lacked the lpg0782_YP_094818.1 gene (encoding a putative O-acetyltransferase). Among the core genes for LPS synthesis, LP14 uniquely contained a gene, alpg2600_YP_ 096605.1, whereas LP11 contained fewer LPS pan genes compared with other serogroups of Legionella (Supplementary Figure S2, available in www.besjournal.com).
Serogroup PAN LPS CORE LPS Core-pan difference LP1 21 19 lpg0777_YP_094813.1, lpg0782_YP_094818.1 LP2 20 19 lpg0782_YP_094818.1 LP3 20 19 lpg0782_YP_094818.1 LP4 19 19 − LP5 19 19 − LP6 20 20 − LP7 20 19 lpg0782_YP_094818.1 LP8 19 19 − LP9 19 19 − LP10 20 19 lpg0920_YP_094954.1 LP11 11 9 lpg0920_YP_094954.1, lpg2799_YP_096794.1 LP12 20 20 − LP13 19 19 − LP14 21 21 − Unknown 21 20 lpg0777_YP_094813.1 Table S2. Numbers of LPS core and pan genes and core-pan differences among Legionella serogroups
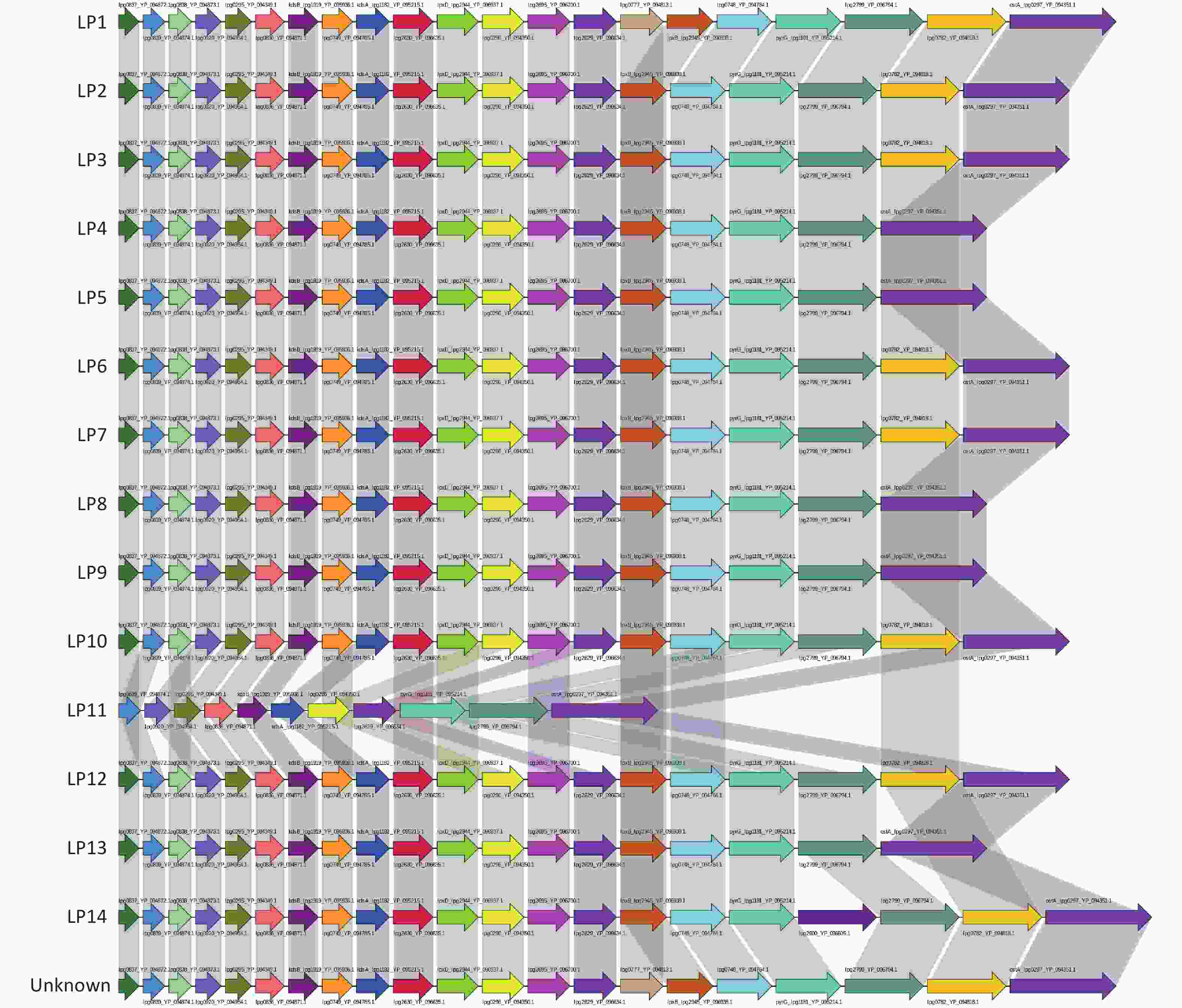
Figure S2. Pan gene LPS structure of Legionella pneumophila each serogroup. The results obtained revealed that there are 15 LPS pan genes in each serogroup. Each arrow represents one pan gene. Identical genes are indicated using the same color.
The sequences of the core and pan genes of each serogroup were then compared with the previously downloaded reference sequences (in BLAST), and filtered based on the following criteria: identity > 80%, and coverage > 80%. Gene clusters were classified according to the LPS sequence length (small to large), and the identical LPS genes of different serogroups were connected by coarse lines. In addition, functional annotation of the LPS genes was performed for each serogroup. The LPS synthesis gene structure of non-L. pneumophila was found to be similar among non-L. pneumophila serogroups. However, among the LPS core genes, the following differences were observed: Legionella_dumoffii_NY_23, Legionella_cherrii_DSM_19213,Legionella_shakespearei_DSM_23087, Legionella_moravica_DSM19234, and Legionella_dumoffii_Tex_KL contained more LPS genes than other non-L. pneumophila serogroups, including lpxB_lpg2945_YP_096938.1 and lpxD_lpg2944_YP_096937.1. Furthermore, Legionella_dumoffii_NY_23, Legionella_dumoffii_Tex_KL, and Legionella_anisa_linanisettee contained the gene lpg0749_YP_094785.1, whereas other non-L. pneumophila serogroups did not (Supplementary Figure S3, available in www.besjournal.com).
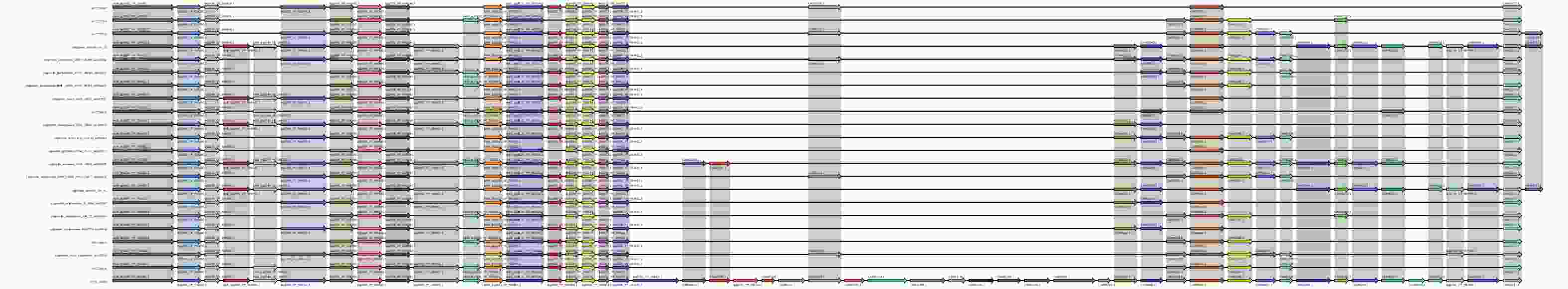
Figure S3. Core gene LPS structure of Legionella non-pneumophila. The results obtained revealed that there are 22 LPS core genes in each serogroup. Each arrow represents one core gene. Identical genes are indicated using the same color.
Among the 15 serogroups of L. pneumophila, LP 14 contained the largest number of LPS genes (21 core genes and 21 pan genes), and L. pneumophila serogroup 11 contained the fewest LPS genes (9 core genes and 11 pan genes). The genes held in common between L. pneumophila and non-L. pneumophila were found to be ostA_lpg0297_YP_094351.1, lpg0296_YP_094350.1, lpg0295_YP_094349.1, lpg2695_YP_096700.1, lpg2630_YP_096635.1, lpg2629_YP_096634.1, kdsA_lpg1182_YP_095215.1, pyrG_lpg1181_YP_095214.1, lpg0839_YP_094874.1, lpg0838_YP_094873.1, lpg0836_YP_094871.1, and CAB65217.1.
Among the non-L. pneumophila serogroups, 14–29 core LPS genes were identified, and these also had a higher distributional difference. L. pneumophila serogroups were found to contain a higher number of LPS synthesis genes compared with the non-L. pneumophila serogroups, and greater LPS gene differences were also detected among non-L. pneumophila serogroups, especially Legionella_dumoffii_NY_23,Legionella_dumoffii_Tex_KL, and Legionella_anisa_linanisettee (Supplementary Figures S2 and S3).
The LPS genes for all Legionella strains were downloaded from the NCBI database, and homologous alignment was subsequently performed for all samples (including 76 strains of L. pneumophila, 21 strains of non-L. pneumophila, and one strain of Coxiella burnetii). Screening samples with the LPS reference sequence annotation, the same information with the LPS reference sequence annotation was chosen. The above results were merged into redundancy, core-pan analysis was performed based on core gene similarity, and a phylogenetic tree was constructed using the neighbor-joining method. Based on the presence of pan genes, a phylogenetic tree was constructed using the unweighted pair-group method with arithmetic means. The phylogenetic tree was constructed according to the LP grouping system using the neighbor-joining method and unweighted pair-group method. After adding the external reference Rickettsia, the results obtained were similar to those without the external reference. The analyzed data revealed a state of gradual evolution of these strains from non-L. pneumophila to L. pneumophila. WZ1519012 (LP11), a strain of L. pneumophila, represented the closest association with non-L. pneumophila, although the distribution of different serogroups of Legionella species was dispersed. According to the phylogenetic tree constructed based on core gene similarity, the evolutionary edge strains included Legionella_shakespearei_DSM_23087_uid199258 and WD-4-1102. WD-4-1102 belongs to LP14 (Figure 1). In the phylogenetic tree based on pan gene similarity, the edge strains included SH003 and Legionella_moravica_DSM_19234_uid185635, although Legionella_shakespearei_DSM_23087_uid199258 and WD-4-1102 were not far apart (Figure 2).
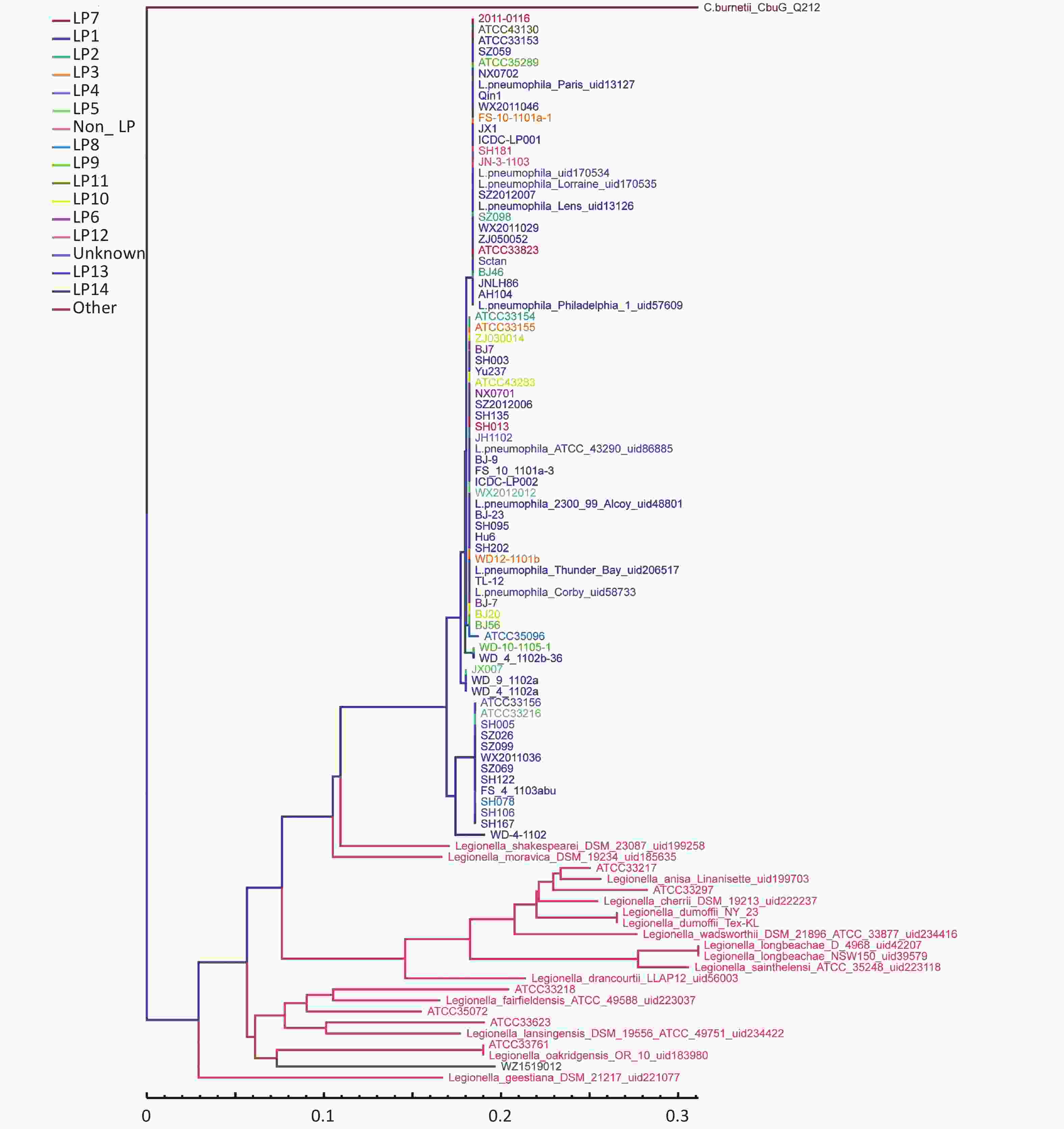
Figure 1. Phylogenetic tree based on core genes. Branch lengths are calculated from the means of the posterior probability density. Values below the nodes represent posterior probabilities. The scale bar represents substitutions per site.
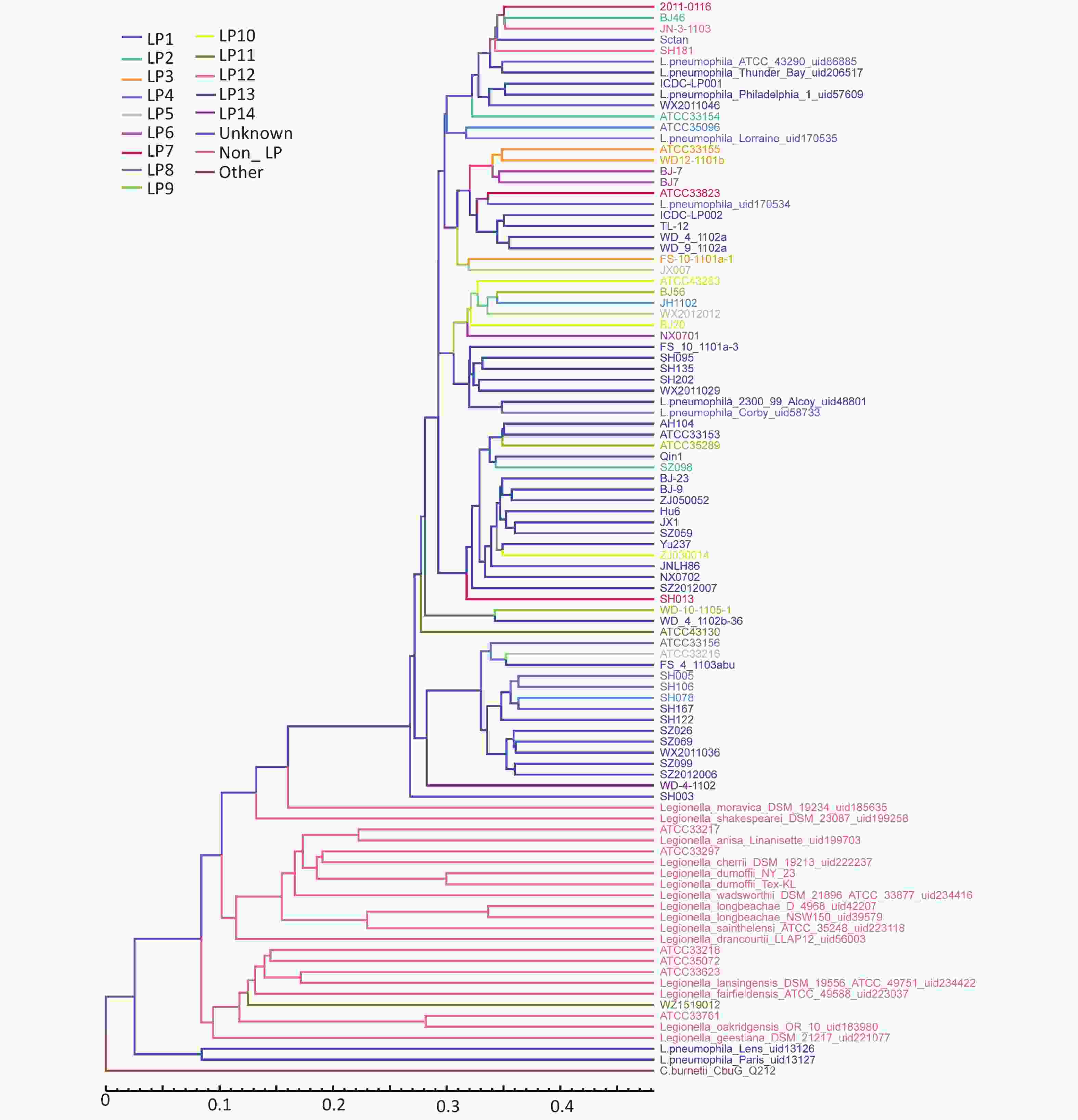
Figure 2. Phylogenetic tree based on pan genes. Branch lengths are calculated from the means of the posterior probability density. Values below the nodes represent posterior probabilities. The scale bar represents substitutions per site.
The difference identified between the two methods was that L. pneumophila_Paris_uid13127 (LP1) and L. pneumophila_Lens_uid13126 (LP1) were clustered together in the pan tree. Whole LPS genes of L. pneumophila_Paris_uid13127 (LP1) and L. pneumophila_Lens_uid13126 (LP1) were similar to those in other samples, although larger numbers of specific genes (264 and 223, respectively) were observed. Evolutionary trees with or without an external reference produced similar results. In the pan tree, SH003 represented the closest association with the evolutionary edge, whereas this was not the case in the core phylogenetic tree (Figure 1 and Figure 2).
The results of phylogenetic analysis of core genes showed that Legionella_dumoffii_NY_23,Legionella_dumoffii_Tex_KL, Legionella_cherrii_DSM_19213, and Legionella_anisa_linanisettee shared a close evolutionary relationship. Based on the phylogenetic analysis results, it was possible to speculate on how evolution may have progressed from non-L. pneumophila to L. pneumophila. WZ1519012 (LP11) represented the closest association with non-L. pneumophila; however, the distribution of serogroups of different Legionella species was more dispersed.
Primer sequences for LP11 and LP14 were tested and screened by PCR based on O-antigen-specific genes. PCR results for hypothetical genes were eliminated in initial screening, and Legionella serogroups 11 and 14 were chosen as the PCR templates. The O-antigen-specific genes for Legionella were repeatedly screened by PCR. Primers were designed according to the specific wzt gene in the gene cluster of Legionella serogroups 11 and 14. Using these primers, bands of the correct (expected) size were obtained after PCR. The PCR reaction mixture comprised, in a total volume of 30 μL the following: 15 μL of Premix Taq (TaKaRa Taq Version 2 plus dye; Takara Biotechnology Co., Ltd.), 1 μL of forward primer (10 μmol/L), 1 μL of reverse primer (10 μmol/L), 11 μL of distilled water (Invitrogen® UltraPure Distilled Water; Thermo Fisher Scientific, Inc.), and 2 μL of template (50 ng). The PCR thermocycling conditions were: 95 °C for 10 min, followed by 35 cycles of 95 °C for 30 s, annealing for 45 s (for the indicated temperatures, see Supplementary Table S3 available in www.besjournal.com), and 72 °C for 1 min, followed by 72 °C for 10 min. The primer sequences for LP11 and LP14, the size of the PCR products, and the annealing temperatures are shown in Supplementary Table S3. With the exception of the positive control group, none of the other groups presented with bands of the correct size. Hence, this demonstrated that the wzt gene was highly specific in Legionella serogroups 11 and 14.
Gene Serogroup Primers Sequence of primers Size of PCR products Annealing temperature wzt LP11 GM0002878-F 5′-TTGGCATGCAGTCTCGGTCAT-3′ 271 bp 54 °C GM0002878-R 5′-GCTCGACAGGAAGTTGGCTAA-3′ Lp14 GM0001175-F 5′-TTACTTATTGCGCCGATGAT-3′ 291 bp 56 °C GM0001175-R 5′- CTGTCCCAACTTACCGCCTAA-3′ Table S3. PCR assay: WZT gene primers, annealing temperature, and size of PCR products
The PCR method was tested for its specificity and sensitivity using agarose gel electrophoresis. The electrophoresis conditions were as follows: the gels were run for 40 min at 100 V in 0.5× TBE (Beijing Solarbio Science & Technology Co., Ltd). The results obtained revealed that LP11 (Figure 3A and 3C) and LP14 (Figure 3B and 3D) displayed bands for the target gene, whereas other serogroups did not. Therefore, the specificity and sensitivity of the primers of LP11 and LP14 were confirmed by agarose gel electrophoresis.
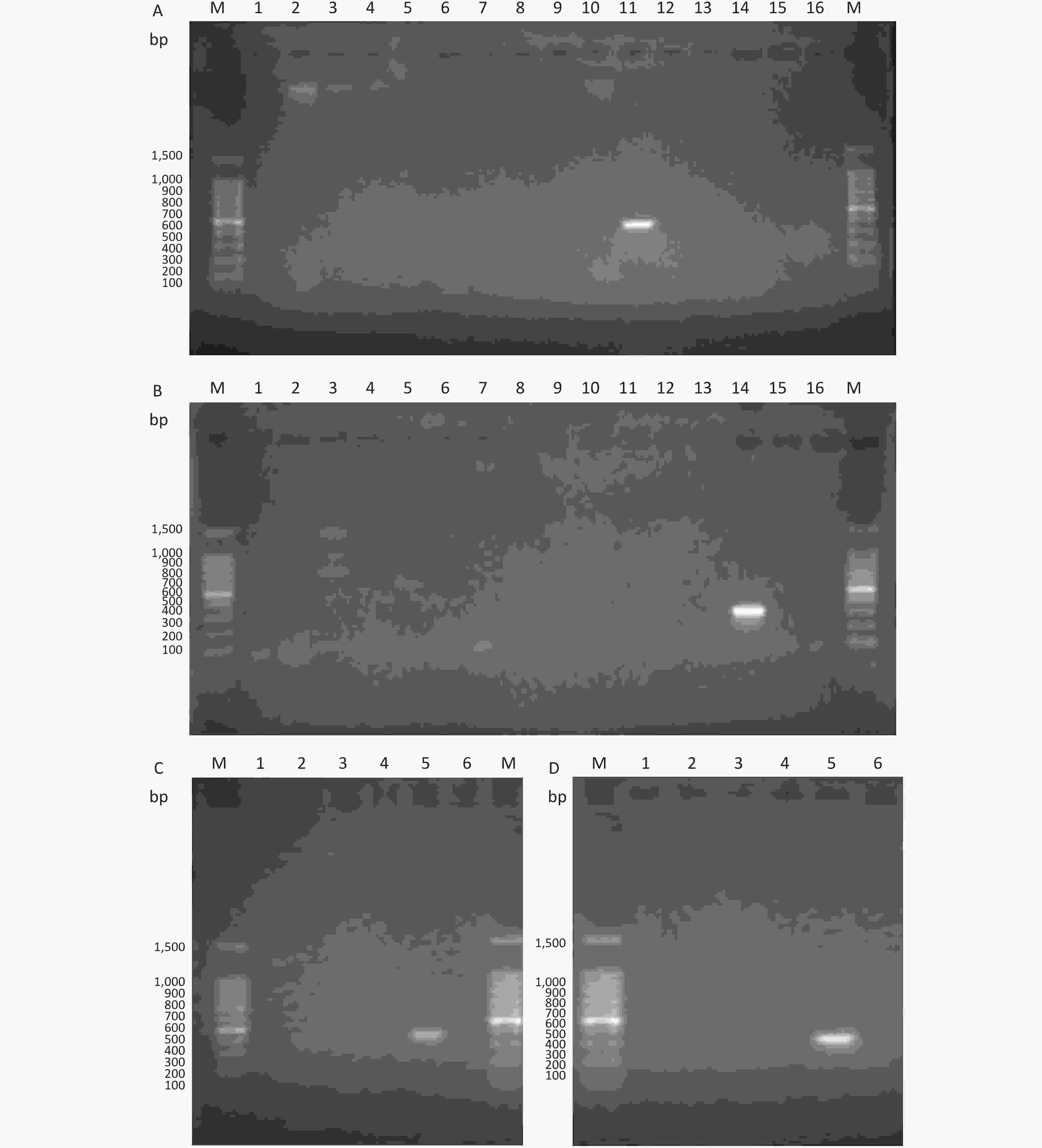
Figure 3. Agarose gel electrophoresis of PCR products. (A) LP11 PCR. PCR using primers designed for LP11: lane M, 100-bp DNA marker; lanes 1–15, L. pneumophila LP1 to LP15; lane 16, negative control. (B) LP14 PCR. PCR using primers designed for LP14: lane M, 100-bp DNA marker; lanes 1–15, L. pneumophila LP1 to LP15; lane 16, negative control. (C) LP11 PCR. PCR using primers designed for LP11: lane M, 100-bp DNA marker; lane 1, Legionella gormanii_ATCC 33297; lane 2, Legionella dumoffii Tex-KL; lane 3, Legionella_cherrii_DSM_19213_uid222237; lane 4, Legionella longbeachae_ATCC33462; lane 5, positive control; lane 6, negative control. (D) LP14 PCR. PCR using primers designed for LP11: lane M, 100-bp DNA marker; lane 1, Legionella gormanii_ATCC 33297; lane 2, Legionella dumoffii Tex-KL; lane 3, Legionella_cherrii_DSM_19213_uid222237; lane 4, Legionella longbeachae_ATCC33462; lane 5, positive control; lane 6, negative control.
In conclusion, the present study has shown that, based on the wzt gene in the LPS cluster, it was possible to establish a rapid and specific method suitable for the identification of L. pneumophila serogroups. Application of this technique enabled identification of the genetic determinants of non-L. pneumophila virulence, and allowed for important comparative studies with other Legionella species to be made. Hence, gene chip- or PCR-based methods can be applied to detect different serogroups of Legionella easily and rapidly.
Phylogenetic Analysis of Legionella Strains and Identification of Serogroups by Lipopolysaccharide- and O-antigen- based PCR Assay
doi: 10.3967/bes2021.065
- Received Date: 2020-08-31
- Accepted Date: 2021-01-21
| Citation: | LI Yi Nan, REN Hong Yu, ZHAO Na, WANG Yan Qing, LI Dai, QIN Tian. Phylogenetic Analysis of Legionella Strains and Identification of Serogroups by Lipopolysaccharide- and O-antigen- based PCR Assay[J]. Biomedical and Environmental Sciences, 2021, 34(6): 483-488. doi: 10.3967/bes2021.065 |







 Quick Links
Quick Links
 DownLoad:
DownLoad:




