HTML
-
Japanese encephalitis (JE) caused by Japanese encephalitis virus (JEV) is mainly prevalent in Asian and Pacific areas[1]. Nearly 3 billion people in 24 countries are at the center of JE endemic and about 67, 900 JE cases occur annually[2-3]. Of JE cases, the fatality rate ranges from 20%-30%[4-5]. JEV is a member of the family Flaviviridae, genus Flavivirus and is transmitted by infected mosquitoes, particularly Culex species[6]. JEV is a single-stranded positive-sense RNA virus with an approximately 11 kb long genome. The genome contains a single long open reading frame (ORF) encoding three structural proteins (C, prM/M, E) and seven non-structural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5)[7]. Based on the E gene sequence, JEV strains have been classified into five genotypes (G1 to G5). Nakayama, isolated in Japan in 1935, is the first JEV-G3 strain dentified[8]. JEV-G3 strains were the most common in areas where Japanese encephalitis was endemic before 2000[9]. M859, isolated in Cambodia in 1967, is the first JEV-G1 strain dentified[10]. However, since 2000, JEV-G1 strains have been increasingly isolated from different vectors (mosquitoes, swine and humans) and have replaced JEV-G3 as the dominant genotype circulating in Asia[11-12]. The JEV-G2 and JEV-G4 strains are mainly found in Malaysia and Indonesia[10]. Up until now, only two JEV-G5 strains (Muar and XZ0934) have been isolated over a 57-year interval[13-15].
Several methods including conventional PCR (nested/semi-nested RT-PCR and multiplex RT-PCR), TaqMan real-time RT-PCR, SYBR Green Ⅰ-based real-time PCR and loop-mediated isothermal amplification (LAMP) have been set up to detect JEV[16-25]. However, these methods were designed mainly for detecting or differentiating JEV-G1 and JEV-G3. Therefore, in this study, we aimed at establishing TaqMan real-time RT-PCR assays that not only detect JEV but also distinguish the five JEV genotypes.
-
The full-length sequences of JEV (G1-G5) were downloaded from GenBank and aligned using ClustalX ver. 2.0. The most conservative regions were identified manually for designing primers and probes. Primer Express 3.0 (Applied Biosystems, Foster City, CA, USA) was used to evaluate the physical properties of the six sets of primers and probes (one set for non-specific JEV detection and five sets for specific JEV genotype detection). The probes were labeled with a 6-carboxy-fluorescein reporter dye at the 5' end and with a 6-carboxyteramethy-rhodamine quencher dye or minor groove binder at the 3' end (Table 1).
Primers/Probes Sequence (5′-3′) Genomic Region Product Size (bp) Universal-JEV-F 5′3588-GCCACCCAGGAGGTCCTT-36053′ Universal-JEV-R 5′3633-CCCCAAAACCGCAGGAAT-36503′ Universal-JEV-MGB-Probe 5′3608-CAAGAGGTGGACGGCC-36233′ NS-1 63 JEV-G1/G3-F 5′4190-GGTCTGCAACCCAAACAAGAA-42103′ JEV-G1/G3-R 5′4299-GCCAGCATGAAGGGTATTGACAT-43213′ JEV-G1-Probe 5′4265-TTGTGGGAGGTCTAGCCGAGTTGG-42883′ NS-2 132 JEV-G3-Probe 5′4264-TCGTAGGTGGTTTGGCCGAGTTG-42863′ NS-2 132 JEV-G2-F 5′612-GAAGACACCATCACCTACGAATGTC-6363′ JEV-G2-R 5′664-CACACCAGCAATCCACATCCT-6843′ JEV-G2-Probe 5′638-CAAGCTCACCACAGGCAATGACCCA-6623′ M 73 JEV-G4-F 5′703-TTCAATATGGACGGTGCACAA-7233′ JEV-G4-R 5′757-CCATGCGTGTGGACAGACA-7753′ JEV-G4-Probe 5′725-AACCTCACACTCCCAGACAAGCAGGAGATC-7743′ M 73 JEV-G5-F 5′681-TGCGACAAACAAGCCGTGTA-7003′ JEV-G5-R 5′741-TTGCACTGACACAGATCTTCTACTTCT-7673′ JEV-G5-Probe 5′714-CGTTGCACGAGGACCAGGCACTC-7363′ M 87 Table 1. Primers and Probes Used for the TaqMan RT-PCR Assay
-
Target gene fragments were cloned into linearized pGEM-T Easy Vectors (Promega, Madison, WI, USA) with T4 DNA ligase. Linearized plasmid DNA templates were recovered, purified and enriched. To recover RNA transcripts, the DNA templates were transcribed with T7 polymerase using the T7 High Efficiency Transcription Kit (TransGen Biotech, Beijing, China) and treated with DNase Ⅰ to remove the residual DNA. Subsequently, the RNA transcripts were stored at -80 ℃ for later use and the RNA copy number was calculated according a described method[17]. JEV-G1 (GZ56 strain, HM366552), JEV-G3 (P3 strain, JEU47032) and JEV-G5 (XZ0934 strain, JF91589) were used as reference standards to establish the methods and conditions in this study.
-
All viral RNAs were extracted from cell cultures or mosquito specimens using the QIAamp Viral RNA Mini Kit (QIAGEN, Dusseldorf, Germany) according to the manufacturer's specifications. Real-time RT-PCR was conducted using AgPath-ID One-step RT-PCR Reagents (Applied Biosystems, Foster City, CA, USA) and performed using Mx3000P Real-Time PCR System (Stratagene, La Jolla, CA, USA). The RT-PCR master mix components were as follows: 2× RT-PCR Buffer (12.5 μL), forward and reverse primers (1 μL of each, 10 pmol/μL), TaqMan probe (1 μL, 5 pmol/μL), 25× RT-PCR enzyme mix (1 μL), RNA template (1 μL) and 7.5 μL of nuclease-free water. The quantitative PCR was performed as follows: 10 min at 45 ℃ for reverse transcription; 10 min at 95 ℃ to activate the DNA polymerase and to deactivate the reverse transcriptase; 40 cycles of 15 s at 95 ℃ and 1 min at 60 ℃. The negative extraction control for the assay consisted of supernatant from uninfected Vero cells. The positive extraction control consisted of supernatant from Vero cells infected with JEV-G3 (P3 strain). The no template control (NTC) was double distilled H2O.
-
RNA transcripts and JEV viral RNAs with ten-fold dilutions were analyzed by qPCR. Each ten-fold dilution was amplified in three replicates. The mean (x), standard deviation (SD) and coefficient of variation (CV) were calculated for the Ct values. Standard curves were generated for each set of primer and probe. Standard curves with slopes between -3.1 and -3.6, corresponding to PCR efficiencies of 90%-110% [efficiency = (10 (-1/slope)-1)][26-27], were regarded as acceptable.
To test the specificity of this detection system, cross-reactivity was examined with Culex flavivirus and Tick-borne encephalitis, West Nile, Zika, Yellow fever, Dengue (serotypes 1-4), Batai, Tahyna, Getah, Sindbis, and Kadipiro viruses.
Design of Primers and the TaqMan Probes
Production of Standard Control for TaqMan qPCR
RNA Extraction, Reverse Transcription and TaqMan Real Time RT-PCR
The Specificity, Sensitivity and Reproducibility of TaqMan Real-time RT-PCR
-
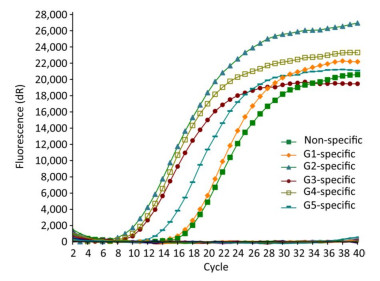
Only the JEV RNA of five genotypes resulted in amplification signals with cycle values < 35, while none of the other arboviruses and no template controls produced a positive amplification signal (Figure 1).
-
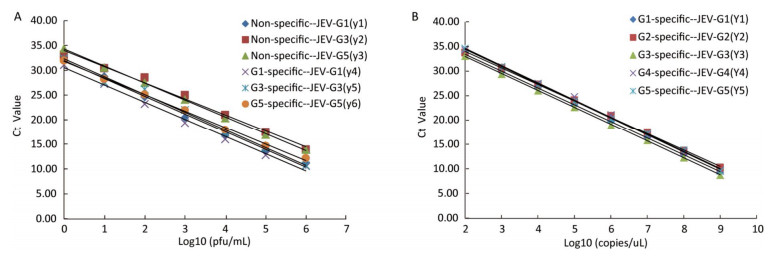
The sensitivities of the JEV titers were observed by serial 10-fold dilution with concentrations ranging from 1 × 106 to 1 × 10-1 pfu/mL. The lowest detection limits of JEV (G1, G3, and G5) titers were 1 pfu/mL (Figure 2A). Similarly, the sensitivities for in vitro-transcribed RNAs were observed by 10-fold serially diluted with concentrations ranging from 1 × 109 to 1 × 100 copies/μL. The detection thresholds of the in vitro-transcribed RNAs with the specific detection systems were 100 copies/μL (Figure 2B).

Figure 2. A. Standard curves of the TaqMan RT-PCR assay generated using a known concentration of Japanese encephalitis virus (JEV) in serial 10-fold dilutions (1 × 106 -1 × 100 pfu/mL). The Ct value (y-axis) was plotted against the log of the known JEV titers (x-axis). JEV-G1, JEV-G3, and JEV-G5 RNA were detected by the non-specific and specific detection systems. The regression equations are as follows: y1 = -3.5948x + 31.976 (R2 = 0.9954); y2 = -3.2522x + 34.017 (R2 = 0.9902); y3 = -3.4284x + 34.311 (R2 = 0.9993); y4 = -3.4894x + 30.551 (R2 = 0.9929); y5 = -3.5947x + 32.321 (R2 = 0.9922); y6 = -3.3504x + 31.823 (R2 = 0.9979). B. Standard curves of the TaqMan RT-PCR assay generated using a known concentration of in vitro-transcribed RNAs by 10 fold serially diluted (1 × 109 -1 × 102 copies/uL). The Ct value (y-axis) was plotted against the log of the known RNA copies (x-axis). JEV-G1, JEV-G2, JEV-G3, JEV-G4 standard and JEV-G5 standard RNA were detected by the specific detection systems. The regression equations are as follows: y1 = -3.4271x + 40.305 (R2 = 0.9999); y2 = -3.3632x + 40.775 (R2 = 0.9992); y3 = -3.4577x + 39.907 (R2 = 0.9998); y4 =-3.5137x + 41.699 (R2 = 0.9978); y5 = -3.5109x + 41.511 (R2 = 0.9992).
To calculate the assay efficiency, standard curves were constructed for each assay, on which Ct values were plotted. A linear correlation was demonstrated between the Ct value and viral concentrations. Statistics for standard curves of the JEV titers and in vitro-transcribed RNAs were calculated. All of the standard curves have slopes ranging from -3.60 to -3.25, corresponding to amplification efficiencies between 90% and 103%, and with R2 values greater than 0.99 (Table 2). Moreover, the standard deviations of the three parallel samples at each serial dilution were < 0.5 and the coefficients of variation were < 2.8%, indicating that these TaqMan assays were reliable.
Samples Viral RNA In Vitro-transcribed RNA Non-specific system Specific system Specific system Genotypes G1 G3 G5 G1 G3 G5 G1 G2 G3 G4 G5 Slope -3.5948 -3.2522 -3.4284 -3.4894 -3.5947 -3.3504 -3.4271 -3.3632 -3.4577 -3.5137 -3.5109 R2 0.9954 0.9902 0.9993 0.9929 0.9922 0.9979 0.9999 0.9992 0.9998 0.9978 0.9992 Efficiency (%) 0.90 1.03 0.96 0.93 0.90 0.99 0.96 0.98 0.95 0.93 0.93 Table 2. Statistics of the Standard Curves and Regression Equations for Viral RNA and in vitro-transcribed RNA Detected by TaqMan RT-PCR
-
TaqMan real-time RT-PCR and semi-nested PCR were performed to verify the accuracy of this detection system. Twenty batches of mosquitoes previously collected in Guizhou province in 2004, Shandong province in 2008 and Tibet Autonomous Region in 2009 were used. Twelve samples were identified as positive (Ct < 35) with this non-specific detection system, while only seven samples were identified as positive with semi-nested PCR (Table 3). Furthermore, the genotypes of these twelve samples were also differentiated with the specific detection system and eight JEV-G1, three JEV-G3 and one JEV-G5 strains were identified. Overall, the results indicated that the sensitivity and accuracy of the TaqMan real-time RT-PCR assay developed in this study were higher than those of semi-nested PCR.
Year Location Strain No. of Mosquitoes Virus Isolation Result Semi-nested PCR Result TaqMan qPCR Non-specific detect system (Ct) Specific detect system (Ct/genotype) 2004 Guizhou GZ04-26 100 Neg Neg 26.55 27.02/G3 Guizhou GZ04-27 100 Neg Neg - Guizhou GZ04-29 100 Pos[9] Pos 28.73 28.15/G3 Guizhou GZ04-32 100 Neg Neg - Guizhou GZ04-36 100 Pos[9] Pos 29.29 29.13/G3 Guizhou GZ04-42 100 Neg Neg - 2008 Shandong SDJN08-10 100 Pos[32-33] Pos 27.51 28.61/G1 Shandong SDJN08-30 100 Neg Neg 27.40 28.60/G1 Shandong SDDY08-15 100 Neg Neg - Shandong SDDY08-17 100 Neg Pos 26.36 26.11/G1 Shandong SDDY08-19 100 Neg Pos 27.34 29.57/G1 Shandong SDDY08-20 100 Neg Pos 28.92 28.44/G1 Shandong SDDY08-22 100 Neg Neg - Shandong SDDY08-26 100 Neg Neg 25.19 26.79/G1 2009 Tibet XZ0922 100 Neg Neg - Tibet XZ0926 100 Neg Neg 31.96 31.90/G1 Tibet XZ0930 100 Neg Neg - Tibet XZ0934 100 Pos[15] Neg 31.57 29.63/G5 Tibet XZ0938 100 Pos[34] Pos 30.31 29.96/G1 Tibet XZ0942 100 Neg Neg - Note. Pos: positive; Neg: negative. Table 3. Comparison of JEV Detection in the Mosquito Pools Examined Using Semi-nested PCR and TaqMan qPCR
Specificity of the TaqMan Real-time RT-PCR Assay
Sensitivity and Standard Curve for TaqMan qPCR
Comparison of Conventional RT-PCR and TaqMan real-time RT-PCR
-
Japanese encephalitis virus, a mosquito-borne zoonotic pathogen, is the leading cause of viral encephalitis in Asia. There are five JEV genotypes: JEV-G1 to JEV-G5. JEV-G1 and JEV-G3 are the predominant genotypes in endemic area. JEV-G5 has recently been detected from mosquitoes in China and South Korea[15, 28-29]. Hence, the establishment of a rapid detection method for distinguishing JEV genotypes is necessary.
To detect JEV rapidly and accurately, conventional RT-PCR, TaqMan real-time RT-PCR, SYBR Green Ⅰ-based real-time PCR and loop-mediated isothermal amplification (LAMP) methods have been developed. However, these methods were designed mainly for detecting or differentiating JEV-G1 and JEV-G3.
In recent years, several new methods were developed for pathogens detecting and sequencing. Next-generation sequencing and third-generation sequencing have been widely used to sequence various pathogens as their long reads, sensitivity and high-throughput[30-31]. Although these methods offer new options for unknown pathogens detection and full-length genome sequencing, they are not applicable to rapid virus screening and genotyping.
In this study, we established TaqMan real-time RT-PCR assays that not only detect JEV but also distinguish the five JEV genotypes. First, a non-specific detection system was established to detect JEV in specimens and then positive samples obtained in the first step were genotyped with the specific detection system.
Seven flaviviruses and five other arboviruses were used to test the specificity of our method. No cross-reactivity was detected between JEV and the other viruses. Besides JEV-G1, JEV-G3, and JEV-G5 strains were also used to verify the sensitivity of detection system, which showed that our detection system had higher sensitivity and similar detection limits for viral isolates. Moreover, all the in vitro-transcribed standard RNAs (JEV-G1 to JEV-G5) had similar limits of detection. However, one limitation of this study was no JEV-G2 and JEV-G4 strains can be obtained to demonstrate the sensitivity of the detection system. Furthermore, we validated this detection system with twenty pools of mosquitoes collected in Guizhou (2004), Shandong (2008), and Tibet (2009). The XZ0934 strain (JEV-G5) was confirmed with this detection system, which was not detected with semi-nested PCR (Table 3). The TaqMan real-time RT-PCR method showed a higher detection rate compared with the semi-nested PCR method.
In summary, TaqMan real-time RT-PCR assay developed in this study showed high specificity, sensitivity and reproducibility and had great potential for application.
-
The TaqMan real-time RT-PCR assay described here for the detection and differentiation of JEV (G1 to G5) exhibited high specificity, sensitivity, and reproducibility and has great potential for laboratory diagnosis, public health surveillance and epidemiological studies in regions where JEV is endemic.
-
No conflict of interest to declare.
Received: January 18, 2018;
Accepted: March 5, 2018







 Quick Links
Quick Links
 DownLoad:
DownLoad: