-
Nosocomial infections caused by antibiotic-resistant bacteria have become increasingly prevalent and are associated with high fatality rates, contributing to a growing disease burden. Antimicrobial resistance stands as one of the top five threats to humanity, with infectious diseases projected to surpass cancer as the leading cause of death by 2050[1]. β-Lactams have traditionally served as the primary treatment for severe infections, with carbapenems emerging as the most effective among them. Their usage is recommended for combating infections caused by extended-spectrum β-lactamase-producing Enterobacteriaceae, particularly Klebsiella pneumonia and Escherichia coli[2,3]. Carbapenemase, the predominant enzyme responsible for Enterobacterales resistance to carbapenems, underscores this challenge. Given that carbapenemase genes are often harbored on mobile genetic elements, facilitating their transfer between bacteria of the same or different species, carbapenemase production has become the primary mechanism driving the emergence of carbapenem-resistant Enterobacteriaceae (CRE). CRE strains typically exhibit resistance to multiple antimicrobials, contributing to their rapid dissemination, heightened lethality, and formidable treatment resistance, thereby posing an enduring global threat.
Salmonella tops the list of bacteria responsible for foodborne illnesses[4]. However, instances of carbapenem-resistant Salmonella strains are rare, with only a few cases documented of blaNDM-1-positive S. enterica isolates in patients[5-7]. Thus far, there have been no reports of healthy carriers of blaNDM-1-carrying Salmonella. Asymptomatic Salmonella carriers shed large quantities of bacteria in their feces, facilitating transmission of the pathogen and its antibiotic resistance genes (ARGs) through contamination of water or food sources[8]. Given the rapid global dissemination of ARGs, impacting humans, wildlife, companion animals, agriculture, and the environment, antibiotic resistance represents a significant "One Health" challenge. Healthy individuals are pivotal in the spread of ARGs within communities, potentially harboring superbugs unknowingly for extended periods[9].
In this study, we present the identification of a carbapenem-resistant S. typhimurium strain isolated from a rectal swab of a healthy food worker. Through a combination of short- and long-read sequencing technologies, we obtained the complete genome, revealing that the strain carries a hybrid plasmid containing blaNDM-1, conferring high resistance to carbapenems. Our investigation delved into the risk of drug resistance transmission, leveraging analyses of genomic and plasmid characteristics.
-
Between January and December 2020, a total of 50,038 healthy food workers, comprising food handlers employed by food and beverage companies, underwent Salmonella screening in Yulin, Guangxi, China. Fecal samples were obtained and subjected to culture to isolate Salmonella spp. following the protocol recommended by the World Health Organization for Salmonella isolation from stool samples.
-
Genomic DNA extraction was performed for each strain through boiling and freeze-thawing methods, with subsequent recovery of the resulting supernatant for PCR templates. Detection of the blaNDM-1 gene was conducted using real-time PCR assays[10].
-
A blaNDM-1-positive strain cultivated in SSI medium (Denmark) underwent serotyping through slide agglutination with commercial antiserum, adhering to the White–Kauffmann–Le Minor scheme for serotype classification[11].
-
The antimicrobial susceptibility testing (AST) of the blaNDM-1-positive strain was evaluated using the broth microdilution method recommended by the Clinical and Laboratory Standards Institute (CLSI). Minimal inhibitory concentrations (MICs) of 29 different antibiotics were determined utilizing the BD Phoenix50 (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) assay. The results were assessed and interpreted based on the criteria outlined in CLSI M100-Ed31 2021. E. coli ATCC 25922 served as the quality control strain.
-
Conjugation experiments were conducted to assess the host range and transferability of the blaNDM-1-positive plasmid, employing E. coli J53 AziR (met pro; azide resistant) as the recipient strain. The efficacy of carbapenem resistance determinant transfer via conjugation was examined on LB agar plates (Oxoid) at initial donor/recipient ratios of 1:1, 1:2, and 1:4. Following incubation at 30 °C and 37 °C for durations of 4 hours and 20 hours, transconjugants were selected on LB agar supplemented with meropenem (4 mg/L) and sodium azide (100 mg/L). Transfer frequency was determined as the number of transconjugants per recipient. Positive transconjugation events were confirmed through PCR targeting blaNDM-1.
-
Genomic DNA extraction was performed utilizing a Wizard Genomic DNA Purification kit (Promega) as per the manufacturer’s instructions, followed by quantification using a Qubit 4 Fluorometer. Subsequently, short-read sequencing was conducted utilizing an MGISEQ-G99 (MGI Tech, China), while nanopore long-read sequencing was carried out using a QNome-3841 platform (Qitan Tech, China), strictly adhering to the manufacturer’s protocols. The complete genome sequence was obtained employing a hybrid de novo assembly strategy that integrated both short and long reads in Unicycler v0.4.4, utilizing default parameters. The long-read assembly tool Flye v2.8.2 was employed to validate the accuracy of complex multidrug-resistance gene regions within the genome[12]. To identify antimicrobial resistance genes, plasmid replicon types, insertion sequences, and genes encoding the type IV secretion system (T4SS), we utilized online tools such as ResFinder v. 4.0, PlasmidFinder v. 2.0.1, ISfinder, and OriFinder[13-16]. The plasmid database was sourced from the National Center for Biotechnology Information (NCBI) database, and local comparisons were conducted using BLAST.
Multilocus Sequence Typing (MLST) of the blaNDM-1-positive strain was performed through EnteroBase online analysis. Additionally, published S. Typhimurium genomes of the same sequence type (ST) from China were retrieved from EnteroBase[17] (accessed on October 17, 2022). The complete chromosomal sequence of the blaNDM-1-positive strain served as a reference for analyzing single nucleotide polymorphisms (SNPs) utilizing MUMmer v3.2.3 (https://mummer.sourceforge.net/), and the polymorphic sites of the genomes were amalgamated based on the reference genome coordinates utilizing a previously published iSNV-calling pipeline (https://github.com/generality/iSNV-calling). Recombination sites were identified using Gubbins v2.4.1[18]. The concatenated sequences of the filtered polymorphic sites conserved in all genomes (core genome SNPs and cgSNPs) were isolated for phylogenetic analysis, employing the maximum likelihood method in IQ-TREE v2.1.2[19].
-
During surveillance of Salmonella infections in the Yulin population in 2020, 979 Salmonella strains were obtained. Among them, one strain (Sa17155) tested positive for the blaNDM-1 gene through PCR analysis. This strain was isolated from a rectal swab of a healthy food worker (a chef) who exhibited no symptoms of Salmonella infection, such as fever, diarrhea, or abdominal pain. Upon further investigation, the isolate was identified as S. Typhimurium using a serum agglutination assay. Antimicrobial Susceptibility Testing (AST) revealed that Sa17155 displayed resistance to carbapenem antibiotics, including meropenem (MIC = 4 mg/L), imipenem (MIC = 8 mg/L), and ertapenem (MIC > 2 mg/L), as well as to several other antimicrobials, such as third-generation cephalosporins, quinolones, and tetracycline. Notably, it was classified as a Multi-Drug Resistant (MDR) bacterium, although it remained susceptible to colistin, tigecycline, aztreonam, and nitrofurantoin (Table 1).
Antimicrobial Sa17155 E. coli J53 transconjugants-E. coli J53 AMK ≤ 8 ≤ 8 ≤ 8 AMC > 32/16 ≤ 8/4 > 32/16 SAM > 16/8 ≤ 4/2 > 16/8 ATM ≤ 2 ≤ 2 ≤ 2 CFZ > 16 ≤ 2 > 16 FEP > 16 ≤ 1 > 16 SCP > 32/8 ≤ 0.5/8 > 32/8 FOX > 16 ≤ 4 > 16 CAZ > 32 ≤ 1 > 32 CRO > 32 ≤ 1 > 32 CXM > 16 ≤ 4 > 16 CHL > 16 ≤ 4 ≤ 4 CIP 1 ≤ 0.5 ≤ 0.5 COL ≤ 1 ≤ 1 ≤ 1 EIP > 2 ≤ 0.25 > 2 FOS ≤ 16 ≤ 16 ≤ 16 GEN > 8 ≤ 2 ≤ 2 IPM 8 ≤ 0.25 8 LVX ≤ 1 ≤ 1 ≤ 1 MEM 4 ≤ 0.125 4 MIN > 16 ≤ 1 ≤ 1 MXF 1 ≤ 0.5 ≤ 0.5 NIT ≤ 16 ≤ 16 ≤ 16 NOR ≤ 2 ≤ 2 ≤ 2 TZP > 64/4 ≤ 4/4 > 64/4 TET > 8 ≤ 2 ≤ 2 TGC ≤ 1 ≤ 1 ≤ 1 TOB > 8 ≤ 2 ≤ 2 SXT > 4/76 ≤ 1/19 ≤ 1/19 Note. MK, amikacin; AMC, amoxicillin-clavulanate; SAM, ampicillin-sulbactam; ATM, aztreonam; CFZ, cefazolin; FEP, cefepime; SCP, cefoperazone-sulbactam; FOX, cefoxitin; CAZ, ceftazidime; CRO, ceftriaxone; CXM, cefuroxime; CHL, chloramphenicol; CIP, ciprofloxacin; COL, colistin; EIP, ertapenem; FOS, fosfomycin w/g6p; GEN, gentamicin; IPM, imipenem; LVX, levofloxacin; MEM, meropenem; MIN, minocycline; MXF, moxifloxacin; NIT, nitrofurantoin; NOR, norfloxacin; TZP, piperacillin-tazobactam; TGC, tetracycline; TOB, tigecycline; TOB, tobramycin; SXT, trimethoprim-sulfamethoxazole. Table 1. Minimum inhibitory concentrations (mg/L) determined via antimicrobial susceptibility testing
-
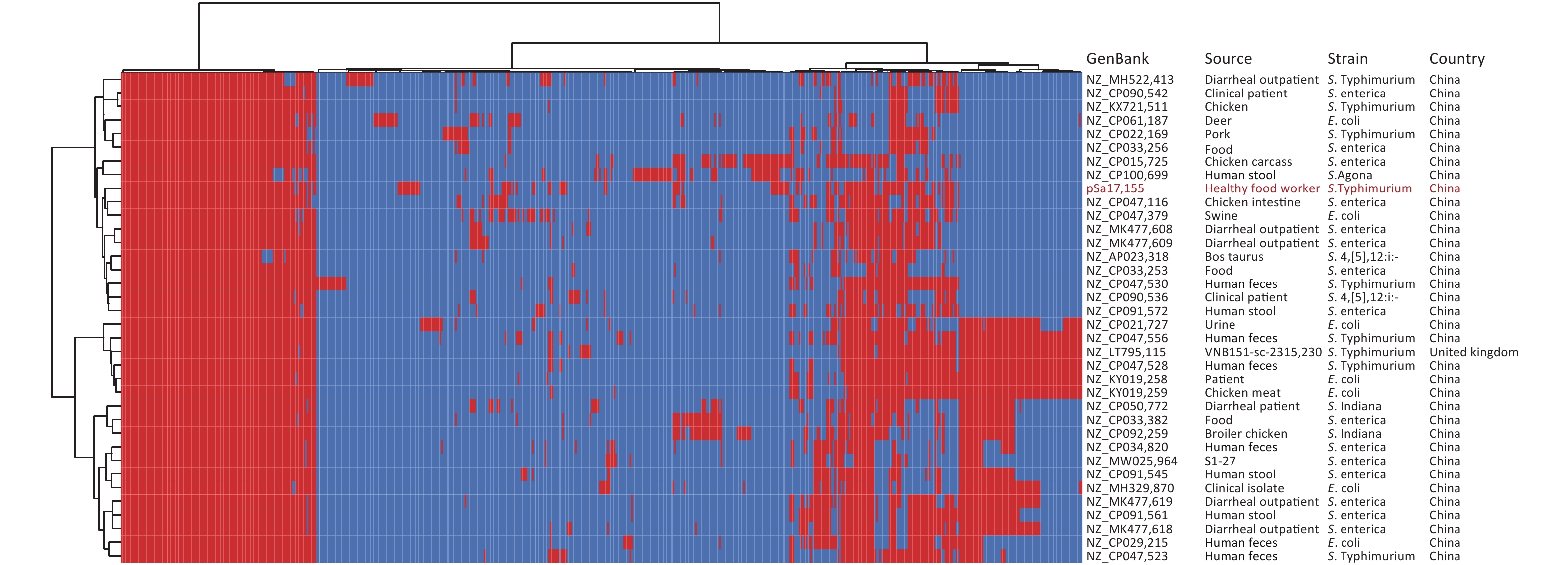
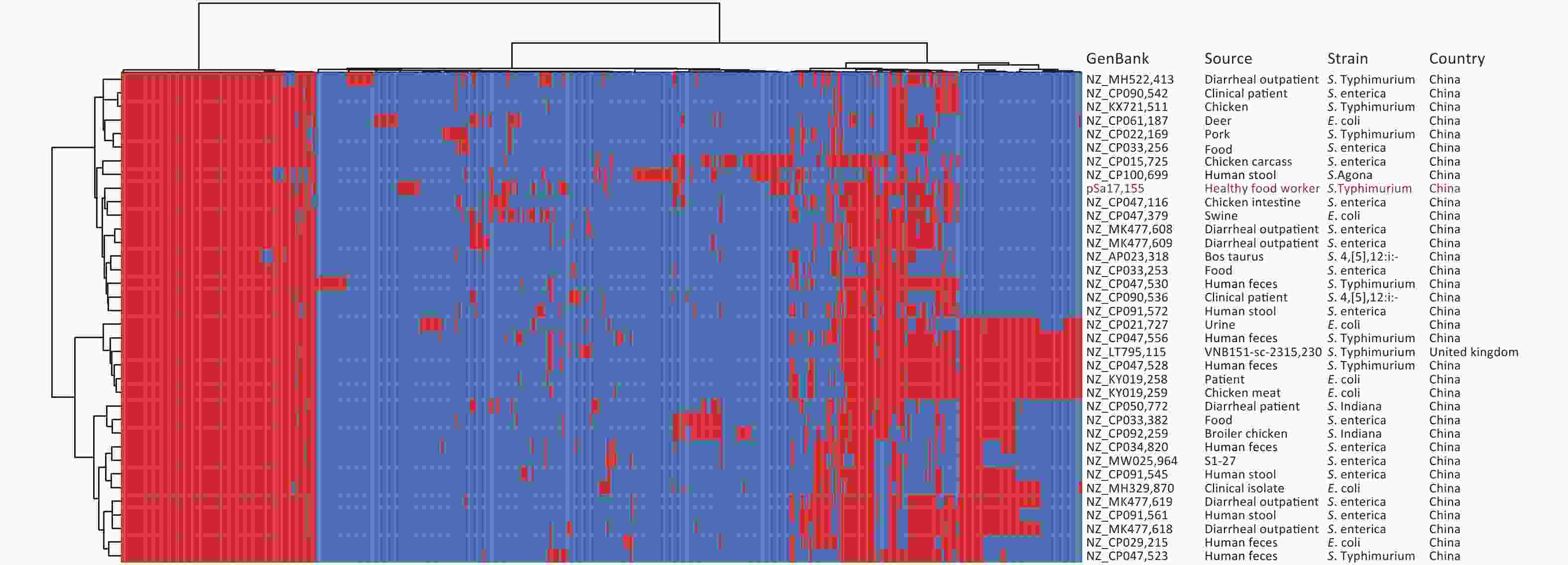
Whole-genome sequencing and de novo genome assembly of strain Sa17155 were conducted using both short- and long-read sequencing techniques. The genome consisted of a circular chromosome (5,041,065 bp, 52.11% GC content) and a circular hybrid plasmid, pSa17155 (198,957 bp, 48.10% GC content). Within the chromosome, multiple resistance genes were identified, including aph(6)-Id, aac(6′)-Iaa, aph(3′′)-Ib, sul2, blaTEM-1B, and tet(B). These findings were consistent with its MDR phenotype, and Sa17155 was designated as ST34. We retrieved 261 Chinese S. Typhimurium ST34 genomes from the EnteroBase database and an additional 100 genomes from a published study[20]. These ST34 S. Typhimurium isolates were sourced from various locations, including humans (197 isolates), livestock farms (43 isolates), poultry (18 isolates), food (13 isolates), wild animals (two isolates), and environmental samples (one isolate). The phylogenetic relationships among these strains, originating from different sources, were analyzed based on 158 recombination-free Single Nucleotide Polymorphisms (SNPs) (Figure 1A). Notably, 23 strains closely resembled Sa17155, forming a subclade with core genome SNP differences ranging from 25 to 68 (Figure 1B). These 23 S. Typhimurium strains were isolated from humans (10 strains), food (11 strains), and animals (two strains) across various provinces in China. Upon comparison of resistance gene repertoires and plasmid types, it was observed that two isolates carried blaNDM-5, while blaNDM-1 was exclusively detected in Sa17155 (Figure 1A).
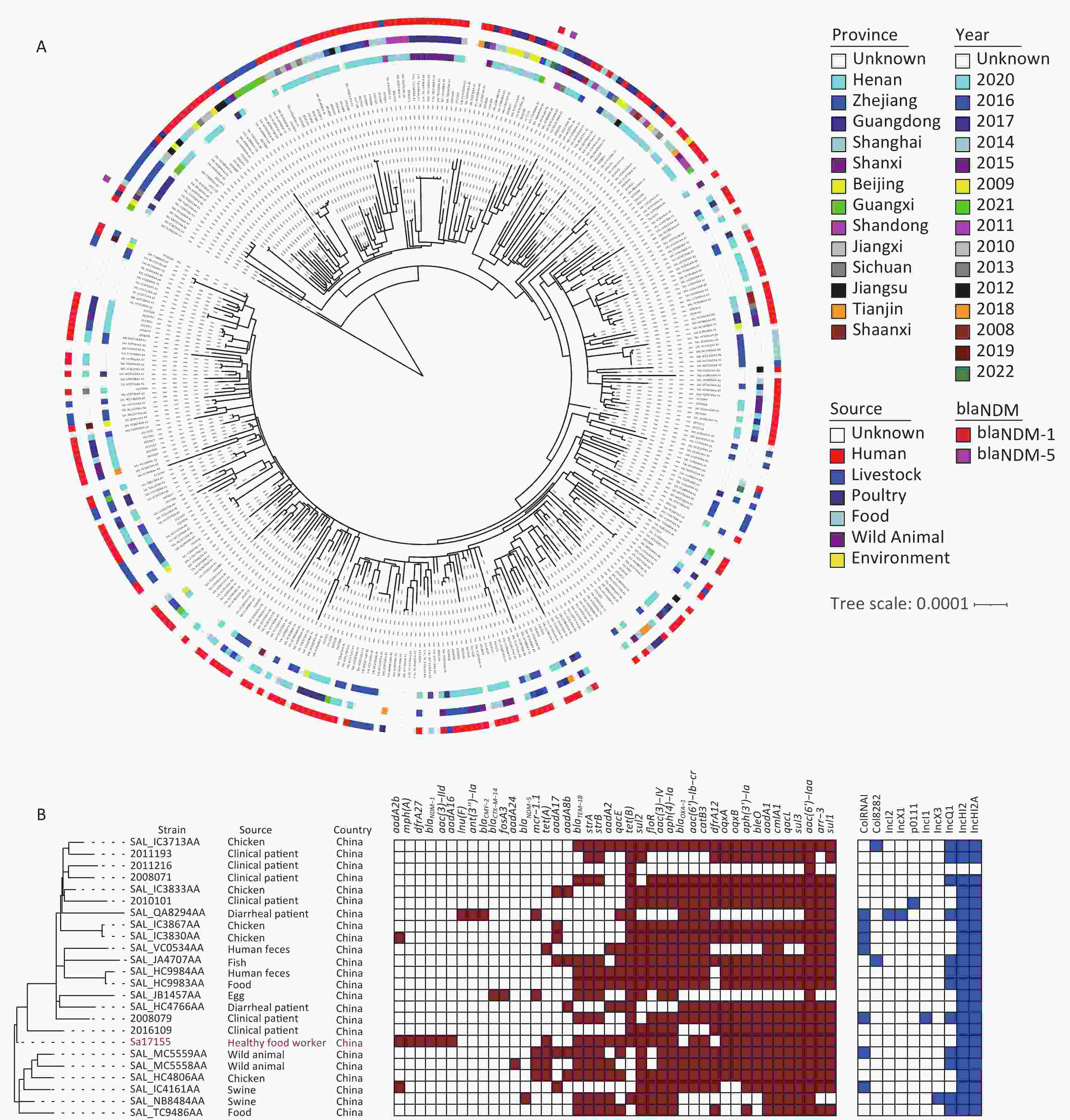
Figure 1. Phylogenetic tree and distribution of published drug-resistance genes and plasmid types of S. Typhimurium type ST34 in strain Sa17155 and China. (A) Phylogenetic analysis of ST34 isolates from China; the rings from inside to outside along the tree represent the geographical origins of the isolates, year of isolation, source of the isolates, and presence of blaNDM-1 and blaNDM-5 genes. (B) Phylogenetic tree depicting the branch in which strain Sa17155 is located, and phylogenetic tree representing distribution of drug-resistance genes and plasmid types of Salmonella ST34 isolated from China in the public database. Phylogenetic tree is shown on the left; heat map of drug-resistance gene carriage is shown in the middle (red indicates carriage of the gene); and distribution of plasmid types is shown on the right (blue indicates presence of plasmid type).
-
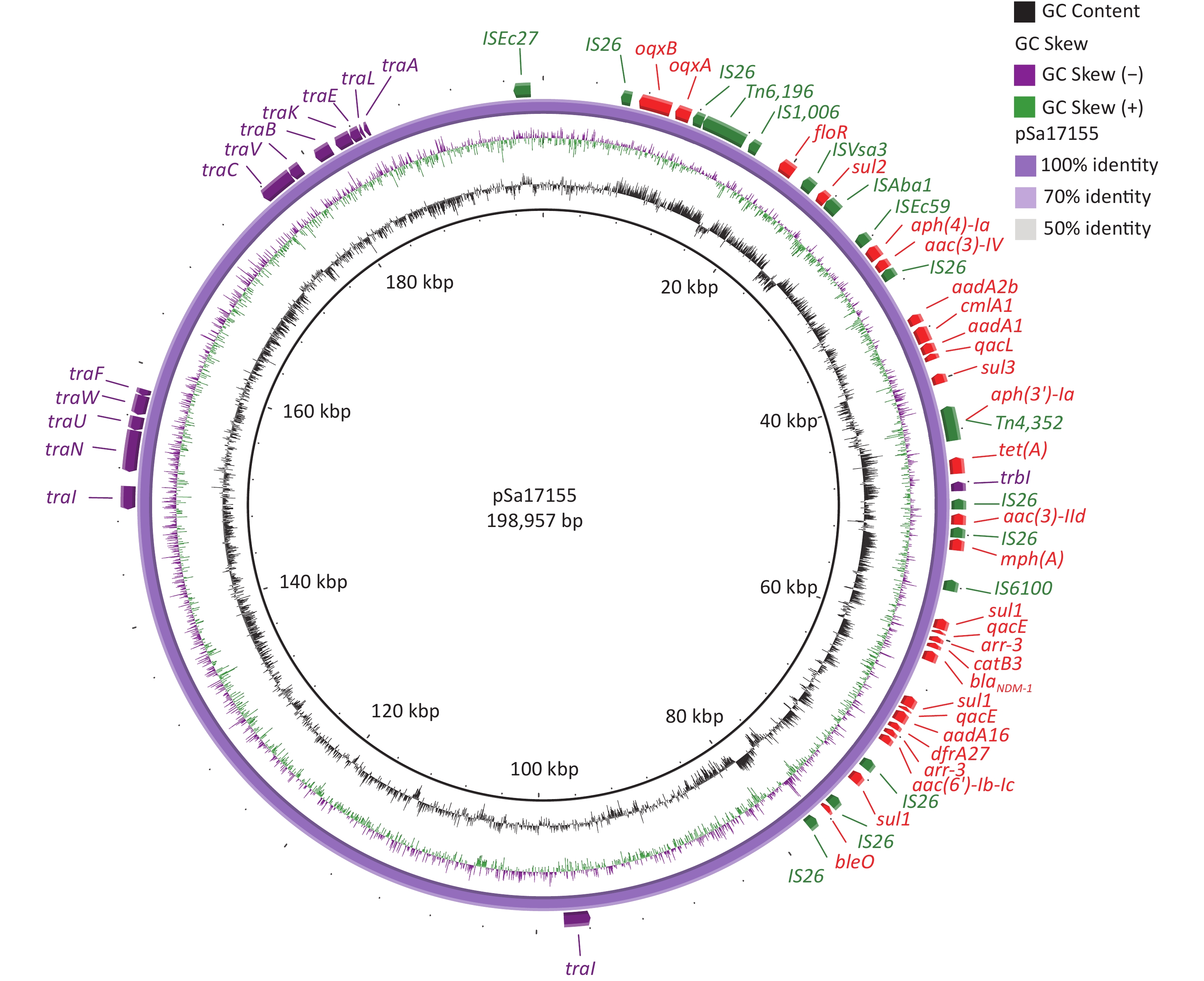
The complete sequence of the blaNDM-1-positive plasmid, designated pSa17155, revealed 370 open reading frames. Analysis of its two replicons, IncHI2 and IncHI2A, indicated that pSa17155 is a hybrid plasmid. This hybrid plasmid harbors a diverse array of genes conferring resistance to various antibiotics, including beta-lactams (blaNDM-1), aminoglycosides (aac(3)-IId, aadA16, aac(6')-Ib-cr, aph(4)-Ia, aac(3)-IV, aph(3')-Ia, bleO, aadA2b, and aadA1), rifamycin (arr-3), quinolones (oqxA and oqxB), tetracycline (tet(A)), folate pathway antagonists (sul3, sul2, sul1, and dfrA27), macrolides (mph(A)), quaternary ammonium compounds (qacE and qacL), and amphenicol (catB3, cmlA1, and floR). The plasmid’s Type IV secretion system (T4SS) includes trbI, traI, traN, traU, traW, traF, traC, traV, traB, traK, traE, traL, and traA, all belonging to type F (Figure 2).
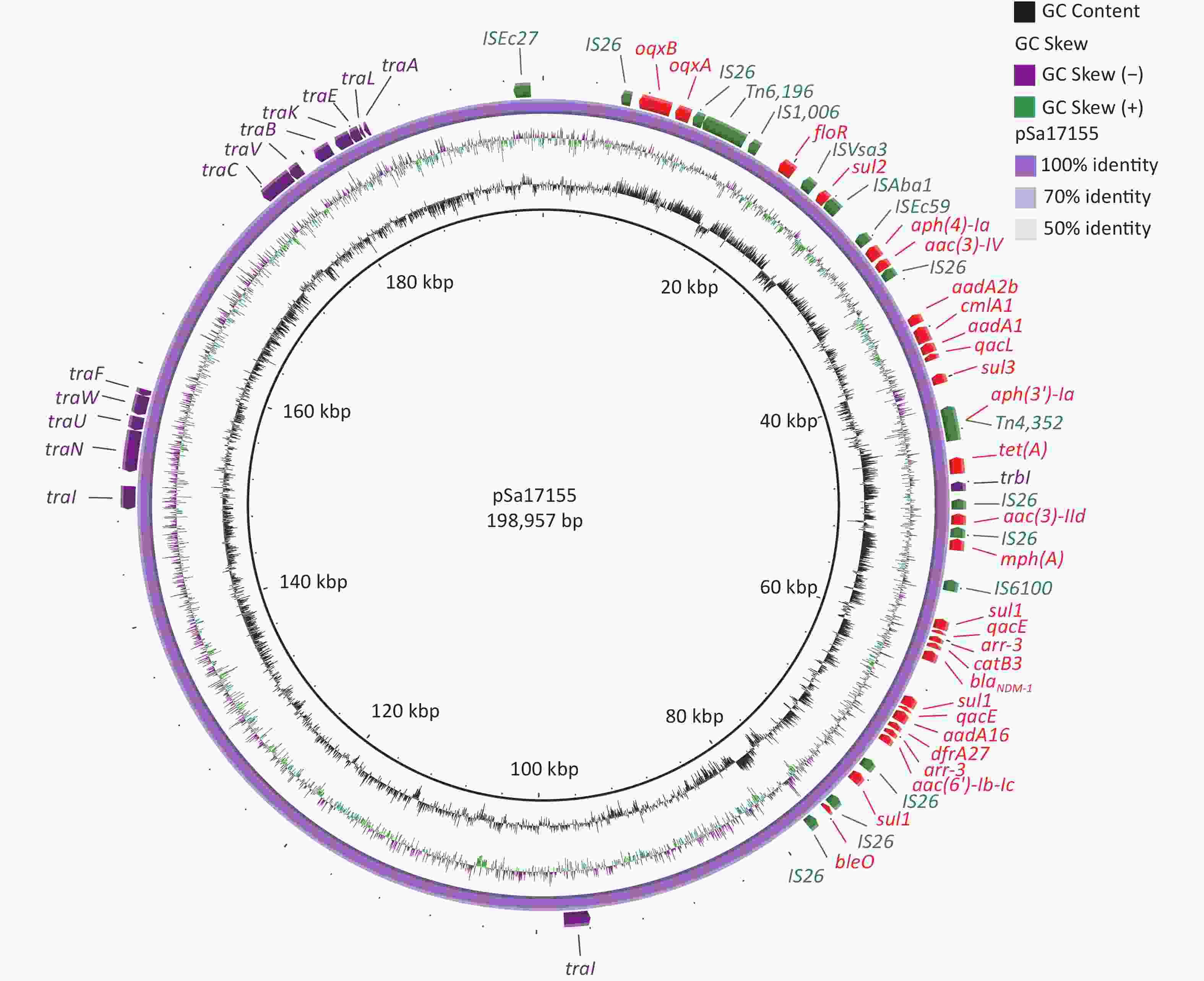
Figure 2. Structure of pSa17155. From inside to outside: ring 1 indicates G+C Content; ring 2 indicates G+C Skew; ring 3 represents pSa17155 ring structure; ring 4 red: drug-resistance gene; green: insertion sequence; purple: T4SS; maroon: plasmid replication.
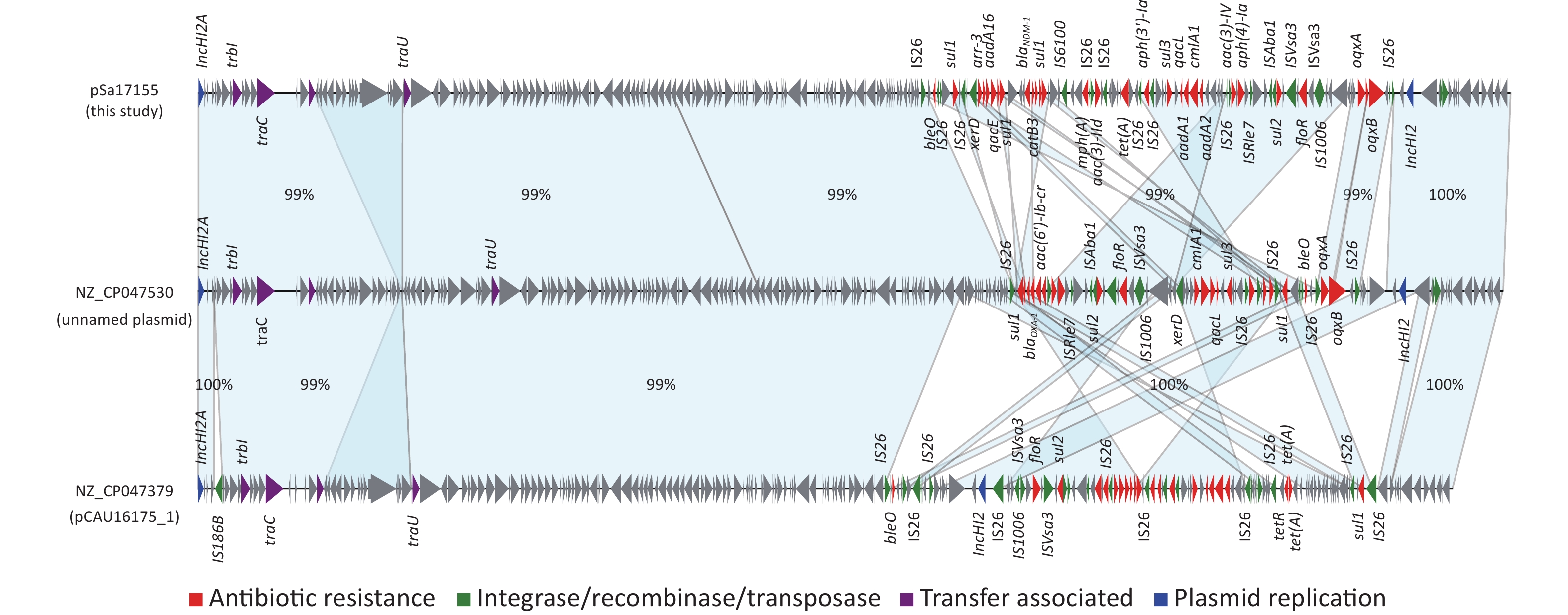
Comparison of the IncHI2/IncHI2A plasmid pSa17155 with the NCBI database via BLASTn revealed significant similarity (> 90% concordance and > 70% coverage) with 36 plasmids sharing a similar backbone but derived from various host bacteria, predominantly Enterobacteriaceae. The majority of these plasmids were isolated from Salmonella (29 strains) and E. coli (seven strains) (Figure 3). Most strains originated from China, with one isolated in the UK. Notably, the blaNDM-1-harboring plasmid pSa17155 exhibited high homology and identity with plasmid NZ_CP047530 from S. Typhimurium strain SJTUF10640 (92% coverage with 98% identity) isolated from human feces in Shanghai, China, and NZ_CP047379 from E. coli in swine in Hunan, China (90% coverage with 98% identity). Although the replicon types of these three plasmids were identical, variable sequences were observed at the positions of the resistance genes, suggesting potential homologous recombination events (Figure 4). Additionally, IS26 was frequently found flanking the antibiotic resistance genes (ARGs), and the genetic context of blaNDM-1 was IS26-mph(A)-IS6100-sul1-qacE-arr-3-catB3-blaNDM-1-sul1-qacE-aadA16-dfrA27-arr-3-aac(6')-Ib-cr-xerD-IS26. The differences between plasmid pSa17155 and the other two plasmids (NZ_CP047530 and NZ_CP047379) mainly involved the insertion of ARGs mediated by IS26, particularly blaNDM-1 carried by pSa17155 (Figures 2 and 4).
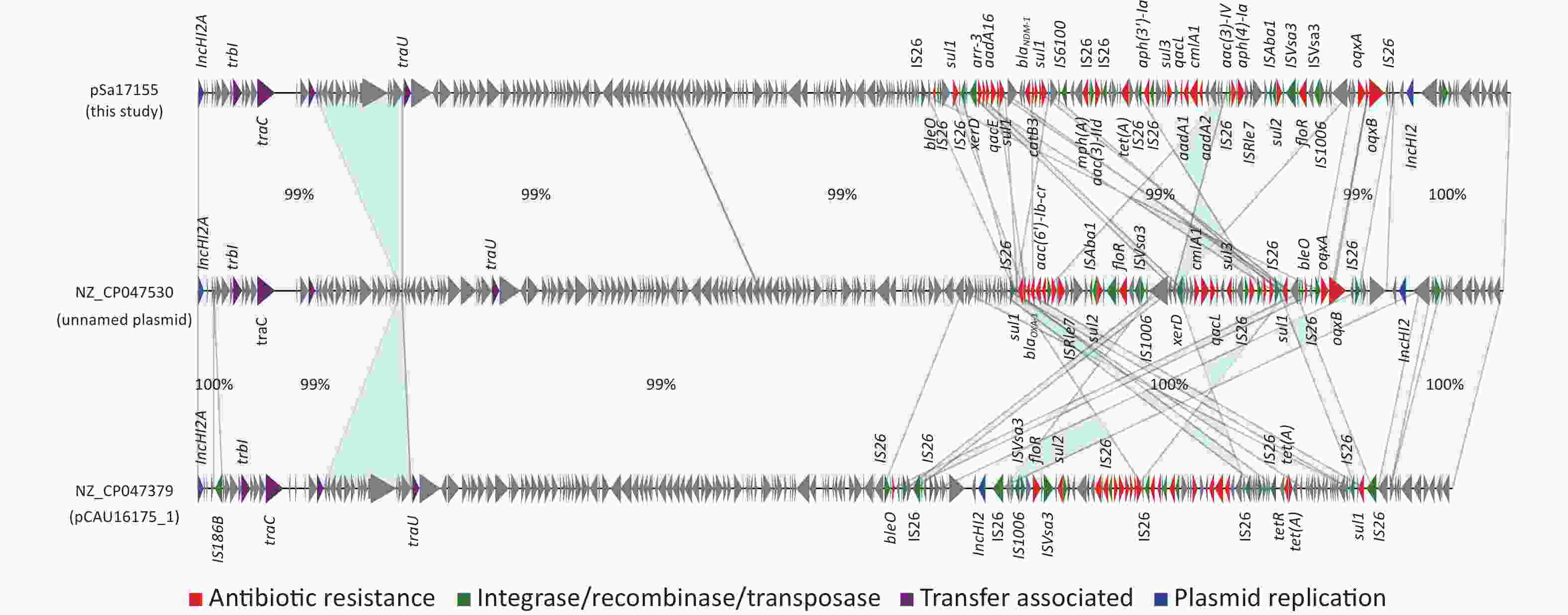
Figure 4. Comparison of plasmids pSa17155, NZ_CP047530, and NZ_CP047379. Detailed sequence comparison of the three plasmids, with the light blue shaded area indicating > 99% homology. Triangles and arrows on the outer circle represent genes of different functional categories (red: antimicrobial resistance; green: integrase, recombinase, transposase; purple: transfer associated; blue: plasmid replication; grey: other functions).
-
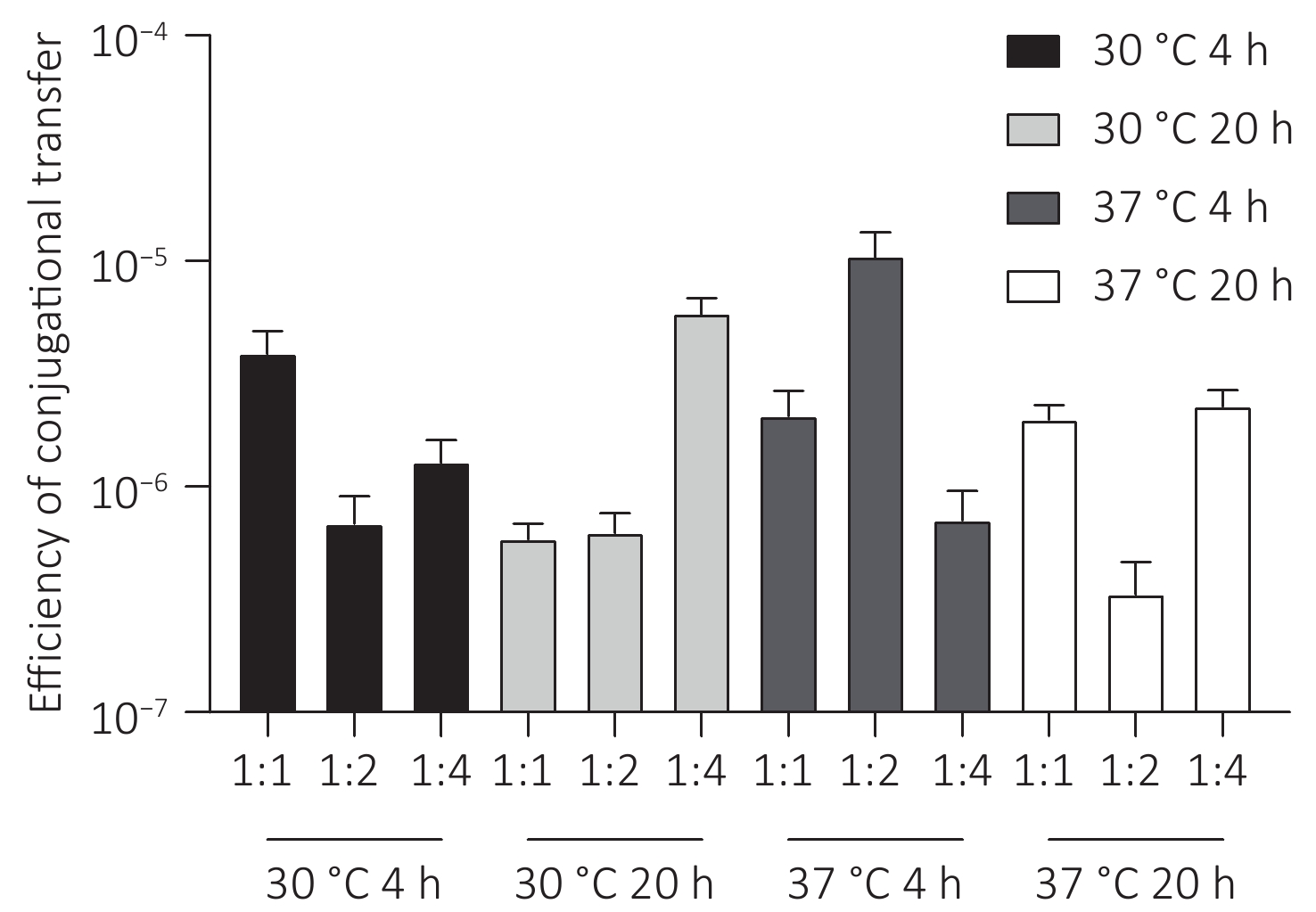
The plasmid pSa17155, carrying the blaNDM-1 gene, was effectively transferred to recipient E. coli J53 with notable conjugation frequencies ranging from 5.0 × 10−6 to 1.5 × 10−5 per recipient strain under various experimental conditions (Figure 5). Particularly, the conjugation frequency was significantly higher when incubated at 37 °C for 4 hours with a donor/recipient bacteria ratio of 1:2 compared to other experimental setups. Furthermore, the conjugants displayed significantly heightened resistance to carbapenems in comparison to the E. coli strain J53. Notably, the meropenem MIC of the conjugants increased by 32-fold (from 0.125 to 4 mg/L). Additionally, transconjugants exhibited acquired resistance to cephalosporins, quinolones, and penicillin (Table 1).
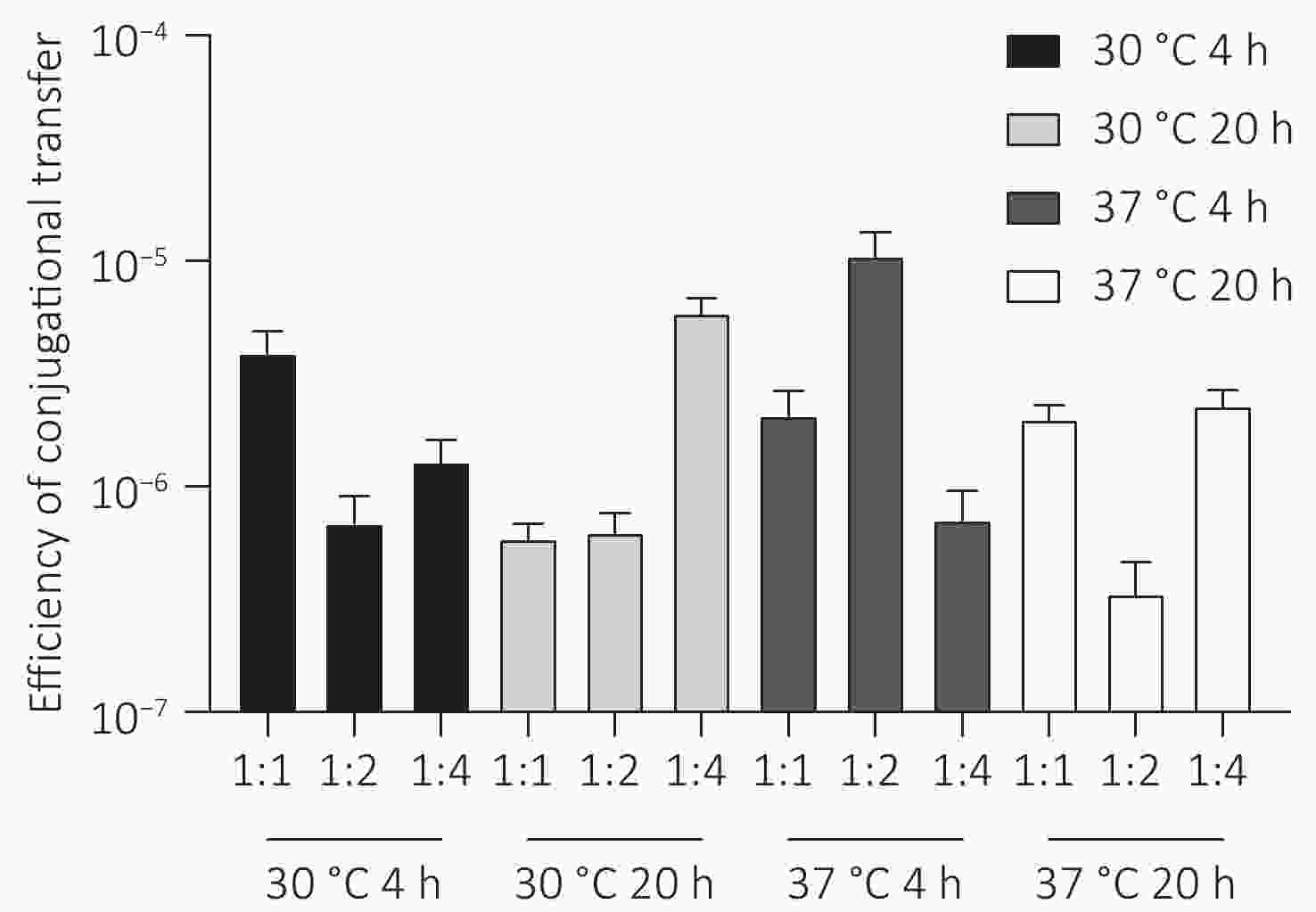
Figure 5. Conjugation frequency (conjugant colony forming units [CFUs]/recipient CFUs) under different experimental conditions. Conjugation experiments were performed at two temperatures (30 °C and 37 °C) for two contact times (4 hours and 20 hours) with donor and recipient broth cultures mixed at ratios of 1:1, 1:2, and 1:4.
-
The “One Health” concept is an evolving philosophy in public health that emphasizes the interconnected health of humans, animals, and the environment, recognizing healthy individuals as crucial components of this interconnected system.
In this study, we detected carbapenem-resistant Salmonella carrying blaNDM-1, a hybrid plasmid, and MDR genes in the intestine of a seemingly healthy individual. The strain was isolated from a chef, raising concerns about asymptomatic carriers potentially serving as reservoirs for infection, silently disseminating Salmonella and ARGs.
Carriage of pathogenic bacteria by food handlers significantly increases the risk of bacterial transmission. For instance, Typhoid Mary was asymptomatic. In recent years, Salmonella-induced food poisoning has been the most prevalent form of bacterial food poisoning in China[21]. Among various bacterial species, S. Typhimurium has been evolving to develop MDR against a broad spectrum of antibiotics, including chloramphenicol, ampicillin, and co-trimoxazole. Currently, quinolones and third-generation cephalosporins are considered effective treatment options for MDR Salmonella in many countries and regions[22]. However, the Sa17155 strain analyzed in this study exhibited resistance to both quinolones and third-generation cephalosporins. Moreover, it carried the blaNDM-1 gene, which confers resistance to carbapenems, posing further challenges to the clinical treatment of Salmonella infections. Phylogenetic analysis suggests that the ST34 strain may have been widely disseminated in China, being transmitted among wild animals, food animals, and humans. There is evidence to suggest that this clone can colonize the intestinal tract of healthy individuals.
Plasmid-mediated horizontal transfer of ARGs is one of the main routes of transmission of resistance genes within human intestines. In this study, pSa17155 was found to be a hybrid IncHI2/IncHI2A plasmid, which is a replicon type found in a variety of bacteria that readily carries ARGs, including β-lactam-resistance genes, especially blaNDM-1[23-25]. BLAST analysis revealed that these three plasmids shared the same replicon type, despite originating from different sources and host bacteria. This particular replicon possesses a distinct advantage in facilitating the spread of drug-resistance genes, particularly among Enterobacteriaceae[3,23,26,27]. Phylogenetic analysis illustrated that the blaNDM-1 gene can undergo horizontal transmission via diverse plasmid types, some of which exhibit broad host range capabilities, thereby significantly accelerating the dissemination of drug-resistance genes both within and across bacterial strains[28]. Conjugation assays further confirmed the horizontal transferability of blaNDM-1 into the recipient E. coli J53, substantiating the broad-host-range nature of the blaNDM-1-harboring IncHI2/IncHI2A plasmid and its potential to disseminate blaNDM-1. Additionally, our investigation revealed a proliferation of antibiotic-resistance genes and a plethora of mobile genetic elements concentrated within a multiple-resistance region spanning approximately 60 kb on plasmid pSa17155. Mobile elements within this region are flanked by IS26, and several studies have implicated IS26-mediated processes, including deletions, insertions, and inversions, in the propagation and spread of resistance genes, particularly blaNDM-1. The global dissemination of CRE poses a grave public health threat, with NDM-producing Enterobacterales being detected across diverse sources, particularly among clinical patients. The blaNDM gene is frequently harbored by IncHI2 plasmids originating from various sources, including patients[29], geese[30], eggs[31], and migratory birds[32]. In this study, the blaNDM IncHI2 plasmid was detected in healthy individuals. Therefore, the NDM-harboring IncHI2 plasmid may underlie transmission of NDM genes.
The frequent transmission of drug-resistance plasmids across various regions and hosts underscores the importance of enhancing surveillance of enteric pathogens and drug-resistant genes in healthy individuals[9,33,34]. Moreover, the establishment of a laboratory surveillance network, utilizing complete genome maps of drug-resistant strains, will be essential for improving monitoring of the prevalence and transmission of drug-resistance-related elements. Therefore, it is imperative to bolster surveillance efforts in healthy populations and devise effective preventive measures to mitigate the global dissemination of antibiotic-resistant genes (ARGs), antibiotic-resistant plasmids, and antibiotic-resistant strains.
-
This study has provided insights into a Salmonella strain carrying blaNDM-1 isolated from a healthy food worker and has expanded our knowledge of the ability of healthy individuals to carry carbapenem-resistant bacteria. The risk of spreading a conjugative hybrid plasmid carrying blaNDM-1 has been highlighted by demonstrating its carriage and transmission in healthy individuals. The transferability and complex genetic structure of blaNDM-1 may increase the diversity of its bacterial hosts, and pose a threat to human health, clinical care, and food safety. Therefore, there is a need to enhance the sequencing surveillance of asymptomatic infected individuals and develop adequate preventive strategies to mitigate the global spread of ARGs and plasmids.
-
In China, according to the Food Safety Law of the People’s Republic of China and the Implementation Rules on Regulations on Public Places Sanitation Administration, practitioners who work in food production and operations and public place services must obtain a health certificate each year. To obtain a certificate, a health examination must be conducted. The health examination, which was performed by departments approved by the local governments, included routine internal and external basic examination, liver function ALT test, and fecal or rectal swab culture.
blaNDM-1 Carried by a Transferable Plasmid in a Salmonella Strain Isolated from Healthy Individuals
doi: 10.3967/bes2024.104
- Received Date: 2024-02-05
- Accepted Date: 2024-05-07
-
Key words:
- Carbapenems /
- NDM-1 /
- IncHI2/IncHI2A plasmid /
- Salmonella Typhimurium /
- Healthy individual
Abstract:
The authors declare they have no competing interests.
&These authors contributed equally to this work.
| Citation: | Wei Zeng, Ming Luo, Pengcheng Du, Zhenpeng Li, Yao Peng, Mengyu Wang, Wenxuan Zhao, Huayao Zhang, Yang Li, Pengjie Luo, Yannong Wu, Jialiang Xu, Xu Li, Xin Lu, Biao Kan. blaNDM-1 Carried by a Transferable Plasmid in a Salmonella Strain Isolated from Healthy Individuals[J]. Biomedical and Environmental Sciences, 2024, 37(11): 1252-1261. doi: 10.3967/bes2024.104 |








 Quick Links
Quick Links
 DownLoad:
DownLoad: