-
Brucellosis is a zoonosis that causes economic losses worldwide, as well as human morbidity and poverty. It is caused by Brucella spp., which are animal pathogens[1]. Among the 13 identified species, Brucella melitensis is the most common and is frequently isolated from both humans and livestock[2]. Human brucellosis is endemic in Gansu Province, China, where Brucella melitensis bv. 1 and bv. 3 have been isolated from sheep samples[3]. However, the genetic diversity and genomic epidemiology of circulating strains in humans in this province remain unclear. The high resolution of whole-genome sequencing (WGS) and single-nucleotide polymorphism (SNP) analysis enables the discrimination among closely related Brucella strains[4]. Therefore, this study aimed to identify the genetic diversity and transmission pattern of the Brucella species circulating in Gansu Province and provide a fundamental reference for improving surveillance and control strategies for human brucellosis.
Forty Brucella spp. strains were isolated from blood samples collected from patients with suspected brucellosis in Gansu Province (Supplementary Table S1). Briefly, blood samples (5–10 mL) were collected from the patients, injected into diphasic medium under a biosafety cabinet, and incubated at 37 °C in the absence and presence of 5%–10% CO2 for at least two weeks, and observed every other day. Conventional biotyping tests were conducted, including phage lysis (TB and BK2), agglutination with anti-A and -M monospecific sera, CO2 requirement for growth, H2S production, and growth in the presence of basic fuchsin and thionine. DNA was extracted from heat-inactivated (80 °C, 10 min) pure cultures using the DNA Extraction and Purification Kits (Qiagen, Heidelberg, Germany) according to the manufacturer’s instructions. DNA samples were fragmented by sonication to a size of 350 bp, the DNA fragments were end-polished, A-tailed, and ligated with full-length adaptors for Illumina sequencing, followed by PCR amplification. Library preparation was performed using the Nextera XT library preparation kit (Illumina Inc., San Diego, CA, USA). High-quality clean data were assembled into contigs and further completed using SOAPdenovo[5]. Using Brucella melitensis 16M (GCA_000007125.1) as the reference genome, core-genome SNPs (cgSNPs) were identified using Snippy v 4.0. Maximum-likelihood trees were constructed using IQ-tree 2 based on the Maximum-Likelihood Phylogenies algorithm with 1,000 bootstrap replicates. Additionally, a nationwide genetic relationship analysis was conducted, comparing 72 strains (32 from GenBank) (Supplementary Table S1) using above-mentioned algorithm.
Among the 40 cases, 33 were men, and 7 were women. The age of the patients ranged from 3–71 years (average 48 years). Regarding occupation, 27 were farmers, 1 was a butcher, and 1 was a child; the remaining cases had unknown occupations. Direct, unprotected exposure to infected animals and/or their products, placental retention intervention, and recurrent encounters with brucellosis-infected farms are the most common modes of transmission[6]. A total of 40 Brucella melitensis strains were isolated and identified, including 5 strains of bv. 1 and 35 strains of bv. 3 (Table 1). These 40 strains were distributed across nine cities (Figure 1). Gansu is a historically endemic region for brucellosis. A total of 109 Brucella strains have been isolated from multiple hosts, including humans, animals, and wildlife, of which 66 B. melitensis strains have been isolated from human blood[7]. These data demonstrate that the human brucellosis epidemic in Gansu is primarily driven by B. melitensis strains, highlighting the need for surveillance and control in small ruminants to curb human brucellosis.
Key Growth feature Agglutination with
monospecific seraPhage lysis tests Number of
strainsInterpretation CO2 H2S BF TH A M R Tb BK2 Wb 544 + + + − + − − CL CL CL 1 B. abortus bv. 1 16M − − + + − + − NL CL NL 1 B. melitensis bv. 1 1330 − ++ − + + − − NL CL CL 1 B. suis bv. 1 Tested
strain− − + + − + − NL CL NL 5 B. melitensis bv. 1 − − + + + + − NL CL NL 35 B. melitensis bv. 3 Note. BF, basic fuchsin; TH, threonine; CL, complete lysis; not lysis; +, positive; −, negative. Table 1. Biotying profiles of the 40 Brucella melitensis strains in this study
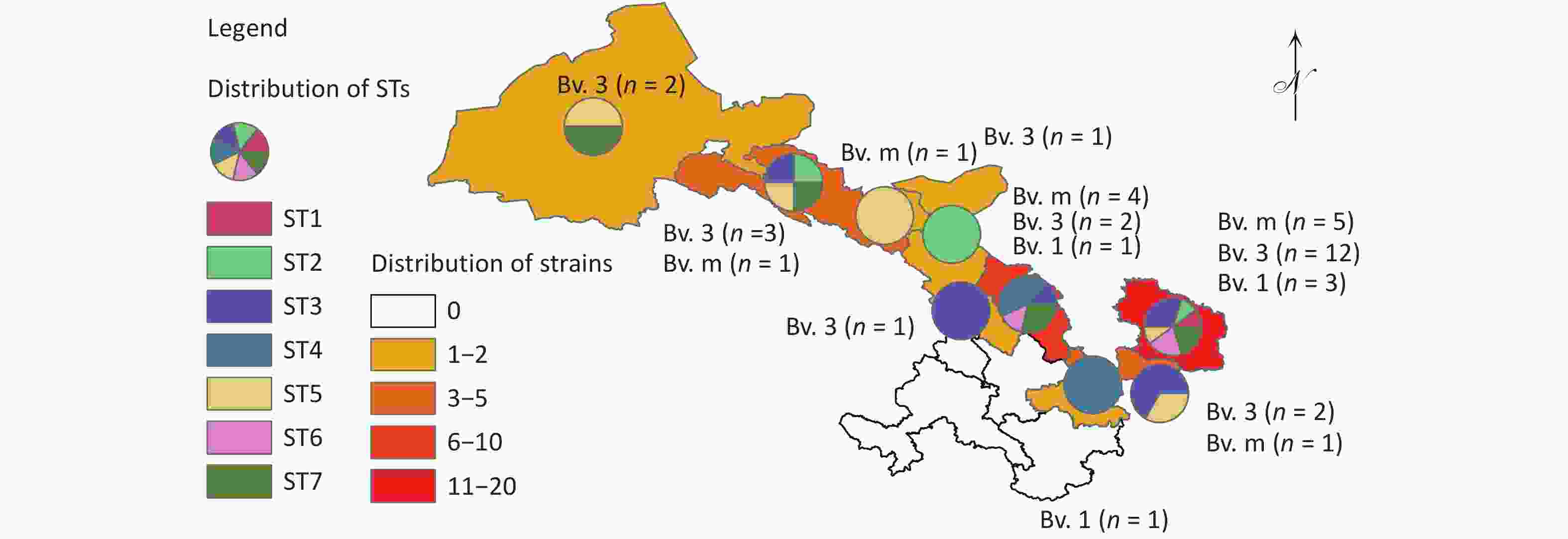
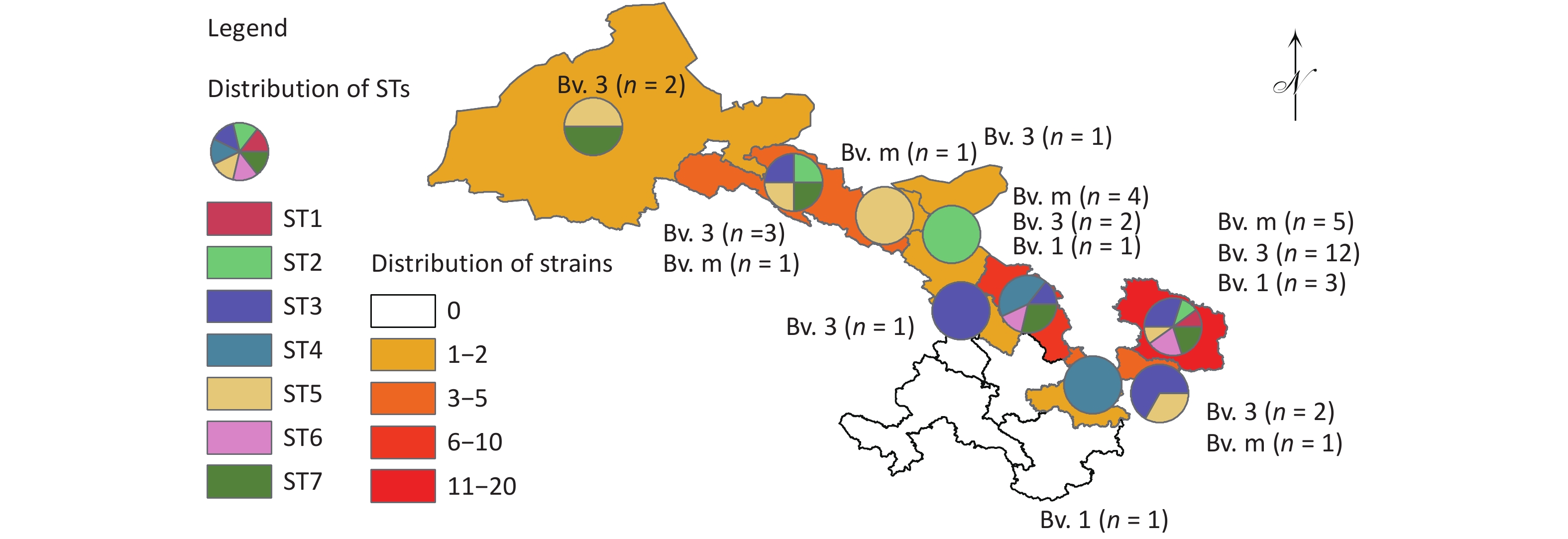
Figure 1. The distribution pattern of the number of strains, species/biovars, and STs of 40 B. melitensis from Gansu Province, China. The red dot indicates the location of the one strain isolated from a goat.
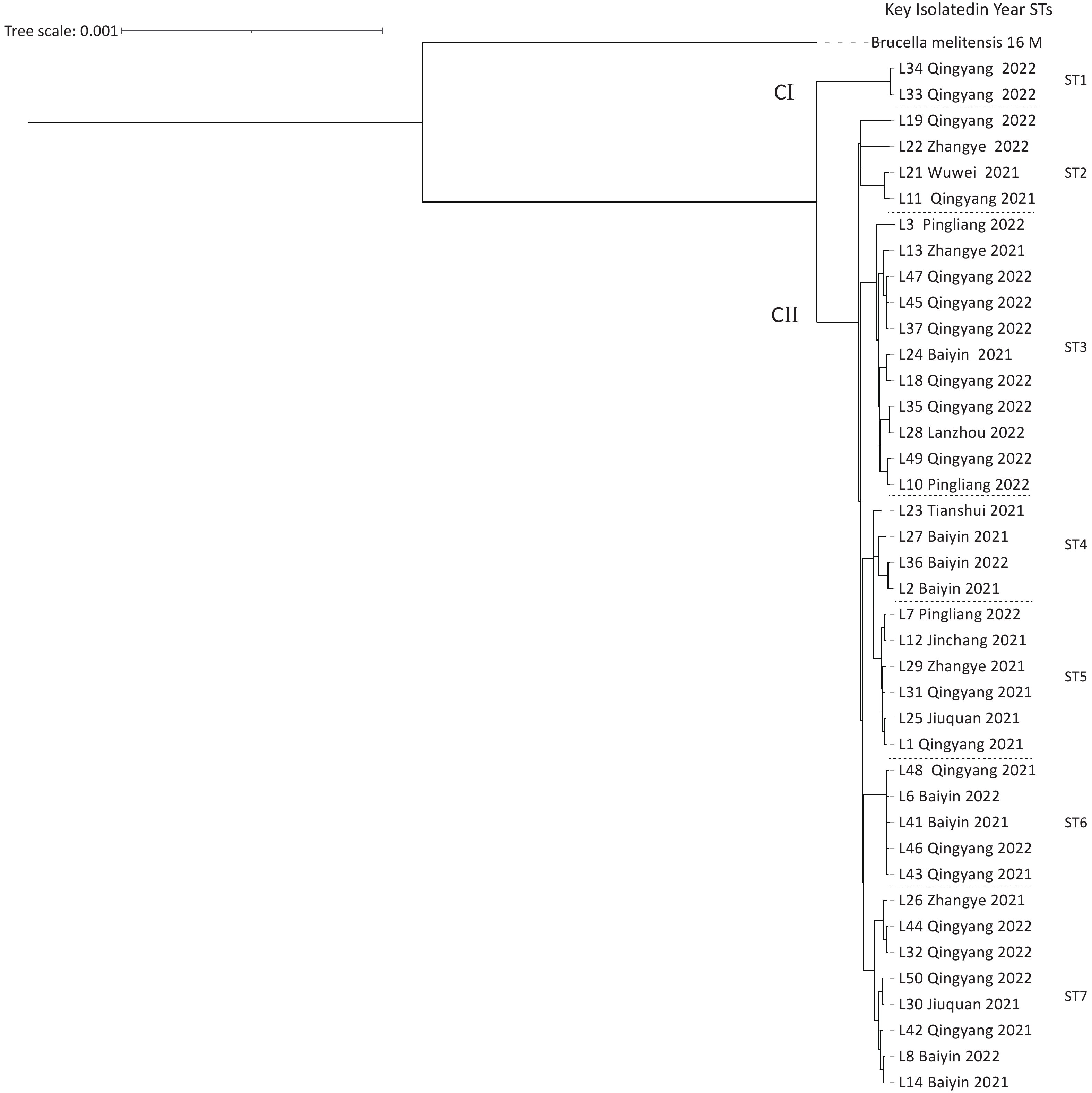
Based on cgSNP analysis, seven SNP genotypes (ST1–7) were identified among the 40 B. melitensis strains. ST1 was distributed in Qingyang; ST2 in Zhangye, Wuwei, and Qingyang; ST3 in Lanzhou, Baiyin, Zhangye, Pingliang, and Qingyang; ST4 in Tianshui and Baiyin; ST5 in Jinchang, Zhangye, Pingliang, Jiuquan, and Qingyang; ST6 in Baiyin and Qingyang; and S7 in Baiyin, Zhangye, Jiuquan, and Qingyang (Figure 2). Notably, six of these STs were observed in Qingyang, followed by four in Baiyin and Zhangye. Among these STs, only ST1 contained two strains from the same location, indicating that two cases were from a common infection source, whereas the remaining six STs each consisted of three to eight strains from different areas. These findings indicated that multiple cross-regional outbreak infection events were caused by multiple common infection sources (Figure 2), implying that the human brucellosis epidemic was primarily driven by cross-regional outbreaks. The results of cgWGS-SNP analysis showed that strains from this study clustered into two branches and seven STs. Six STs were identified in Qingyang, which is adjacent to Shaanxi province, where the number of human brucellosis cases has continuously increased. This increased incidence was related to the development of large-scale small ruminant farms in Guanzhong and some southern Shaanxi regions[8]. CII consisted of six STs that included 38 strains from different cities, implying that the dominant cause of the epidemic pattern was multiple cross-regional outbreak events caused by multiple common sources of infection.
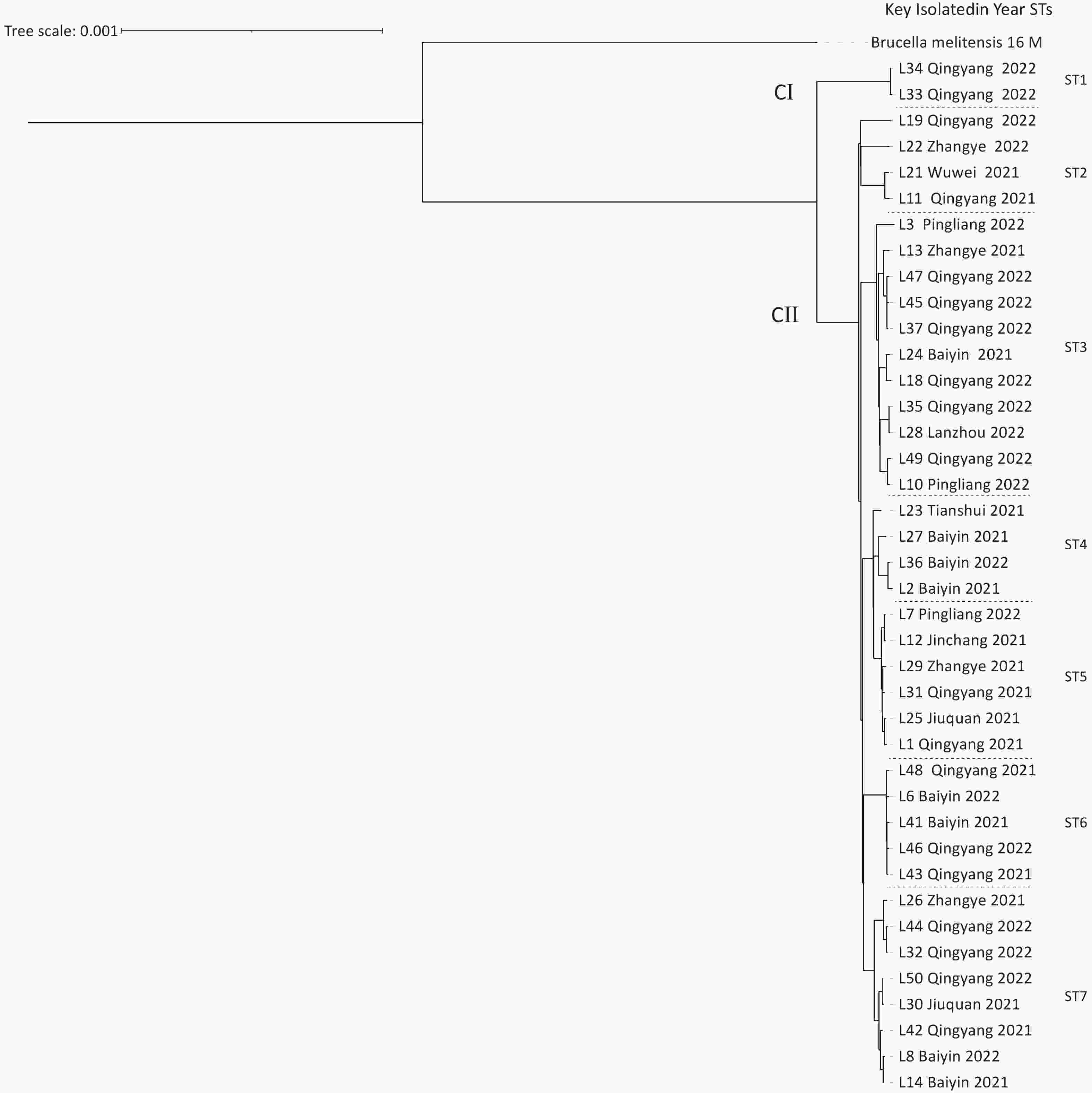
Figure 2. The maximum-likelihood tree generated using the cgSNP matrix of 40 B. melitensis on the county scale. The phylogeny trees of the 40 B. melitensis strains were created using TreeBeST based on the maximum-likelihood phylogeny (PHYML) algorithm with 1,000 bootstrap replicates.
To understand the genetic and phylogenetic profiles of B. melitensis strains on a national scale the phylogenetic analysis was conducted on the 40 strains from this study. The results showed that all 40 strains clustered into SNP genotype II. The strains exhibited genetic homogeneity with strains from Xinjiang, Qinghai, and Inner Mongolia (Supplementary Figure S1), indicating the uncontrolled movement and transfer of infected animals in northwestern China. Two previously reported strains (QY1 and TZ), which were isolated from sheep, shared the same STs (ST2 and ST7) with strains isolated from humans, indicating that identical ST lineages were persistently transmitted between animals and humans (Supplementary Figure S1). In Qinghai, the results of the WGS and SNP analyses of 54 B. melitensis strains showed that the cross-regional transmission events were caused by locally circulating B. melitensis lineages, and the predominant circulating genotypes were endemic in different regions[9]. A B. melitensis TZ strain was isolated from female Tibetan sheep, and QY1 was isolated from sheep in Gansu. The two strains share identical SNP genotypes with the human strains in this study, indicating that these strains were continuously circulating between humans and sheep, and sheep were the source of human cases. Similarly, different B. melitensis lineages circulate in Kyrgyzstan according to the cgSNP and cgMLST analyses, all of which belong to the Eastern Mediterranean group of the global Brucella phylogeny and show the highest similarity to strains from Turkmenistan, Iran, and Turkey[10].
Although phylogenetic analyses revealed a high degree of genetic homogeneity between strains from this study and those from Xinjiang, Qinghai, and Inner Mongolia—regions where brucellosis is severely public health concern—the direction of infection sources remains unclear. Further investigation is required to identify the transmission patterns of B. melitensis strains in northwest China. B. melitensis QH61 strain, which was isolated from a yak undergoing an abortion in 2015 in Qinghai, China, clustered into the same clade as the strains from Inner Mongolia and Gansu Province, implying that the strains from this genotype have been continuously circulating among different hosts in northwest China. However, this hypothesis requires more evidence for verification. Therefore, genome sequence and phylogenetic analyses of more strains from different hosts are urgently needed to uncover the transmission pattern across a wider geographic area.
The present study performed a regional phylogenetic comparison of B. melitensis strains from Gansu, improving our understanding of their transmission patterns and genetic diversity in northwestern China. Over the past decade, extensive research on the genetic diversity of B. melitensis has greatly expanded our knowledge about the transmission of the disease at the molecular level. However, further genomic epidemiological investigations of B. melitensis strains across northwestern China and on a national (or Asia-wide) will provide the additional evidence required to reveal the transmission pathways and situation of B. melitensis strains.
Whole-Genome Phylogeny of Brucella melitensis Isolates from Gansu Province, China
doi: 10.3967/bes2025.084
- Received Date: 2024-12-30
- Accepted Date: 2025-02-28
The authors declare that this study was conducted in the absence of any commercial or financial relationships that could be construed as conflicts of interest.
This study was conducted in accordance with the principles of the Declaration of Helsinki. The research protocol was reviewed and approved by the Ethics Committee of the Gansu Provincial Centre for Disease Control and Prevention (No: GJL2018001). This study was conducted in accordance with local legislation and institutional requirements.
| Citation: | Xiaoyan Zhou, Pinggui Wang, Qingqing Xu, Yu Feng, Dingsheng Wang, Qi Zhao, Lixia Niu, Minghui Ma, Aiwei He, Hai Jiang. Whole-Genome Phylogeny of Brucella melitensis Isolates from Gansu Province, China[J]. Biomedical and Environmental Sciences. doi: 10.3967/bes2025.084 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: