-
Getah virus (GETV) was first isolated from Culex samples collected in Malaysia in 1955. The prototype virus strain was MM2021[1]. GETV belongs to the genus Alphavirus of the family Togaviridae and is a mosquito-transmitted arbovirus[2]. To date, GETV has been identified in about 10 countries or regions, including Australia[2], Malaysia[1], Japan[3], China[4], Mongolia[5], and Russia[5]. GETV can cause fever, body rashes, and leg edema in horses[3], as well as fetal death and reproduction disorders in pigs[6]. Thus, GETV is an important animal pathogen. Although antibodies neutralizing GETV have been identified in human serum samples in Malaysia, northern Australia, and Hainan Province in China[4,7], GETV has not been reported to cause human diseases.
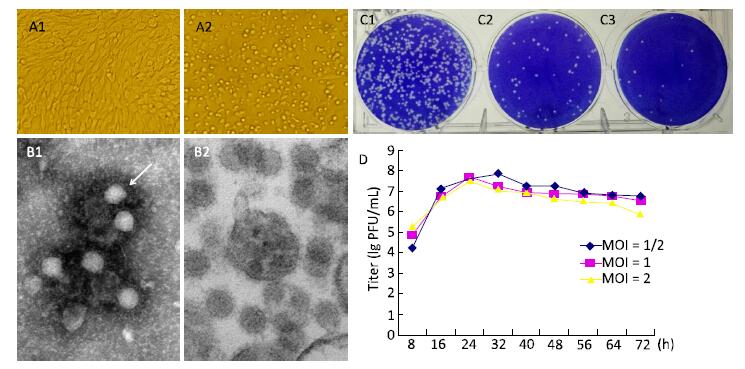
An arbovirus investigation was conducted in the China-Laos border region (longitude 100°5'E, latitude 21°69'N) in August 2012. Specimens of Armigeres subalbatus collected during this investigation were ground and centrifuged. The obtained supernatant was utilized to inoculate BHK cells and the resulting cytopathic effect (CPE) was monitored[8]. After 32 h, BHK-21 cells infected with YN12031 exhibited an obvious CPE, including rounding up, aggregation, and exfoliation (Figure 1A). This CPE progressed rapidly to the '+++' level (i.e., 75% of the cells became cytopathic) about 48 h post-infection.
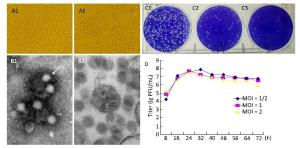
Figure 1. Biological characteristics of YN12031. (A) Cytopathic effect of YN12031 in BHK-21 cells (200 × magnification). (A1) Control (uninfected BHK-21 cells; 48 h). (A2) Infected BHK-21 cells 48 h post-infection showing rounding and exfoliation. (B) Electron micrographs of YN12031 particles negatively stained with 2% potassium phosphotungstate. (B1) Black arrow indicates an intact particle. (B2) Morphology of virus particles. (C) Plaques formed after inoculation of (C1) 10-4, (C2) 10-5, and (C3) 10-6 dilutions of YN12031. (D) Growth curve of YN12031 in BHK-21 cells.
BHK cells inoculated with YN12031 were visualized by transmission electron microscopy (TEM)[8]. YN12031 exhibited a typical alphavirus morphology; the virus particles were spherical, 0-70 nm in diameter, enveloped, and featured surface fibers (Figure 1B1). After infection, the BHK cells were centrifuged, sectioned, and visualized by TEM. Virus particles were evident, and the majority were located in cytoplasmic vesicles. Virus particles contained a core with a high electron density (Figure 1B2).
To understand the plaque morphology and proliferation of YN12031 in tissue culture cells, we first observed the formation of virus plaques in BHK-21 cells and then detected dynamic changes in virus multiplication by plaque assay[9]. The plaques were 1.09 mm (1.15 ± 0.35 mm, n = 10, 2d) in diameter and regular in shape with distinct edges (Figure 1C). Following infection of BHK-21 cells at multiplicity of infeetion (MOIs) of 1 and 2, YN12031 proliferated rapidly about 8-24 h post-infection and reached peak titers of 1 × 107.69 and 1 × 107.55 pfu/mL, respectively. These titers decreased thereafter, reaching minima of 1 × 106.77 and 1 × 105.95 pfu/mL, respectively, at 72 h. Following infection at an MOI of 0.5, the YN12031 titer increased rapidly from 8 h to 16 h. The titer increased slowly from 16 h to 32 h, at which point it peaked (1 × 107.86 pfu/mL). The YN12031 titer decreased slowly afterward, reaching a level comparable with that following infection at an MOI of 1 (1 × 106.76 pfu/mL) at 72 h. These results are shown in Figure 1D.
BHK-21 cells infected with GETV exhibited an obvious CPE 32 h post-infection, and the CPE level reached '+++' after 48 h. The virus titer peaked 32 h post-infection [1 × 107.86 pfu/mL (MOI = 1/2)] and then decreased gradually (Figure 1D). Previous studieshave reported that BHK-21 cells infected with Sindbis virus (YN87448), the model virus of alphavirus, can exhibit an obvious CPE at 24 h and that the virus titer peaks (1 × 109.5 pfu/mL) at 36 h[10]. By comparison, the CPE caused by infection with Japanese encephalitis virus, a flavivirus with linear positive-sense single-stranded RNA, appears at 48-72 h, and the highest viral titer could reach 1 × 107.1 pfu/mL[9]. These findings reveal that alphaviruses can cause CPEs in tissue culture cells faster than other virus types can.
Whole-genome amplification of YN12031 was performed using the GETV gene amplification primers described in Table 1. The amplified products were examined by agarose gel electrophoresis, purified using a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA USA), and then sequenced directly. The sequences obtained were assembled, edited, and corrected using SeqMan in DNASTAR[8]. The coding region of YN12031 is 11, 166 nt in length and encodes 3, 720 amino acids. The length of the non-structural gene is 7, 404 nt and located in the region between 79 and 7, 482 nt. The non-structural gene codes four non-structural proteins (NSP1-NSP4) with a total of 2, 467 amino acids. The structural gene is 3, 762 nt in length and located in the region between 7, 527 and 11, 288 nt. This gene encodes a variety of structural proteins (E1, E2, E3, 6K, and capsid protein) with a total of 1, 253 amino acids.
Table 1. Primers Used in This Study
Primer Amplity Region Orientation Sequence (5'-3') Site in Genome GETV-F1 5'UTR-Nsp1 Sense ATGGCGGACGTGTGACATCAC 1-21 GETV-R1 Antisense GTAACCTTCGCATGACACCACC 909-930 GETV-F2 Nsp1-Nsp2 Sense GGCATTTACCTTCCGTGTTTC 848-868 GETV-R2 Antisense TGTGCTTGCGGTGTAACCTTC 1710-1730 GETV-F3 Nsp2 Sense TAGTGAGCGGCTCTTGTGCTG 1610-1630 GETV-R3 Antisense CCGCACAGTACTACCTTACCTGAC 2493-2516 GETV-F4 Nsp2 Sense GATGAGGCGTTCGCGTGTCACT 2434-2455 GETV-R4 Antisense GGTAACCGACGATTGGATGGGACT 3480-3503 GETV-F5 Nsp2-Nsp3 Sense TCTACGTGGCAACATGAACTCG 3399-3420 GETV-R5 Antisense CGTGAATAAGTGGTTCAAGGACTGC 4440-4464 GETV-F6 Nsp3 Sense GCTGTGGCTAGCATAATTAGTACC 4345-4368 GETV-R6 Antisense TGGGATAGCGCGTATGTCTGT 5308-5328 GETV-F7 Nsp3-Nsp4 Sense GTCGCCCAACTTAGACAGG 5212-5230 GETV-R7 Antisense TGGTTGGTGGTATGCGTGG 6171-6189 GETV-F8 Nsp4 Sense CCGATGAGTATGACGCTTATCTGG 6071-6094 GETV-R8 Antisense ACTTCCATGTTGACCCAACTC 7098-7118 GETV-F9 Nsp4-C Protein Sense CGCTGCTGAACATTGTCATAG 6965-6985 GETV-R9 Antisense GGTTGTCGATCACACCTTTG 7967-7986 GETV-F10 C Protein-E2 Sense GATTGCATCTTCGAGGTCAAGC 7881-7922 GETV-R10 Antisense GTGCGTGTTTGTACTGCACTTTG 8897-8919 GETV-F11 E2 -6K Protein Sense TGCGCTATTCGAGGCACGAT 8793-8812 GETV-R11 Antisense ATGATTATGGCAGCGAGCGG 9861-9880 GETV-F12 E2-E1 Sense CCGGTAACACTAGGAGTACTATGC 9744-9767 GETV-R12 Antisense TTGTCATTCAGCGACGTGCCT 10712-10732 GETV-F13 E1-3'UTR Sense CCTCAAGTTGTCAAGACCTTCGTC 10625-10648 GETV-R13 Antisense GTAAAATATTAAAAAAACAAATTAGACGCC 11661-11690 Nucleotide and amino acid homology analyses of the GETV coding region (excluding 5' and 3' non-coding sequences) were conducted using MegAlign in DNASTAR (version 5.00) and BioEdit (version 7.0.5.3), respectively[8]. The nucleotide homology of YN12031 with 13 other GETVs obtained from GenBank ranged from 95.9% (Korean isolate South Korea) to 97.7% (Russian isolate LEIV/16275/Mag), and its amino acid homology ranged from 95.4% (Korean isolate South Korea) to 97.0% (Russian isolate LEIV/16275/Mag). The coding region of GETV was clearly relatively conserved. Both the nucleotide and amino acid homology between various strains varied between 95% and 98%, and YN12031 showed the closest relationship with the LEIV/16275/Mag isolate obtained in Russia among other GETV strains studied.
Twenty-six GETV strains isolated from both China and abroad were selected for homology analysis of the nucleotides and amino acids of the E2 gene. The nucleotide homology between YN12031 and 25 other strains ranged from 93.8% (Malaysia prototype strain MM2021) to 97.4% (Russian isolate LEIV/16275/Mag), and its amino acid homology ranged from 96.0% (Malaysia prototype strain MM2021) to 98.1% (Russian isolate LEIV/16275/ Mag). The homology of YN12031 with the Russian isolate LEIV/16275/Mag was the closest found. GETV isolates used in this study and the sequence identities of the E2 gene are provided in Supplementary Tables 1 and 2, respectively, which are available in BES online.
Table 1. GETV Isolates Analyzed in This Study
Strain Date Country Host Genebank Accession No complete
sequencesE2 Capsid MM2021 1955 Malaysia C.gelidus - AF339484 AF339484 GETV-SAGV 1956 Japan mosquito AB032553 AB032553 AB032553 GETV-SAGV-Original 1956 Japan - - AF339483 AF339483 Kochi/01/2005 2005 Japan Sus scrofa AY702913 AB859822 AB859822 14-I-605-C2 2014 Japan Equus caballus LC079089 LC079089 LC079089 14-I-605-C1 2014 Japan Equus caballus LC079088 LC079088 LC079088 MI-110-C2 1978 Japan Equus caballus LC079087 LC079087 LC079087 MI-110-C1 1978 Japan Equus caballus LC079086 LC079086 LC079086 South Korea 2004 South Korea swine AY702913 AY702913 AY702913 LEIV 16275 Mag 2000 Russia Aedes sp EF631998 EF631998 EF631998 LEIV 17741 MPR 2000 Mongolia Culex sp EF631999 EF631999 EF631999 M1 1964 China, Hainan Culex sp EU015061 EU015061 EF375826 YN0540 2005 China, Yunnan Armigeres subalbatus EU015063 EU015063 EU015063 HB0234 2002 China, Hebei Culex tritaeniorhynchus Giles EU015062 EU015062 EF375825 SC1210 2012 China, Sichuan Armigeres subalbatus LC107870 LC107870 LC107870 HB0215-3 2002 China, Hebei Culex tritaeniorhynchus Giles - EU015065 EF375824 QIAG9303 1993 South Korea swine - KR081240 KR081240 QIAG9302 1993 South Korea swine - KR081239 KR081239 QIAG9301 1993 South Korea swine - KR081238 KR081238 GS10-2 2006 China, Gansu Armigeres subalbatus - EU015070 - SH05-17 2005 China, Shanghai Culex tritaeniorhynchus - EU015069 - SH05-16 2005 China, Shanghai Culex tritaeniorhynchus - EU015068 - SH05-15 2005 China, Shanghai Culex tritaeniorhynchus - EU015067 - SH05-6 2005 China, Shanghai Culex tritaeniorhynchus - EU015066 - YN0542 2005 China, Yunnan Armigeres subalbatus - EU015064 - YN12031* 2012 China, Yunnan Armigeres subalbatus MI-110 1978 Japan Equus caballus - - LC012886 Miho2014 2014 Japan Equus caballus - - LC012884 YN08 2008 China, Yunnan Aedes albopictus - - JN578104 Note.*First report of the Getah virus in this study. '-':not available in GenBank. Table 2. Sequence ldentities for the E2 Gene of GETV

We constructed a phylogenetic tree using the coding region sequences of YN12031 and 10 other alphaviruses-Western equine encephalitis virus, Eastern equine encephalitis virus, Sindbis virus, Chikungunya virus, several Maya Luo virus, BarmahForestvirus, O'Nyong-nyongvirus, Semlikiforestvirus, RossRivervirus and GETV-using the neighbor-joining method (Figure 2A). YN12031 and MM2021, the prototype strain of GETV first isolated in Malaysia in 1955, were included in the same clade, which means YN12031 could be a GETV.
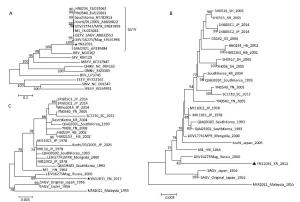
Figure 2. Phylogenetic analysis of YN12031. (A) Phylogenetic tree constructed using the coding region sequences of YN12031 and 10 other alphavirus strains. (B) Phylogenetic analysis of the E2 gene sequences of GETV isolates from Malaysia, Japan, South Korea, Russia, and China. (C) Phylogenetic tree constructed using the nucleotide sequences of the GETV capsid protein gene.
Further phylogenetic analysis of the E2 gene sequences of GETV isolates from Malaysia, Japan, South Korea, Russia, and China demonstrated that the Malaysia prototype strain MM2021 was located at the root of phylogenetic tree. The newly isolated strain YN12031 was identified in the same clade as the LEIV/16275/Mag isolate obtained in Russia (Figure 2B). Phylogenetic analysis of the GETV capsid gene sequences of these strains yielded similar results (Figure 2C).
In this study, the structural gene (E2) nucleotide and amino acid homologies between the GETV prototype strain isolated in Malaysia in 1955 and other GETVs isolated during the next 60 years were 93.8%-100% and 96.0%-100%, respectively, thus suggesting that the GETV gene is conserved. Phylogenetic analysis of E2 and capsid genes showed that GETVs isolated from different mosquito vectors, pigs, and horses in different countries (or regions), such as China, Japan, Korea, Mongolia, and Russia, from 1964 to 2014 were clustered in the same evolutionary clade. All of the GETVs isolated in Russia (60°N latitude) and Hainan province in China (19°N latitude) where thousands of kilometers apart from each other, and no matter GETVs isolated from specimens of mosquito or host animals (horses, pigs), they were all located at the different positions in the evolutionary branches. No vector, host specificity and geographic distribution characteristic were observed in the distribution of GETVs in the phylogenetic tree.
GETV was previously isolated form A. subalbatus in Yunnan Province[7], but the newly isolated strain (YN12031) is not located in the same evolutionary branch as two other stains (YN0540, YN0542) isolated from samples of A. subalbatus in the same province (Figure 2B-C), this finding suggests that diverse GETV populations with different evolutionary states exist in A. subalbatus in Yunnan Province. The YN12031 and LEIV/16275/Mag isolates obtained from mosquito specimens in Russia (60°N latitude) are in the same evolutionary branch, and homology analysis of the GETV coding region and E2 gene revealed the very close relationship between YN12031 and LEIV/16275/Mag. These results suggest that GETV could adapt to both tropical environments (Yunnan, China) and frigid zone environments (Russia) to survive and evolve; this characteristic could enable the spread of equine diseases[11] endemic in tropical regions to a wider range or higher latitude (such as Russia). Therefore, strengthening the detection and monitoring of GETV and its infections in temperate and northern frigid zones is essential.
GETV is an important animal pathogen, and epidemics of GETV infection among horses and pigs have emerged several times across Asia[3,6,11]. A number of GETV strains in China have been geographically isolated across the latitudes 19°N (Hainan Province) to 42°N (Liaoning Province) and longitudes 97°E (Gansu Province) to 124°E (Liaoning Province). GETV can be isolated from Culex[1,7], Aedes[1], Armigeres[7], and Anopheles[1]. Considering this prevalence, GETV and GETV infections present an increasing threat to animal health, particularly horses and pigs, and strengthening the detection and monitoring of GETV in animals is necessary to prevent development of the related animal diseases in China and reduce economic losses.
doi: 10.3967/bes2017.028
-
Abstract: In this study, we isolated a virus strain (YN12031) from specimens of Armigeres subalbatus collected in the China-Laos border. BHK-21 cells infected with YN12031 exhibited an evident cytopathic effect (CPE) 32 h post-infection. The virus particles were spherical, 70 nm in diameter, and enveloped; they also featured surface fibers. Molecular genetic analysis revealed that YN12031 was closely related to alpha viruses such as Chikungunya virus and Sindbis virus, and located in the same clade as MM2021, the prototype of Getahvirus (GETV) isolated in Malaysia in 1955. Phylogenetic analysis of the E2 and capsid genes further revealed that YN12031 was located in the same clade as the Russian isolate LEIV/16275/Mag. Analysis of the homology of nucleotides and amino acids in the coding area and E2 gene demonstrated that the YN12031 isolated from the China-Laos border (tropical region) was related closest to the LEIV/16275/Mag isolate obtained in Russia (North frigid zone area) among other isolates studied. These results suggest that GETV can adapt to different geographical environments to propagate and evolve. Thus, strengthening the detection and monitoring of GETV and its related diseases is very crucial.
-
Key words:
- Getah virus /
- China-Laos border /
- Phenotypic characteristics /
- Molecular evolution
-
Figure 1. Biological characteristics of YN12031. (A) Cytopathic effect of YN12031 in BHK-21 cells (200 × magnification). (A1) Control (uninfected BHK-21 cells; 48 h). (A2) Infected BHK-21 cells 48 h post-infection showing rounding and exfoliation. (B) Electron micrographs of YN12031 particles negatively stained with 2% potassium phosphotungstate. (B1) Black arrow indicates an intact particle. (B2) Morphology of virus particles. (C) Plaques formed after inoculation of (C1) 10-4, (C2) 10-5, and (C3) 10-6 dilutions of YN12031. (D) Growth curve of YN12031 in BHK-21 cells.
Figure 2. Phylogenetic analysis of YN12031. (A) Phylogenetic tree constructed using the coding region sequences of YN12031 and 10 other alphavirus strains. (B) Phylogenetic analysis of the E2 gene sequences of GETV isolates from Malaysia, Japan, South Korea, Russia, and China. (C) Phylogenetic tree constructed using the nucleotide sequences of the GETV capsid protein gene.
Table 1. Primers Used in This Study
Primer Amplity Region Orientation Sequence (5'-3') Site in Genome GETV-F1 5'UTR-Nsp1 Sense ATGGCGGACGTGTGACATCAC 1-21 GETV-R1 Antisense GTAACCTTCGCATGACACCACC 909-930 GETV-F2 Nsp1-Nsp2 Sense GGCATTTACCTTCCGTGTTTC 848-868 GETV-R2 Antisense TGTGCTTGCGGTGTAACCTTC 1710-1730 GETV-F3 Nsp2 Sense TAGTGAGCGGCTCTTGTGCTG 1610-1630 GETV-R3 Antisense CCGCACAGTACTACCTTACCTGAC 2493-2516 GETV-F4 Nsp2 Sense GATGAGGCGTTCGCGTGTCACT 2434-2455 GETV-R4 Antisense GGTAACCGACGATTGGATGGGACT 3480-3503 GETV-F5 Nsp2-Nsp3 Sense TCTACGTGGCAACATGAACTCG 3399-3420 GETV-R5 Antisense CGTGAATAAGTGGTTCAAGGACTGC 4440-4464 GETV-F6 Nsp3 Sense GCTGTGGCTAGCATAATTAGTACC 4345-4368 GETV-R6 Antisense TGGGATAGCGCGTATGTCTGT 5308-5328 GETV-F7 Nsp3-Nsp4 Sense GTCGCCCAACTTAGACAGG 5212-5230 GETV-R7 Antisense TGGTTGGTGGTATGCGTGG 6171-6189 GETV-F8 Nsp4 Sense CCGATGAGTATGACGCTTATCTGG 6071-6094 GETV-R8 Antisense ACTTCCATGTTGACCCAACTC 7098-7118 GETV-F9 Nsp4-C Protein Sense CGCTGCTGAACATTGTCATAG 6965-6985 GETV-R9 Antisense GGTTGTCGATCACACCTTTG 7967-7986 GETV-F10 C Protein-E2 Sense GATTGCATCTTCGAGGTCAAGC 7881-7922 GETV-R10 Antisense GTGCGTGTTTGTACTGCACTTTG 8897-8919 GETV-F11 E2 -6K Protein Sense TGCGCTATTCGAGGCACGAT 8793-8812 GETV-R11 Antisense ATGATTATGGCAGCGAGCGG 9861-9880 GETV-F12 E2-E1 Sense CCGGTAACACTAGGAGTACTATGC 9744-9767 GETV-R12 Antisense TTGTCATTCAGCGACGTGCCT 10712-10732 GETV-F13 E1-3'UTR Sense CCTCAAGTTGTCAAGACCTTCGTC 10625-10648 GETV-R13 Antisense GTAAAATATTAAAAAAACAAATTAGACGCC 11661-11690 Table 1. GETV Isolates Analyzed in This Study
Strain Date Country Host Genebank Accession No complete
sequencesE2 Capsid MM2021 1955 Malaysia C.gelidus - AF339484 AF339484 GETV-SAGV 1956 Japan mosquito AB032553 AB032553 AB032553 GETV-SAGV-Original 1956 Japan - - AF339483 AF339483 Kochi/01/2005 2005 Japan Sus scrofa AY702913 AB859822 AB859822 14-I-605-C2 2014 Japan Equus caballus LC079089 LC079089 LC079089 14-I-605-C1 2014 Japan Equus caballus LC079088 LC079088 LC079088 MI-110-C2 1978 Japan Equus caballus LC079087 LC079087 LC079087 MI-110-C1 1978 Japan Equus caballus LC079086 LC079086 LC079086 South Korea 2004 South Korea swine AY702913 AY702913 AY702913 LEIV 16275 Mag 2000 Russia Aedes sp EF631998 EF631998 EF631998 LEIV 17741 MPR 2000 Mongolia Culex sp EF631999 EF631999 EF631999 M1 1964 China, Hainan Culex sp EU015061 EU015061 EF375826 YN0540 2005 China, Yunnan Armigeres subalbatus EU015063 EU015063 EU015063 HB0234 2002 China, Hebei Culex tritaeniorhynchus Giles EU015062 EU015062 EF375825 SC1210 2012 China, Sichuan Armigeres subalbatus LC107870 LC107870 LC107870 HB0215-3 2002 China, Hebei Culex tritaeniorhynchus Giles - EU015065 EF375824 QIAG9303 1993 South Korea swine - KR081240 KR081240 QIAG9302 1993 South Korea swine - KR081239 KR081239 QIAG9301 1993 South Korea swine - KR081238 KR081238 GS10-2 2006 China, Gansu Armigeres subalbatus - EU015070 - SH05-17 2005 China, Shanghai Culex tritaeniorhynchus - EU015069 - SH05-16 2005 China, Shanghai Culex tritaeniorhynchus - EU015068 - SH05-15 2005 China, Shanghai Culex tritaeniorhynchus - EU015067 - SH05-6 2005 China, Shanghai Culex tritaeniorhynchus - EU015066 - YN0542 2005 China, Yunnan Armigeres subalbatus - EU015064 - YN12031* 2012 China, Yunnan Armigeres subalbatus MI-110 1978 Japan Equus caballus - - LC012886 Miho2014 2014 Japan Equus caballus - - LC012884 YN08 2008 China, Yunnan Aedes albopictus - - JN578104 Note.*First report of the Getah virus in this study. '-':not available in GenBank. Table 2. Sequence ldentities for the E2 Gene of GETV

-
[1] Karabatsos N. International catalogue of arthropod-borne viruses. 3rd ed. American Society for Tropical Medicine and Hygiene, San 4 Antonio (TX), 1985, 418-9. [2] Fukunaga Y, Kumanomido T, Kamada M. Getah virus as an equine pathogen. Vet Clin North Am Equine Pract, 2000, 16: 605-17. https://www.ncbi.nlm.nih.gov/pubmed/11219353 [3] Nemoto M, Bannai H, Tsujimura K, et al. Getah Virus Infection among Racehorses, Japan, 2014. Emerg Infect Dis, 2015, 21: 883-5. doi: 10.3201/eid2105.141975 [4] Li XD, Qiu FX, Yang H, et al. Isolation of Getah virus from mosquitos collected on Hainan Island, China, and results of a serosurvey. Southeast Asian J Trop Med Public Health, 1992, 23: 730-4. https://www.ncbi.nlm.nih.gov/pubmed/1338481 [5] L' vov SD, Gromashevski' i VL, Aristova VA, et al. Isolation of Getah virus (Togaviridae, Alfavirus) strains in North-Eastern Asia. Vopr Virusol, 2000, 45: 14-8. https://www.ncbi.nlm.nih.gov/pubmed/11107647 [6] Yago K, Hagiwara S, Kawamura H, et al. A fatal case in Newborn piglets with Getah virus infection:isolation of the virus. Jpn J Vet Sci, 1987, 49: 989-94. doi: 10.1292/jvms1939.49.989 [7] Zhai YG, Wang HY, Sun XH, et al. Complete sequence characterization of isolates of Getah virus (genus Alphavirus, family Togaviridae) from China. J Gen Virol, 2008, 89: 1446-56. doi: 10.1099/vir.0.83607-0 [8] Lei WW, Guo XF, Fu SH, et al. Isolation of Tibet orbivirus, TIBOV, from Culicoides Collected in Yunnan, China. PLoS one, 2015, 10: e0136257. doi: 10.1371/journal.pone.0136257 [9] Cao L, Fu SH, Gao XY, et al. Low Protective Efficacy of the Current Japanese Encephalitis Vaccine against the Emerging Genotype 5 Japanese Encephalitis Virus. PLoS Negl Trop Dis, 2016, 10: e0004686. doi: 10.1371/journal.pntd.0004686 [10] Zhu WY, Wang LH, Yang YL, et al. Interaction of E2 Glycoprotein with Heparan Sulfate Is Crucial for Cellular Infection of Sindbis Virus. PLoS One, 2010, 5: e9656. doi: 10.1371/journal.pone.0009656 [11] Broen CM, Timoney PJ. Getah virus infection of Indian horses. Trop Anita Health Prod, 1998, 30: 241-52. doi: 10.1023/A:1005079229232 -




 下载:
下载:



 Quick Links
Quick Links