-
The coronavirus disease 2019 (COVID-19) is an infection of the respiratory tract which first broke out in Wuhan, China, in December 2019. The etiologic agent of this disease was identified as a betacoronavirus related to severe acute respiratory syndrome coronavirus (SARS-CoV). Thus, the International Committee on Virus Classification named the new coronavirus severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)[1]. Coronaviruses consist of enveloped virions (virus particles) that measure approximately 120 nm in diameter[2]. Viral particles can be round or oval, but are usually polymorphic. Recent studies have shown that SARS-CoV-2 displays > 85% homology with a bat SARS-like coronavirus (bat-SL-CoVZC45, accession no. MG772933.1)[3]. When cultured with cells in vitro, SARS-CoV-2 could be observed in human respiratory epithelial cells after about 96 h, while in Vero E6 and Huh-7 cell lines the detection of SARS-CoV-2 took about 6 d[4].
Nucleic acid testing, being conveniently rapid and simple, is one of the main techniques for SARS-CoV-2 testing[5, 6]. In this process, samples from nasopharyngeal swabs, sputum, and other secretions of the lower respiratory tract, as well as from blood and feces, are first collected. Next, the laboratory testing personnel use quantitative real-time polymerase chain reaction (qPCR) to detect nucleic acids in the specimens. With an increasing number of tested people, the risk of infection for the testing personnel in the environment of the testing laboratory rapidly increases. Nevertheless, only a few people are aware of this problem, and related studies were not conducted. In this study, the environment of a SARS-CoV-2 nucleic acid testing laboratory in Tianjin, China, was evaluated. This work provides useful guidance and helpful indications for the environmental biosafety of SARS-CoV-2 nucleic acid testing laboratories.
Sampling swabs and sampling liquid were purchased from Youkang Hengye Biotechnology (Beijing) Co., Ltd, China. They were delivered in a plastic package, which had been sterilized with gamma rays before leaving the factory. The nucleic acid extraction kit for virus DNA/RNA (16T/kit) was produced by Xi’an Tianlong Science and Technology Co., Ltd, China. The testing kit for SARS-CoV-2 nucleic acid was produced by Shanghai BioGerm Medical Technology Co., Ltd., China. Nucleic acid extraction was carried out with the Tianlong automatic nucleic acid extractor (NP968-C, Xi’an Tianlong Science and Technology Co., Ltd). Nucleic acids were detected by qPCR (7500 Real-Time PCR System, Thermo Fisher Scientific, USA). A chloride dioxide air disinfector (HYF-201) was kindly provided by ZYUNICON (Tianjin) Science & Technology Co. Ltd, China. The hydrogen peroxide disinfector (HTY-SUPER SD2) used was manufactured by Zhejiang Tailin Bioengineering Co., Ltd, China.
The Pathogen Detection Laboratory of the Tianjin Center for Disease Control and Prevention is a laboratory for nucleic acid testing of SARS-CoV-2. This laboratory includes a preparation room, a sample processing room (for nucleic acid extraction), an amplification room, and an analysis room. The laboratory has a testing capacity of 800 samples/day.
The Sample Processing Room is equipped with a chlorine dioxide (ClO2) air disinfector for continuous air disinfection. The disinfector is based on an ultra-low-ClO2-concentration protection technology, which uses an ultraviolet light source to stimulate slow release of gaseous ClO2 from a gel as core disinfectant. In the room, the ClO2 concentration was always maintained lower than 0.3 ppm (0.9 mg/m3). Such concentration meets the biosafety standards of the US National Institute for Occupational Safety and Health (NIOSH)[7].
Moreover, terminal disinfection with hydrogen peroxide vapor was carried out in the whole laboratory whenever there was a spill from the laboratory area of primary containment, if SARS-CoV-2 nucleic acid was detected in the environment, or after the laboratory ran continuously for a week. To maintain proper concentration and distribution of hydrogen peroxide vapor, the room supply and exhaust vents were closed and the biosafety cabinet was locked throughout the disinfection period. The hydrogen peroxide disinfector was used for the final disinfection of the laboratory without testing personnel. The core disinfectant of the disinfector was 35% hydrogen peroxide (H2O2). The disinfection procedures included preheating for 5 min, spraying of disinfectant for 45 min (flow, 4 g/min), and spraying of catalase for 1 min by a fast-decomposing protease apparatus. The laboratory was then closed for 15 min. Afterwards, the personnel could enter and work again in the laboratory.
SARS-CoV-2 nucleic acid sampling was carried out on the handle of the biosafety chamber, on the surface of instruments, air, etc. in the Preparation Room, Sample Processing Room, Amplification Room, and Analysis Room of the laboratory.
For sampling on the surface of objects, a sampling swab made of artificial fibers wrapped with plastic rods was first soaked in the sampling liquid. Then, the swab was rubbed back and forth and rotated at the same time on the surface of the object. After the sampling was finished, the swab was dipped in the sample fluid and oscillated for 5 s. Nucleic acid extraction was performed with a nucleic acid extractor.
On the other hand, a liquid impact sampler loaded with PBS solution was used for air sampling, after which nucleic acid extraction was carried out.
Nucleic acid extraction was based on the principle of magnetic bead adsorption, occurring through adsorption by magnetic rods, transfer to magnetic beads, and release of nucleic acids. The sample volume was 300 μL and the nucleic acid extraction process was automatically completed in 16 min by the nucleic acid extractor. According to the kit instructions, the extraction process included pyrolysis (5.5 min), first washing (2 min, 90 °C), second washing (2 min, 90 °C), elution (5.5 min, 90 °C), and demagnetization (1 min). SARS-CoV-2 nucleic acids were detected by qPCR with specific primers and Taqman probes targeting the open reading frame ORF1ab and the nucleoprotein (N) gene of SARS-CoV-2. Five microlitres of extracted nucleic acids were added into a PCR tube containing 20 μL of amplification buffer. The reaction was carried out as follows: reverse transcription: one cycle at 50 °C for 10 min; pre-denaturation: one cycle at 95 °C for 5 min; amplification: 40 cycles of denaturation at 95 °C for 10 s and annealing/extension/detection of fluorescence at 55 °C for 40 s. Disinfection was performed in parallel when the experimental utensils were transferred by hand throughout the whole procedure.
Results were judged according to the following criteria: when the threshold cycle (Ct) values of the FAM channel (ORF1ab) and the VIC channel (N) were lower than 38, the sample was considered positive; when the Ct values of the FAM channel (ORF1ab) and the VIC channel (N) could not be calculated or were greater than 38, the sample was identified as negative. If the object surface and the air were sampled in the sampling processing room, and the results were positive, then the contamination should have derived from viruses in patient specimens or from the nucleic acid extraction kit. On the other hand, if the object surface or the air was sampled in the amplification room, and the results were positive, then the contamination should be attributed to the contact with viral nucleic acid or amplification products. For quality control, negative controls were run for the FAM and the VIC channels; in addition, positive controls were prepared for the FAM, VIC, and ROX channels [internal reference RNase P (RNP)]. Good quality was confirmed with no amplification in negative controls and Ct values lower than 30 for positive controls[8].
The surface of the following objects: test bed, handle of the biosafety chamber, counter of the biosafety chamber, nucleic acid extractor, pipette, test tube rack, oscillator, centrifuge, refrigerator handle, and door handle, as well as the air from the SARS-CoV-2 nucleic acid testing laboratory, were sampled 10 times, and the presence of viral nucleic acid was tested. The results are shown in Table 1, Figure 1A and 1B. In Table 1 it is reported that, among 10 samples from the test tube rack, refrigerator handle, and door handle, two resulted positive for the RNP gene. There are two possible explanations for this phenomenon. The first is that the laboratory staff did not wear gloves while touching these objects. However, due to the strict laboratory rules, it was impossible for the personnel to work without wearing gloves. Another possibility is that the laboratory staff wore gloves that came in contact with negative specimens, and then touched and contaminated these surfaces.
Table 1. Results of environment monitoring of a SARS-CoV-2 testing laboratory
Room Monitoring site Number of samples Positive samples FAM channel (ORF1ab) VIC channel (N) ROX channel (internal
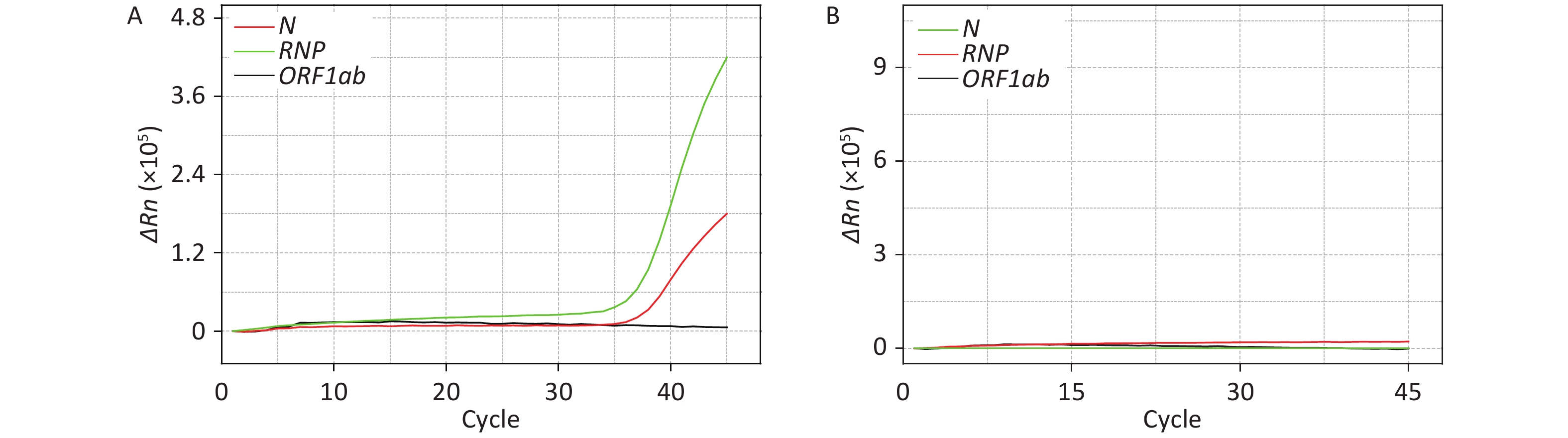
reference RNP)Preparation room Test bed 10 0 0 0 Handle of biosafety chamber 10 0 0 Sample processing room Counter of biosafety chamber 10 0 0 Handle of biosafety chamber 10 0 0 Nucleic acid extractor 10 0 0 Pipette 10 0 0 Test tube rack 10 0 0 2 Oscillator 10 0 0 Centrifuge 10 0 0 Refrigerator handle 10 0 0 2 Door handle 10 0 0 2 Air 10 0 0 Amplification Room qPCR instrument 10 0 0 Computer keyboard of qPCR instrument 10 0 1 1 Air 10 0 0 Analysis room Computer keyboard 10 0 0 Positive control 10 10 10 10 Negative Control 10 0 0 0 Note. The template for the positive control was a mixture of plasmids containing the ORF1ab and N genes and virus-like particles with a specific fragment of the RNase P (RNP) gene; the negative control was a saline solution without genetic material. 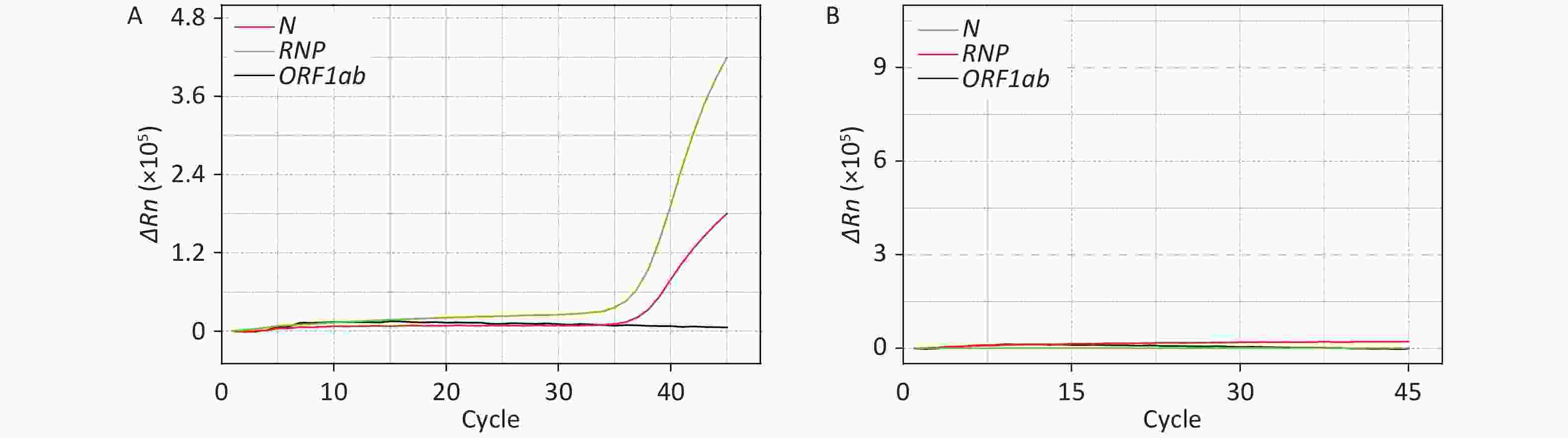
Figure 1. Detection and absence of SARS-CoV-2 nucleic acid on the computer keyboard of the qPCR instrument in the Amplification Room. ΔRn is product quantity of qPCR.
It is worth noting that the computer keyboard of the qPCR instrument in the Amplification Room exhibited one positive sample among the 10 tested. However, by comparing Figure 1 with Supplementary Figure S1 available in www.besjournal.com, it is evident that this sample resulted positive for the RNP and N genes, while resulting negative for the ORF1ab gene. Since the contamination occurred after the process of nucleic acid extraction and analysis, this cannot be due to live virus. Therefore, the computer keyboard was probably contaminated by the extracted SARS-CoV-2 nucleic acid of a positive sample. Although the staff strictly followed the operative guidelines during testing, it is still possible that the extracted nucleic acids of positive samples contaminated the outer wall of the test tube or other testing appliances. Then, if the operator had touched the outer wall of the tube, the gloves would have been contaminated. When the personnel analyzed nucleic acids using the qPCR instrument, they probably did not change into new gloves because of the heavy workload, and thus the contaminated glove further spread the contamination to the computer keyboard of the qPCR instrument.
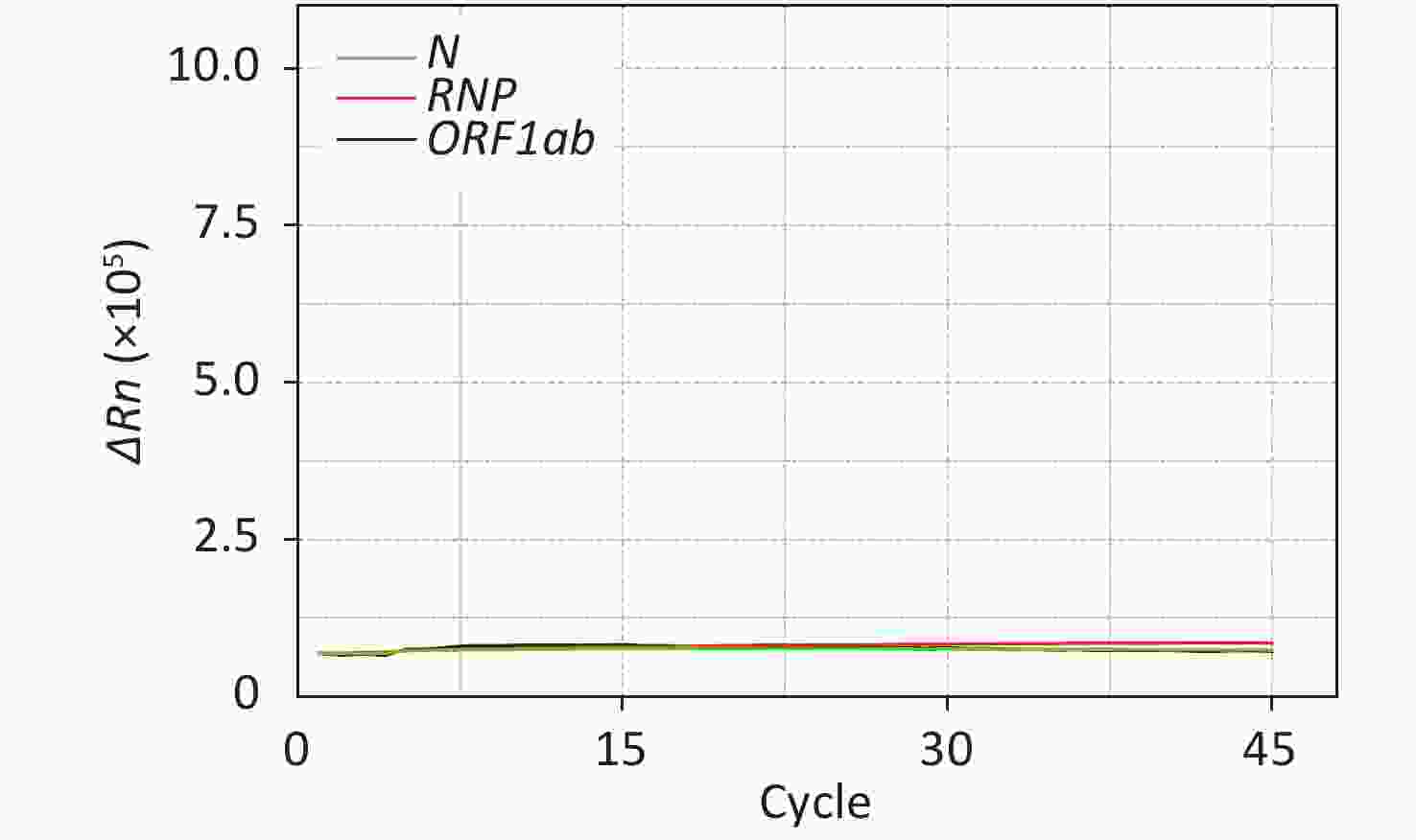
Figure S1. Negative control of SARS-CoV-2 nucleic acid amplification on the computer keyboard of the qPCR instrument in the Amplification Room. ΔRn is product quantity of qPCR.
After the sample from the computer keyboard resulted positive for SARS-CoV-2, the testing work was immediately suspended and terminal disinfection with hydrogen peroxide vapor was carried out in the whole laboratory. The computer keyboard was then re-sampled, and the sample resulted negative for the N, RNP, and ORF1ab genes (Figure 1B). This demonstrates that terminal disinfection is an effective method to eliminate contaminations of SARS-CoV-2 nucleic acid in a testing laboratory. In addition, it is worth mentioning that after the computer keyboard resulted positive for the N gene, all specimens from patients resulting positive only for the N gene on the same day were re-collected and re-tested in order to avoid false positives.
A study by Guo et al.[9] has demonstrated that SARS-CoV-2 nucleic acid was widely distributed in the air and on the surfaces of objects in ICUs and common infection wards were new coronavirus patients were cared for. Similarly, the extracted nucleic acid of SARS-CoV-2 might contaminate the testing laboratory environment. Consequently, the staff of nucleic acid testing laboratories might come in contact with the extracted nucleic acid of SARS-CoV-2, and thus they should strictly follow the laboratory rules and standardize operations to avoid potential exposure to the virus. In particular, terminal disinfection can significantly eliminate SARS-CoV-2 contaminations.
In summary, the environment of a SARS-CoV-2 nucleic acid testing laboratory was examined. The surface of the objects frequently touched in the testing laboratory could be easily contaminated by nucleic acid of the new coronavirus. Therefore, the laboratory personnel should be more attentive to the operational details of sample handling such as sample transfer and handover. In addition, the personnel should change gloves multiple times to reduce the risk of contamination with SARS-CoV-2 nucleic acid in the testing laboratory environment.
The authors have no conflict of interest to declare.
doi: 10.3967/bes2020.102
-
-
Table 1. Results of environment monitoring of a SARS-CoV-2 testing laboratory
Room Monitoring site Number of samples Positive samples FAM channel (ORF1ab) VIC channel (N) ROX channel (internal
reference RNP)Preparation room Test bed 10 0 0 0 Handle of biosafety chamber 10 0 0 Sample processing room Counter of biosafety chamber 10 0 0 Handle of biosafety chamber 10 0 0 Nucleic acid extractor 10 0 0 Pipette 10 0 0 Test tube rack 10 0 0 2 Oscillator 10 0 0 Centrifuge 10 0 0 Refrigerator handle 10 0 0 2 Door handle 10 0 0 2 Air 10 0 0 Amplification Room qPCR instrument 10 0 0 Computer keyboard of qPCR instrument 10 0 1 1 Air 10 0 0 Analysis room Computer keyboard 10 0 0 Positive control 10 10 10 10 Negative Control 10 0 0 0 Note. The template for the positive control was a mixture of plasmids containing the ORF1ab and N genes and virus-like particles with a specific fragment of the RNase P (RNP) gene; the negative control was a saline solution without genetic material. -
[1] Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol, 2020; 5, 536−44. doi: 10.1038/s41564-020-0695-z [2] The Editors of Encyclopaedia Britannica. Coronavirus. https://www.britannica.com/science/coronavirus-virus-group. [2020-07-06]. [3] Ji JS. Origins of MERS-CoV, and lessons for 2019-nCoV. Lancet Planet Health, 2020; 4, E93. doi: 10.1016/S2542-5196(20)30032-2 [4] Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature, 2020; 579, 265−9. doi: 10.1038/s41586-020-2008-3 [5] World Health Organization. Clinical management of Severe Acute Respiratory Infection (SARI) when COVID-19 disease is suspected: interim guidance, 13 March 2020. Geneva: World Health Organization, 2020. [6] Centers for Disease Control and Prevention. Research use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primers and probes. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. [2020-04-24]. [7] NIOSH. Chlorine Dioxide. NIOSH pocket guide to chemical Hazards, 2020. https://www.cdc.gov/niosh/npg/npgd0116.html. [2020-04-24]. [8] Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem, 2020; 66, 549−55. doi: 10.1093/clinchem/hvaa029 [9] Guo ZD, Wang ZY, Zhang SF, et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis, 2020; 26. doi: 10.3201/eid2607.200885 -




 下载:
下载:



 Quick Links
Quick Links