-
The emergence and spread of a novel human coronavirus, SARS-CoV-2 have resulted in over 774,000,000 confirmed cases of coronavirus-19 disease (COVID-19), including over 7,000,000 deaths as of March 3, 2024. In China alone, there were 99,336,751 confirmed COVID-19 cases and 121,993 deaths. Over 292,000 new COVID-19 cases and 6,200 deaths have been reported between February 5 and March 3, 2024. Currently, reported cases do not accurately represent infection rates owing to a reduction in testing and reporting globally[1].
The 2019-nCoV caused by SARS-COV-2 infection, has propagated worldwide. Rapid and accurate viral nucleic acid testing is one of the main methods used to control the spread of the virus. RT-PCR is the gold standard for the diagnosis of 2019-nCoV[2]. It is highly sensitive and specific but requires professional operators, reagents, instruments, and testing laboratories. Furthermore, this method is time-consuming. The development of on-site rapid detection methods is needed for field screening of outbreak infections or in remote areas[3]. Point-of-care testing (POCT) offers a more suitable solution for primary medical units to conduct nucleic acid detection[4]. POCT comprises a compact detection instrument and a corresponding cartridge. Users simply add the sample to be tested into the cassette, while the instrument handles sample processing, amplification, result interpretation, and other essential steps, significantly simplifying nucleic acid detection. Previous studies have shown that nucleic acid amplification reactions are subject to interference by substances in the sample, such as hemoglobin, mucin, and drug residues. Therefore, the nucleic acids must be isolated before amplification.
In this study, we designed an integrated disposable device to achieve the switching of different liquids by rotating the solution chambers and mixing the solution by moving a syringe up and down. High-purity nucleic acids were retained via silicon film absorption. Similarly, nucleic acid amplification reagents are mixed by moving the syringe up and down and are transferred by rotating the solution chambers. The proposed method is simple and practical. In conjunction with rapid nucleic acid amplification reagents, high-quality nucleic acid extraction and amplification can be completed within 40 min. This can be performed by non-laboratory personnel with simple training at the sample collection site. It is suitable for remote locations, such as airports and communities, where access to laboratory-testing infrastructure is limited.
-
Lysis solution: 4 mol/L guanidine hydrochloride, 1% NP-40, 50 mmol/L Tris/HCl buffer, 20 mmol/L sodium citrate, pH 8.0; wash buffer 1:20% absolute ethanol, 900 mmol/L GITC, 10 mmol/L Tris/HCl buffer, pH 7.5; wash buffer 2: acetone and absolute ethanol at a ratio of 3:1; elution buffer: TE buffer (10 mmol/L Tris/HCl buffer, 1 mmol/L EDTA, pH 8.0).
-
QIAamp MinElute Virus Spin Kit (57704, Qiagen); 2019-nCoV nucleic acid detection kit (Fluorescent RT-RAA) (F02R13, Jiangsu Qitian Gene Biotechnology Co., Ltd.); One Step PrimeScript™ RT-PCR Kit (Perfect Real Time) (RR064A, TAKARA); One-step Digital RT-PCR Mix (Sniper Medical Technology Co., Ltd.).
-
Sniper DQ24 Digital PCR System (Sniper Medical Technology Co., Ltd.) and Archimed X6 time-resolved real-time fluorescent quantitative PCR system (Rocgene (Beijing) Technology Co., Ltd.) were used.
-
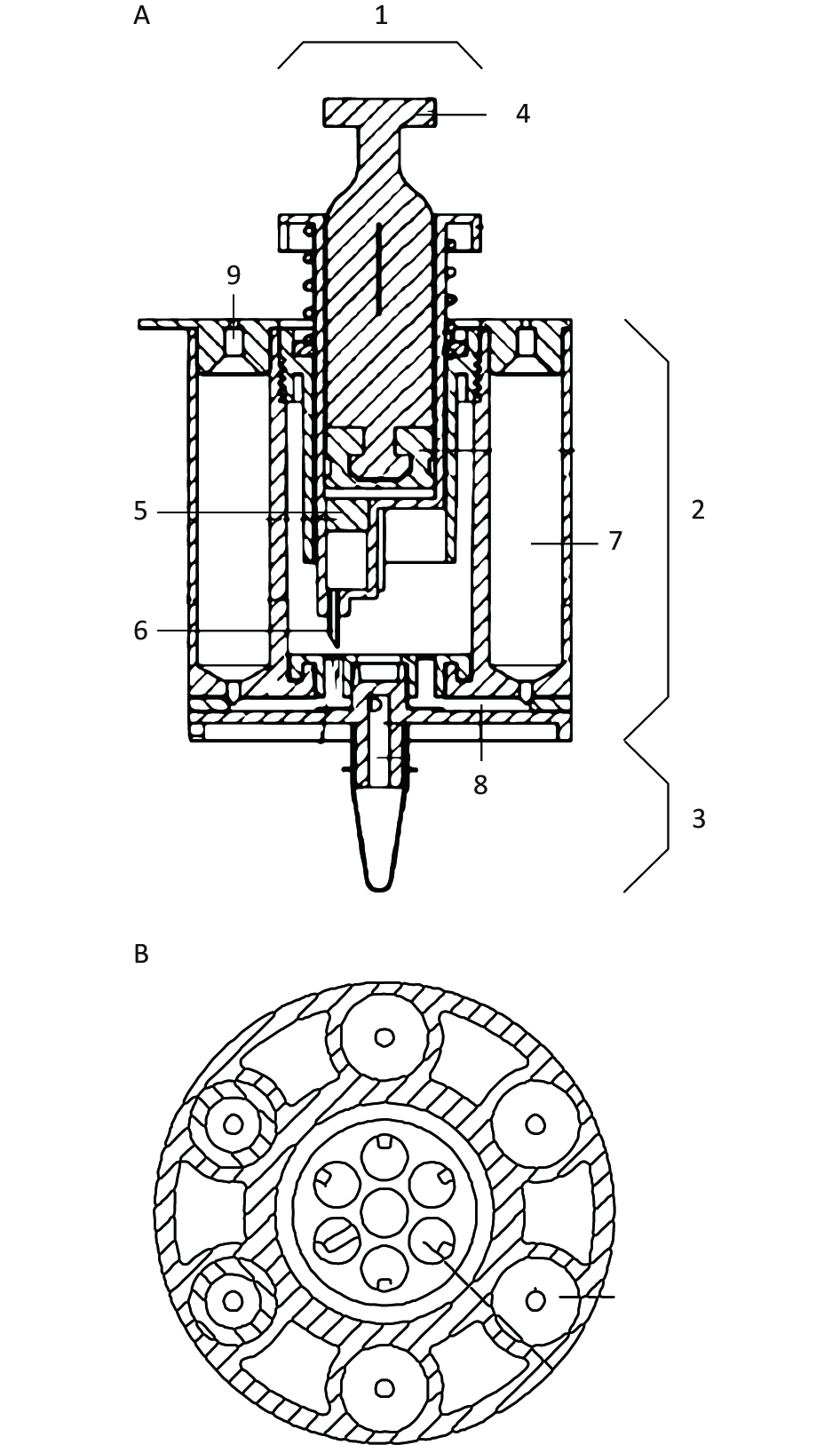
An integrated disposable device consisting of a nucleic acid extraction unit and nucleic acid amplification unit was created. The nucleic acid extraction unit included a syringe with a silicone film, a spring, a sealing ring, microtubes, and solution chambers. The nucleic acid amplification unit consisted of an amplification tube, temperature control module, and fluorescence detection module.
The silicone film was placed in a syringe, and the nucleic acid was absorbed. A syringe needle was positioned at the top of the microtube opening at equal intervals and sealed using a silicone rubber plug. The solution chambers were axially parallel to each other and arranged in a wheeled shape. The solution chambers were connected to a silicone rubber plug to seal the top of the solution chamber, and the microtubes were connected to the other end. The relative rotation between the syringe and solution chambers, along with the upward and downward movement of the syringe needle, facilitated the switching between different liquids. The solution was mixed by moving the push-pull rod up and down. The combination of these processes resulted in the purification and amplification of nucleic acids.
The five solution chambers of the nucleic acid extraction unit were pre-filled with the lysis solution, wash buffer 1, wash buffer 2, elution buffer, and nucleic acid amplification reaction mix. Once the sample was added to the first solution chamber of the integrated disposable device, the nucleic acid extraction was initiated. The syringe needle was positioned at the opening of the microtube linked to the first solution chamber by mechanical rotation and by pushing it down on the syringe, which was connected to the first solution chamber. By moving the push-pull rod up and down, the lysis buffer containing the sample was mixed, and the nucleic acid was absorbed onto the silicone film. After the syringe was emptied, it was drawn up, removed, and connected to the other solution chambers, and the previous up-and-down movements were repeated. Finally, 50 µL of mixed amplification reaction solution in the fifth solution chamber was injected into the amplification tube. Nucleic acid amplification was performed according to a predetermined program wherein the temperature control module provided the appropriate temperature and reaction time, and the fluorescence detection module monitored the fluorescence signals. The resulting data were displayed in a concise format.
-
After adding 500 µL deionized water to the first solution chamber of the integrated disposable device, the syringe was connected to the first chamber. Precise solution suction and discharge of the syringe transfer were measured using a weighing method, with set volumes of 20, 30, and 60 µL respectively. Each measurement was tested separately 10 times and the mean value was calculated from the resulting values.
-
A temperature of 60 °C and 95 °C was set, and the temperature of a 50 µL mixed amplification reaction solution in the amplification tube was measured using a thermocouple thermometer. The mean value was calculated from ten measured values.
-
The reagents were added to the solution chambers or amplification tubes, as shown in Table 1. The openings of the solution chambers were sealed with silicone rubber plugs to complete the integrated disposable device for SARS-CoV-2 nucleic acid extraction and amplification.
Table 1. Reagents used for the integrated disposable SARS-CoV-2 nucleic acid extraction and amplification device
Number of solution chambers
or amplification tubeSolution 1 300 µL of lysis buffer and 250 µL of absolute ethanol 2 800 µL of wash buffer 1 3 1,200 µL of wash buffer 2 4 200 µL of TE Buffer 5 SARS-Cov-2 nucleic acid amplification solution Amplification tube SARS-Cov-2 nucleic acid amplification mix (powder) -
A recombinant lentivirus containing the N gene 28273-29557 sequence of SARS-CoV-2 (Accession Number: NC_045512) was constructed. The sequence was then synthesized and inserted into the plvx-puro plasmid. The correct plasmid clone (plvx-SARS2-N) was selected using restriction enzyme digestion and sequencing. The plvx-SARS2-N and lentiviral backbone plasmids were co-transfected into 293T cells. The culture medium was replaced at 8 h post-transfection. The cell culture supernatants were collected at 48 and 72 h. The cell culture supernatant was centrifuged at 3,000 ×g for 30 min, filtered through a 0.45 µmol/L syringe filter, and centrifuged at 80,000 ×g for 1.5 h. The pellet was resuspended in PBS and residual nucleic acid was removed using BeyoZonase™ Super Nuclease. The recombinant lentivirus was aliquoted and stored at −80 °C.
-
Digital RT-PCR of SARS-CoV-2 nucleic acids was performed using a One-step Digital RT-PCR Mix (Sniper Medical Technology Co., Ltd.). For each reaction, master mix (11 μL), enzyme mix (2 μL), forward and reverse primers (1 μL each, 500 nmol/L), and probe (0.5 μL, 250 nmol/L) were added to 6.5 μL RNA. The reaction conditions were as follows: 50 °C for 5 min and 95 °C for 3 min for reverse transcription, followed by 40 cycles of denaturation at 95 °C for 30 s and annealing/extension at 60 °C for 45 s. The copy number of SARS-CoV-2 nucleic acids was calculated and used as a quantitative standard for the absolute quantitative RT-PCR detection of SARS-CoV-2 nucleic acids.
-
After adding 600 µL of the recombinant lentivirus carrying the SARS-COV-2 N gene into the first solution chamber of the integrated disposable device, nucleic acid extraction was done as described above as follows: 1) The syringe was connected to the first solution chamber and the lysis buffer containing the sample was mixed and absorbed onto the silicone film by moving a push-pull rod up and down 40 times; 2) The syringe was connected to the second solution chamber and the push-pull rod was moved up and down 10 times; 3) The syringe was connected to the third solution chamber and the push-pull rod was moved up and down 10 times; and 4) The syringe was connected to the fourth solution chamber and the push-pull rod was moved up and down 10 times to obtain the SARS-CoV-2 nucleic acid.
-
Nucleic acids were extracted from an equivalent amount of recombinant lentivirus carrying the SARS-COV-2 N gene based on the manufacturer’s instructions (QIAamp MinElute Virus Spin Kit, Qiagen).
-
Fluorescent RT-qPCR amplification of the SARS-CoV-2 nucleic acid was done using a one-step RT-qPCR reagent (TAKARA). For each reaction, 2× One-Step RT-PCR Buffer III (10 μL), TaKaRa Ex Taq HS (0.4 μL), PrimeScript RT Enzyme Mix II (0.4 μL), forward and reverse primers (0.4 μL each, 200 nmol/L), and probe (0.8 μL, 400 nmol/L) were added to 7.6 μL RNA. The reaction conditions were as follows: 50 °C for 5 min and 95 °C for 3 min for reverse transcription, followed by 40 cycles of denaturation at 95 °C for 30 s and annealing/extension at 60 °C for 45 s. Nucleic acids detected by digital RT-PCR were used to establish a standard curve, and the copy numbers of the nucleic acids extracted by the two methods were calculated.
-
The quantified SARS-CoV-2 nucleic acids were diluted to 10,000, 1,000, 500, 250, and 50 copies/mL. Nucleic acid extraction and amplification were performed using an integrated disposable device assembled as described above. Nucleic acids were extracted as previously described. Then, 50 μL of mixed amplification reaction solution in the fifth solution chamber was injected into the amplification tube to dissolve the RAA mix powder. The temperature control module maintained the amplification reaction solution at 42 °C for 600 s and the fluorescence detection module collected measured signals every 10 s. The resulting data were displayed in a concise format. The mean value was calculated from three measurements.
-
The structure of the integrated disposable device is shown in Figures 1 and 2.
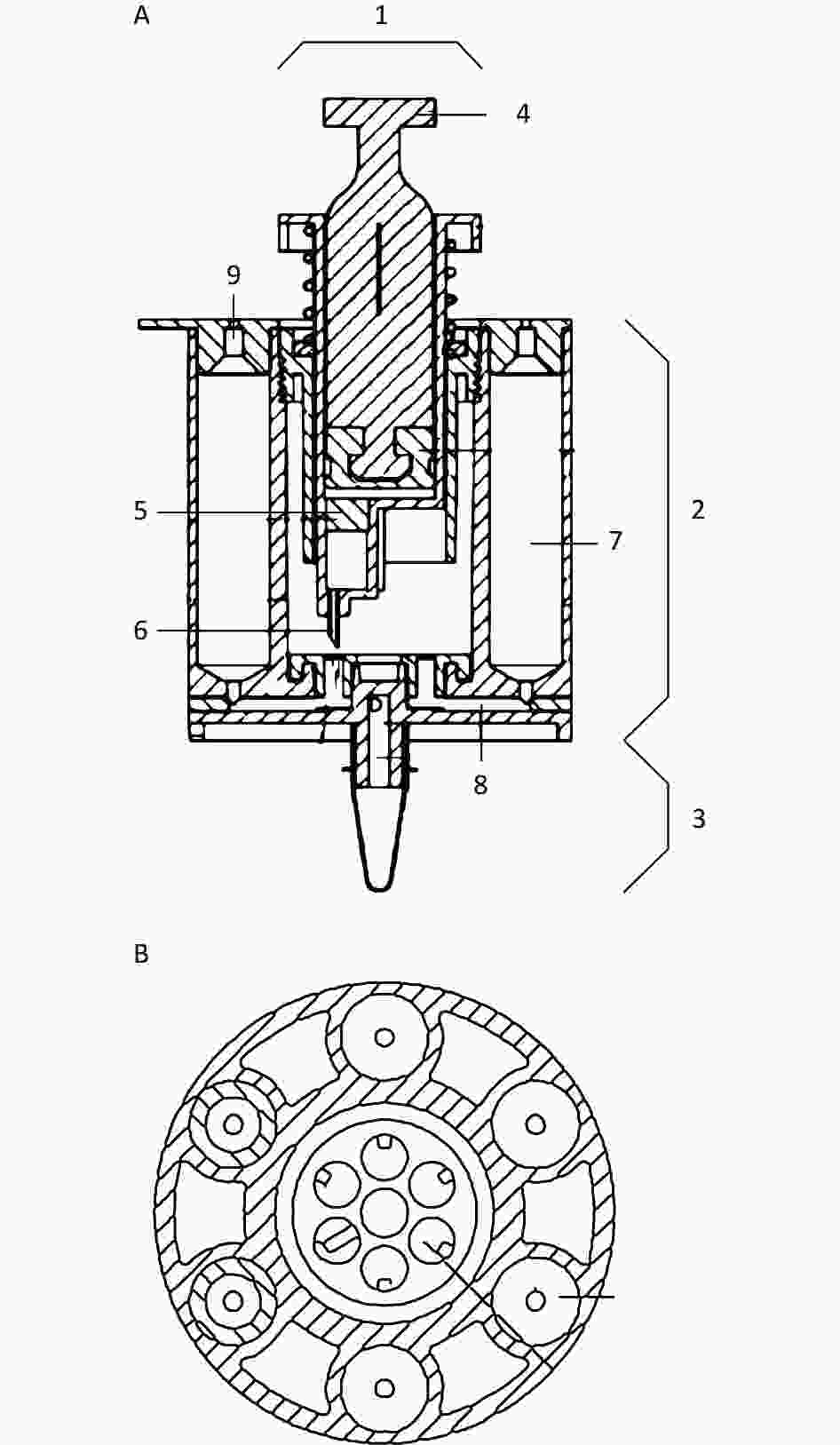
Figure 1. The schematic view and top view of the integrated disposable device. A) a schematic view of the integrated disposable device; B) a top view of the integrated disposable device. 1: syringe; 2: nucleic acid extraction unit; 3: nucleic acid amplification unit; 4: push-pull rod; 5: silicone film; 6: syringe needle; 7: solution chamber; 8: microtube; 9: silicone rubber plug.
-
The solution volumes for syringe suction and discharge were set to 20, 30, and 60 μL. The mean values calculated from the measurements were 19.2, 32.2, and 57.2 μL, respectively. The standard deviation is < 10% (Table 2).
Table 2. The measured values of liquid transfer
Set volume (μL) Measured volume (μL) Mean ± SD (μL) 1 2 3 4 5 6 7 8 9 10 20 22 20 18 17 18 21 19 22 17 18 19.2 ± 1.9 30 30 32 34 34 34 31 30 31 33 33 32.2 ± 1.6 60 55 57 57 62 57 56 57 62 50 59 57.2 ± 3.5 -
The temperature of the amplification reaction mix was set to 60 °C and 95 °C. The mean values calculated from ten measurements were 60.0 °C and 95.1 °C, respectively. The standard deviation is < 1% (Table 3).
Table 3. The measured values of temperature
Set value
(°C)Measured value (°C) Mean ± SD
(°C)1 2 3 4 5 6 7 8 9 10 60 60.0 60.0 60.0 60.0 60.0 60.0 60.0 60.0 60.0 60.0 60.0 ± 0.0 95 95.2 95.3 94.9 95.2 95.0 94.9 95.1 95.0 95.3 94.9 95.1 ± 0.2 -
SARS-CoV-2 nucleic acid was measured at 1.12 × 108 copies/mL by digital RT-PCR (Figure 3) and used as a quantitative standard for absolute quantitative RT-PCR.
-
The copy numbers of SARS-CoV-2 nucleic acids from the two nucleic acid extraction methods were determined using absolute quantitative RT-PCR. The nucleic acid extracted by the integrated disposable device was 7.10 × 106 copies/mL, whereas that extracted by the commercial kit was 2.98 × 106 copies/mL (Table 4 and Figure 4). The amount of nucleic acid extracted by the integrated disposable device was higher than that extracted using a commercial kit.
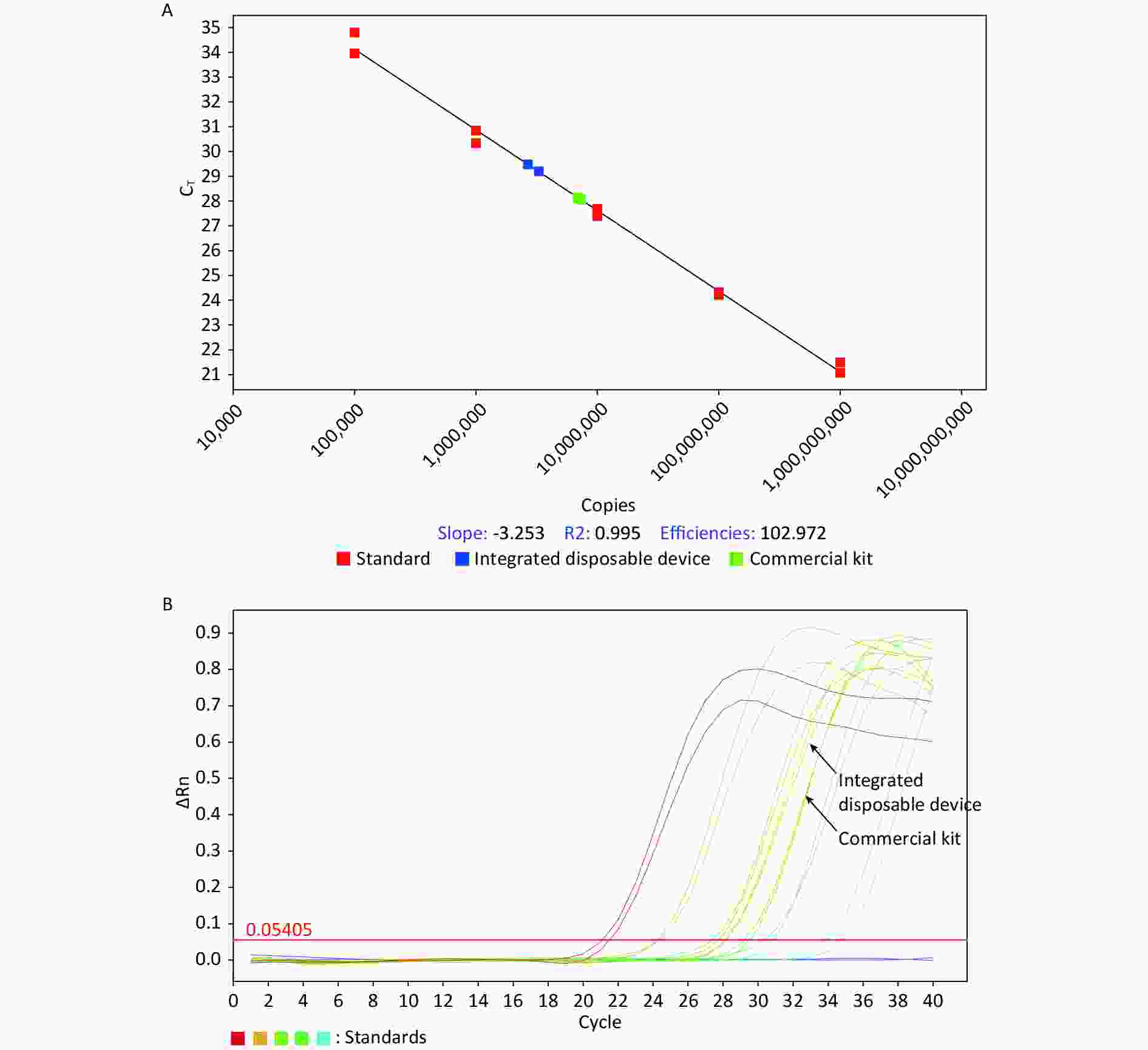
Figure 4. Absolute quantitative RT-PCR detection of SARS-CoV-2 nucleic acid extracted by the integrated disposable device and a commercial kit.
Table 4. Copy number of SARS-CoV-2 nucleic acid extracted by the integrated disposable device and commercial kit
Extraction method Copy number 1 (copies/mL) Copy number 2 (copies/mL) Mean value (copies/mL) Integrated disposable device 7.12 × 106 7.08 × 106 7.10 × 106 Commercial kit 3.28 × 106 2.68 × 106 2.98 × 106 -
The mean times to complete SARS-CoV-2 nucleic acid extraction and amplification were 36 min and 45 s (2,187 s), respectively. Amplification curves were obtained with an initial concentration ranging from 250 to 1 × 104 copies/mL, and two of the three reactions at 50 copies/mL. No curves were observed at the initial concentration of 0 (Figure 5 and Table 5). The lower limit of detection was 250 copies/mL.
Table 5. The result of the absolute quantitative RT-PCR test of SARS-CoV-2 nucleic acid
Number Initial Concentration
(copies/mL)Result† 1 2 3 1 1 × 104 + + + 2 1 × 103 + + + 3 5 × 102 + + + 4 250 + + + 5 50 + - + 6 0 - - - Note. †Each test was repeated three times. Positive results are marked as “+”, and negative results are marked as “-.” -
The SARS-CoV-2 virus epidemic that occurred in the winter of 2019 has brought unprecedented challenges to pathogen detection. Rapid and accurate SARS-CoV-2 testing is necessary to establish appropriate measures for epidemic prevention. In China, nucleic acid detection is the primary method for diagnosing novel coronavirus pneumonia[5] and laboratory RT-qPCR is the gold standard for SARS-CoV-2 nucleic acid detection[6]. This method enables high-throughput detection under laboratory conditions by using automated equipment. However, potential exposure to the virus is a concern, as the environment of a SARS-CoV-2 nucleic acid testing laboratory could be contaminated by the virus’s nucleic acids[7]. In scenarios requiring rapid testing, such as airports, train stations, and emergency departments, the POCT testing method proves to be a superior solution. Health Canada has approved a new coronavirus POCT test method for use in symptomatic individuals and has provided testing guidelines[8]. Recombinase-mediated isothermal nucleic acid amplification technology (recombinase-aided amplification, RAA) uses recombinase, single-stranded binding proteins, and DNA polymerase to amplify nucleic acids under isothermal conditions[9-10]. At optimal reaction temperatures, efficient amplification of the target gene can be achieved within 5–20 min of template synthesis, whereas the detection limit of most pathogens is 500 copies/mL[11-13]. However, the RAA reaction system is highly sensitive to interfering substances and requires high-purity nucleic acids. Nucleic acids obtained by extraction-free processing methods used for most POCT usually contain high concentrations of surfactants or salt ions. These substances affect the amplification efficiency of the RAA reaction, resulting in missed detections. Therefore, the use of magnetic beads or silica gel membranes to purify nucleic acid samples is necessary for RAA amplification systems.
Five regions in the SARS-CoV-2 genome are widely used for designing primers, including the nucleocapsid (N), envelope (E), RNA polymerase-dependent RNA (RdRp), ORF1ab, and Spike (S). It has been proven that ORF1ab, N, and RdRp have higher sensitivity, specificity, and positive predictive values than the others[14]. Furthermore, the N gene is generally considered a suitable target for in vitro diagnostic detection because of the conservation of its sequence[15]. The N gene is one of the targets selected for in vitro diagnostic detection by the Chinese Center for Disease Control and Prevention and most commercial companies[16]. We chose the N gene as the target to establish a method for SARS-CoV-2 nucleic acid.
In the present study, we integrated silica gel membrane nucleic acid extraction and rapid amplification of RAAs into an integrated cartridge that automatically extracted nucleic acids after adding the sample. It was designed to have a hermetically sealed structure and allow safe handling of potentially infectious samples. Purified nucleic acids were used for the subsequent RAA rapid amplification. Thus, the SARS-CoV-2 nucleic acid detection method established in this study could complete the entire nucleic acid extraction and amplification process within 40 min. It can effectively detect 250 copies/mL of a recombinant virus containing the SARS-Cov-2 nucleic acid fragments. This is comparable to the results of multiple commercial SARS-Cov-2 nucleic acid POCTs[17].
Thus, nucleic acid extraction is a major concern. Its quantity and quality can be considered vital factors for diagnosis. The integrated disposable device used a silica gel membrane to adsorb nucleic acids, similar to the QIAamp MinElute Virus Spin Kit (control kit). Both methods included four steps: lysis, binding, washing, and elution. In the present study, the amount of nucleic acid extracted by the device was higher than that extracted using the control kit (7.10 × 106 copies/mL and 2.98 × 106 copies/mL, respectively). This may be due to small losses of nucleic acids in the fluid shift from lysis to binding in the control kit, whereas it is not necessary to transfer nucleic acids in the device because lysis and binding are the same steps.
SARS-CoV-2 has exhibited high genetic diversity in the last few years[18], which may cause diagnostic escape in SARS-CoV-2. Most assays use several targets, including the N gene, to achieve high sensitivity. This is the work what we should to do next. Subsequently, the clinical samples were tested.
SARS-CoV-2 will not be the last global pandemic pathogen encountered by humans. In the long term, stockpiling rapid pathogen detection technologies will be an effective way to manage future pandemics. The rapid detection method established in this study is not only suitable for the on-site rapid detection of SARS-Cov-2, but also provides a basis for next-generation POCT detection methods.
doi: 10.3967/bes2024.070
Development of an Integrated Disposable Device for SARS-CoV-2 Nucleic Acid Extraction and Detection
-
Abstract:
Objective To develop a highly sensitive and rapid nucleic acid detection method for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Methods We designed, developed, and manufactured an integrated disposable device for SARS-CoV-2 nucleic acid extraction and detection. The precision of the liquid transfer and temperature control was tested. A comparison between our device and a commercial kit for SARS-Cov-2 nucleic acid extraction was performed using real-time fluorescence reverse transcription polymerase chain reaction (RT-PCR). The entire process, from SARS-CoV-2 nucleic acid extraction to amplification, was evaluated. Results The precision of the syringe transfer volume was 19.2 ± 1.9 μL (set value was 20), 32.2 ± 1.6 (set value was 30), and 57.2 ± 3.5 (set value was 60). Temperature control in the amplification tube was measured at 60.0 ± 0.0 °C (set value was 60) and 95.1 ± 0.2 °C (set value was 95) respectively. SARS-Cov-2 nucleic acid extraction yield through the device was 7.10 × 106 copies/mL, while a commercial kit yielded 2.98 × 106 copies/mL. The mean time to complete the entire assay, from SARS-CoV-2 nucleic acid extraction to amplification detection, was 36 min and 45 s. The detection limit for SARS-CoV-2 nucleic acid was 250 copies/mL. Conclusion The integrated disposable devices may be used for SARS-CoV-2 Point-of-Care test (POCT). -
Key words:
- An integrated disposable device /
- SARS-Cov-2 /
- Nucleic acid detection
&These authors contributed equally to this work.
注释:1) AUTHOR CONTRIBUTIONS: -
Figure 1. The schematic view and top view of the integrated disposable device. A) a schematic view of the integrated disposable device; B) a top view of the integrated disposable device. 1: syringe; 2: nucleic acid extraction unit; 3: nucleic acid amplification unit; 4: push-pull rod; 5: silicone film; 6: syringe needle; 7: solution chamber; 8: microtube; 9: silicone rubber plug.
Table 1. Reagents used for the integrated disposable SARS-CoV-2 nucleic acid extraction and amplification device
Number of solution chambers
or amplification tubeSolution 1 300 µL of lysis buffer and 250 µL of absolute ethanol 2 800 µL of wash buffer 1 3 1,200 µL of wash buffer 2 4 200 µL of TE Buffer 5 SARS-Cov-2 nucleic acid amplification solution Amplification tube SARS-Cov-2 nucleic acid amplification mix (powder) Table 2. The measured values of liquid transfer
Set volume (μL) Measured volume (μL) Mean ± SD (μL) 1 2 3 4 5 6 7 8 9 10 20 22 20 18 17 18 21 19 22 17 18 19.2 ± 1.9 30 30 32 34 34 34 31 30 31 33 33 32.2 ± 1.6 60 55 57 57 62 57 56 57 62 50 59 57.2 ± 3.5 Table 3. The measured values of temperature
Set value
(°C)Measured value (°C) Mean ± SD
(°C)1 2 3 4 5 6 7 8 9 10 60 60.0 60.0 60.0 60.0 60.0 60.0 60.0 60.0 60.0 60.0 60.0 ± 0.0 95 95.2 95.3 94.9 95.2 95.0 94.9 95.1 95.0 95.3 94.9 95.1 ± 0.2 Table 4. Copy number of SARS-CoV-2 nucleic acid extracted by the integrated disposable device and commercial kit
Extraction method Copy number 1 (copies/mL) Copy number 2 (copies/mL) Mean value (copies/mL) Integrated disposable device 7.12 × 106 7.08 × 106 7.10 × 106 Commercial kit 3.28 × 106 2.68 × 106 2.98 × 106 Table 5. The result of the absolute quantitative RT-PCR test of SARS-CoV-2 nucleic acid
Number Initial Concentration
(copies/mL)Result† 1 2 3 1 1 × 104 + + + 2 1 × 103 + + + 3 5 × 102 + + + 4 250 + + + 5 50 + - + 6 0 - - - Note. †Each test was repeated three times. Positive results are marked as “+”, and negative results are marked as “-.” -
[1] Epidemic and Pandemic Preparedness and Prevention (EPP) TEAM. COVID-19 epidemiological update – 15 March 2024. https://www.who.int/publications/m/item/covid-19-epidemiological-update-15-march-2024. [2024-3-15] [2] Eftekhari A, Alipour M, Chodari L, et al. A comprehensive review of detection methods for SARS-CoV-2. Microorganisms, 2021; 9, 232. doi: 10.3390/microorganisms9020232 [3] Gupta N, Augustine S, Narayan T, et al. Point-of-care PCR assays for COVID-19 detection. Biosensors, 2021; 11, 141. doi: 10.3390/bios11050141 [4] Luppa PB, Müller C, Schlichtiger A, et al. Point-of-care testing (POCT): current techniques and future perspectives. TrAC Trends Anal Chem, 2011; 30, 887−98. doi: 10.1016/j.trac.2011.01.019 [5] The Office of the National Health Commission, Office of the State Administration of Traditional Chinese Medicine. Diagnosis and treatment plan for novel coronavirus pneumonia (trial version 9). National Health Office Medical Letter [2022] No. 71. https://www.gov.cn/zhengce/zhengceku/2022-03/15/content_5679257.htm. [2022-03-14]. (In Chinese [6] The State Council Responds to the Epidemic Situation of Novel Coronavirus Pneumonia, Joint Prevention and Control Mechanism Medical Treatment Team. Work manual for novel coronavirus nucleic acid testing in medical institutions (trial version 2). Joint Prevention and Control Mechanism Medical [2020] No. 313. [2020-12-28]. (In Chinese [7] Ning PY, Yu AP, Wang Y, et al. Environmental monitoring of a laboratory for new coronavirus nucleic acid testing. Biomed Environ Sci, 2020; 33, 771−4. [8] Taher J, Randell EW, Arnoldo S, et al. Canadian Society of Clinical Chemists (CSCC) consensus guidance for testing, selection and quality management of SARS-CoV-2 point-of-care tests. Clin Biochem, 2021; 95, 1−12. doi: 10.1016/j.clinbiochem.2021.05.010 [9] Zhang XY, He XY, Zhang YB, et al. A new method for the detection of Mycobacterium tuberculosis based on the CRISPR/Cas system. BMC Infect Dis, 2023; 23, 680. doi: 10.1186/s12879-023-08656-4 [10] Lü B, Cheng HR, Yan QF, et al. Recombinase-aid amplification: a novel technology of in vitro rapid nucleic acid amplification. Sci Sin Vitae, 2010; 40, 983−8. (In Chinese doi: 10.1360/zc2010-40-10-983 [11] Zhao KC, Cui LB, Ge YY, et al. Detection of a novel avian-origin influenza A (H7N9) virus by recombinase aided amplification. Jiangsu J Prev Med, 2016; 27, 524−30. (In Chinese [12] Zhou DG, Luo J, Chen JH, et al. Development and application of a RT-RAA assay to detect middle east respiratory syndrome coronavirus. Chin J Virol, 2018; 34, 45−51. (In Chinese [13] Wei Y, Guo LC, Zhang XP, et al. Establishment of Recombinase aided amplification assay for group A streptococcus pyogens detection. Chin J Frontier Health Quar, 2018; 41, 314−6,323. (In Chinese [14] Mollaei HR, Afshar AA, Kalantar-Neyestanaki D, et al. Comparison five primer sets from different genome region of COVID-19 for detection of virus infection by conventional RT-PCR. Iran J Microbiol, 2020; 12, 185−93. [15] Dutta NK, Mazumdar K, Gordy JT. The nucleocapsid protein of SARS-CoV-2: a target for vaccine development. J Virol, 2020; 94, e00647−20. [16] Chinese Center for Disease Control and Prevention. Chinese Center for Disease Control and Prevention, technical guidelines for laboratory testing of novel coronavirus pneumonia. https://www.chinacdc.cn/jkzt/crb/zl/szkb_11803/jszl_11815/202003/t20200309_214241.html. (In Chinese [17] Tang YN, Jiang DD, Wang XJ, et al. Recent progress on rapid diagnosis of COVID-19 by point-of-care testing platforms. Chin Chem Lett, 2024; 35, 108688. doi: 10.1016/j.cclet.2023.108688 [18] Liang D, Wang T, Li JJ, et al. Genomic epidemiology of imported cases of COVID-19 in Guangdong province, China, October 2020 - May 2021. Biomed Environ Sci, 2022; 35, 393−401. -




 下载:
下载:




 Quick Links
Quick Links