-
Anesthesiologists play a decisive role in determining the mode of anesthesia, maintaining the stability of patients' intraoperative vital signs and implementing postoperative analgesia, and also in reducing healthcare-associated infections. Several studies have shown that anesthesiologists may still lack awareness in operating room infection control and self-monitoring of hand hygiene [1-6]. In China, very few studies concentrated on the effectiveness of hand hygiene among anesthesiologists or microorganisms contamination in anesthesia environments. In 2012 and 2019, China issued “Regulation of disinfection technique in healthcare settings” (WS/T 367-2012) and “Specification of hand hygiene for healthcare workers” (WS/T 313-2019) [7,8], respectively, which stipulated the maximum number of total viable counts monitored after environment disinfection and hand disinfection. Although the regulations were issued, not every health worker can comprehend and implement properly, nor the monitor is timely. In particular, the specific colony count also requires a lot of work to do, and not every hospital can meet the standard. Obtaining the baseline levels of microorganisms' growth on the hands of health workers and in the environment before and after disinfection is really important for the local government to set or adjust the standards or guidelines, but the data in China are still unclear. In this study, we only focused on the neglected population anesthesiologists, and tried to establish the baseline levels of microorganisms' growth on the hands of anesthesiologists and in the anesthesia environment from a single center’s point of view.
To characterize the baseline levels of contamination on anesthesiologists' hands and in the anesthesia environment, we conducted a prospective observational study in nine operating rooms and on 25 anesthesiologists at a cancer hospital in 2021. We further studied the effectiveness of the decontamination on providers’ hands and in the anesthesia environment to help understand the necessity and practical frequency of decontaminating behaviors.
-
The study was approved by the appropriate Institutional Review Board (IRB), and the requirement for written informed consent was waived by the IRB.
-
This was an observational trial on a convenience sample performed in nine operating rooms and among 25 anesthesiologists at a cancer hospital over 2 consecutive months (June and July 2021). Sampling of the hands of these anesthesiologists and the anesthesia environment was performed at a ready-to-use operating room before patient contact began. After decontamination of hands (Apply a palmful of 70% alcohol and cover all surfaces of the hands. Rub hands more than 15 seconds until dry) and anesthesia environments (using a 5.5%–6.5% chlorine solution), sampling was repeated in 2 minutes after dry to help understand the effectiveness of decontaminating behaviors.
A designated investigator sampled and recorded in the present study, he was notified to enter the operating room (randomly chosen by lot) in the morning of working days before all anesthesiologists and cleaning staff were informed. Considering the influence of different operation times on the results, only the first operation was employed for observation, and the operation time should be greater than 4 hours because the working table should be monitored at least 4 hours. The arrangement of the anesthesiologists and the operating room is carried out according to the normal schedule, some unexpected situations such as temporary shifts, cancellation of surgery, which leading to a situation where the same anesthesiologist entered the same operating room twice in different observational days should be excluded in analysis to make sure that all anesthesiologists and operating rooms were sampled and analyzed only once, but the inoculation on agar plates was duplicated. All anesthesiologists and cleaning staff could access his/her result and the best practice guidance[7, 8] after the investigation was finished.
-
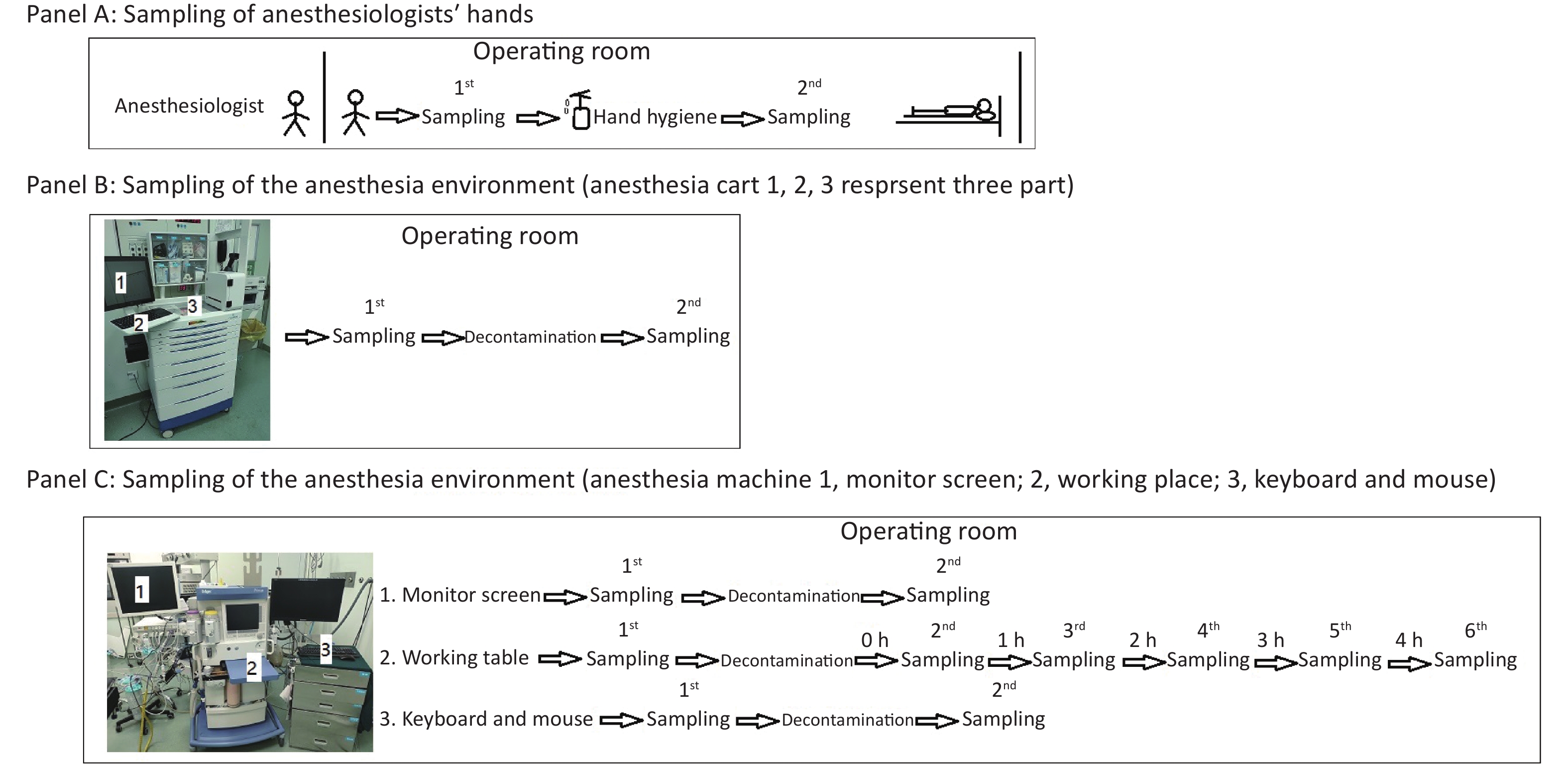
As depicted in Figure 1, we first obtained bacterial or fungal cultures from the hands of anesthesiologists when they entered the operating room before contacting any surfaces to determine if they had performed active decontamination on their hands (WS/T 313-2019[8], handwashing at least 15 seconds) before entering the operating room (Figure 1, Panel A). After that, their hands were decontaminated with 70% alcohol in the standard manner from WS/T 313-2019 and WHO hand-hygiene guidelines (Apply a palmful of 70% alcohol and cover all surfaces of the hands. Rub hands more than 15 seconds until dry)[8], and then participants’ hands were sampled again for bacterial or fungal cultures to determine the effectiveness of decontaminating behavior in this situation (Figure 1, Panel A).
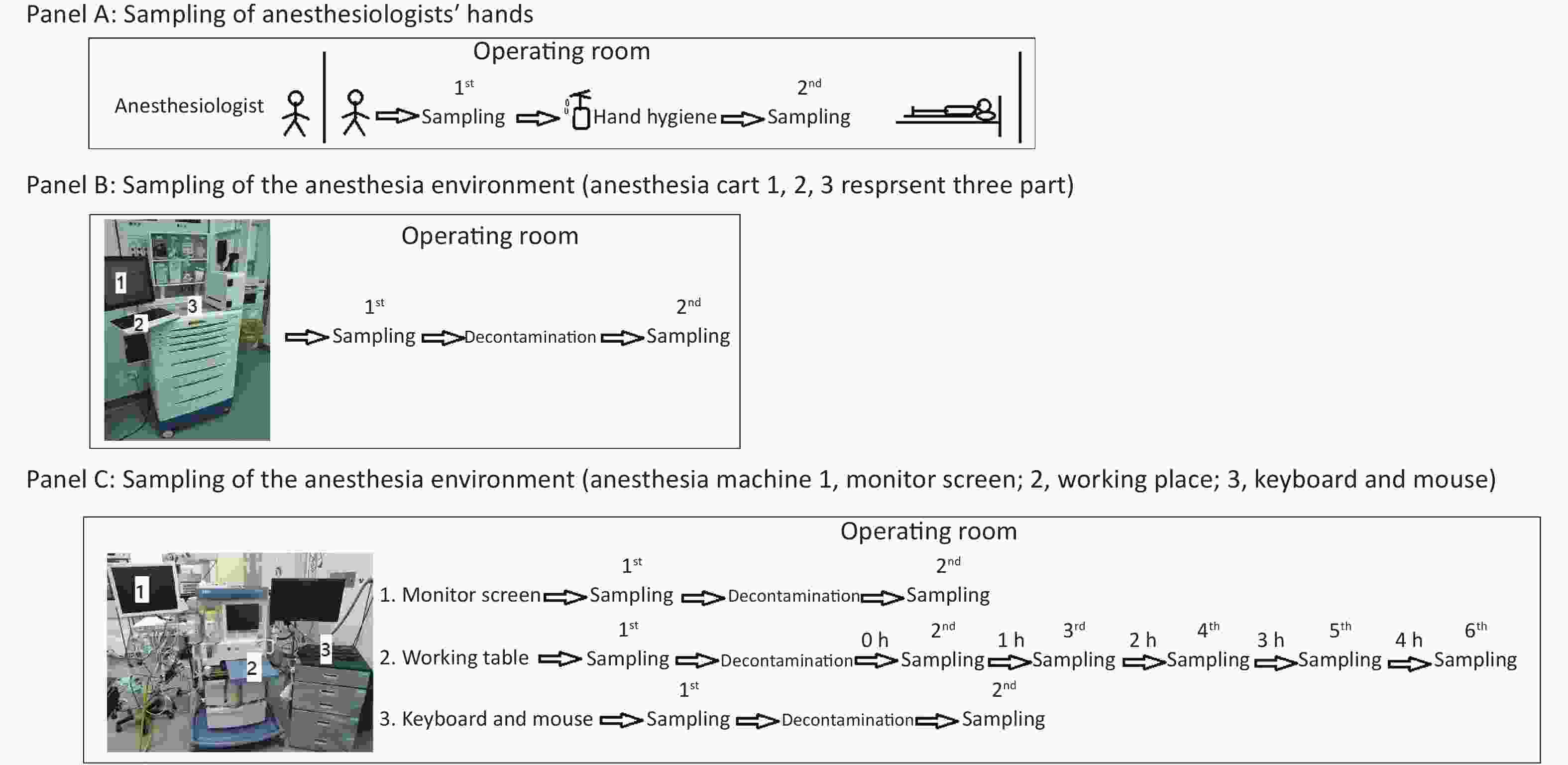
Figure 1. Protocol for sampling anesthesiologists’ hands (A), the anesthesia environment (B), and the anesthesia machine (C).
Secondly, in ready-to-use operating rooms, we also sampled for bacterial or fungal cultures three parts of the anesthesia cart (Figure 1, Panel B) that anesthesiologists typically touched very often. We sampled these three parts of the anesthesia cart again 30 minutes after decontamination with 5.5%–6.5% chlorine solution, to determine the necessity and effectiveness of decontaminating behavior in this situation (Figure 1, Panel B).
Thirdly, we sampled the monitor screen (Figure 1, Panel C 1), working table (Figure 1, Panel C 2), and keyboard and mouse (Figure 1, Panel C 3) of the anesthesia machine, the areas most frequently contacted by anesthesiologists. Thirty minutes after decontaminating them with 5.5%–6.5% chlorine solution, we sampled the three parts again to assess the necessity and effectiveness of decontaminating behavior in this situation (Figure 1, Panel C). In this study, we also monitored the microorganisms' growth sampled from the working table of the anesthesia machines at different time points to determine how soon there should be a repeat cleaning (Figure 1, Panel C).
-
The method of sampling the hands of anesthesiologists was based on the health industry standards of the People’s Republic of China: “Specification of hand hygiene for healthcare workers” (WS/T 313-2019)[8].
Per these specifications, the five fingers of the tested anesthesiologists are held close together, and a sterile fiber test piece soaked with sterile normal saline is rubbed back and forth on the finger surface of both hands from the finger root to the finger end twice (the wiping area of one hand is 30 cm2), and then the sampling swab is rotated, the test piece is immersed in the sampling tube containing preservation solution, the hand contact part is bent off, and the tube is covered. After the sampling tube is thoroughly shaken, 0.2 mL of eluent with different dilution times is inoculated on BHI agar plates (duplicates), smeared evenly with a sterilized L rod, then placed in a 35–37 °C incubator for 48 hours. The number of colonies is calculated in the sampling area of a square centimeter (cm2). For hand disinfection, the total number of bacterial or fungal colonies monitored should be ≤ 10 CFU per cm2 (WS/T 313-2019)[8].
-
The method of sampling the anesthesia environment was based on WS/T 367-2012 [7]. Per these specifications, for sampling surfaces less than 100 cm2, all surfaces are taken; for surfaces > 100 cm2, 100 cm2 is sampled. One sterile fiber swab soaked with sterile normal saline is applied to the surface to be tested, applied vertically and back and forth five times within the range of 5 cm × 5 cm, and then the swab is rotated to continuously sample 1 to 4 areas. The swab is immersed into the sampling tube containing preservation solution, the hand contact part is bent off, and the tube is covered. After the sampling tube is thoroughly shaken, 0.2 mL of eluent with different dilution times is inoculated on BHI agar plates (duplicates), smeared evenly with a sterilized L rod, then placed in a 35–37 °C incubator for 48 hours. The number of colonies is calculated within a sampling area of a square centimeter. For operating rooms, the total number of bacterial or fungal colonies observed should be ≤ 5 CFU/cm2 (WS/T 367-2012) [7].
-
Microorganisms recovered from anesthesiologists’ hands and the anesthesia environment were identified by MALDI-TOF MS as previously described[9]. The acquisition and analysis of mass spectra were performed by the M-Discover 100 MS (MS-ID version v3.2, Zhuhai Meihua Medical Technology Co., Ltd. China). Per the instructions of the M-Discover 100 MS [9], identification scores of ≥ 90 indicated species-level identification, scores of 60–90 indicated genus-level identification, and scores of ≤ 60 were considered “not reliable” (NRI).
-
Microorganisms’ growth from samples of anesthesiologists’ hands and anesthesia environments before and after disinfection were statistically evaluated employing the two-tailed Mann-Whitney test (Figures 2–3). Comparison of microorganisms’ growth of monitor screen, working table, and keyboard and mouse before and after disinfection were statistically evaluated using one-way ANOVA test (multiple comparisons) (Figures 4–5). A P-value < 0.05 was considered significant.
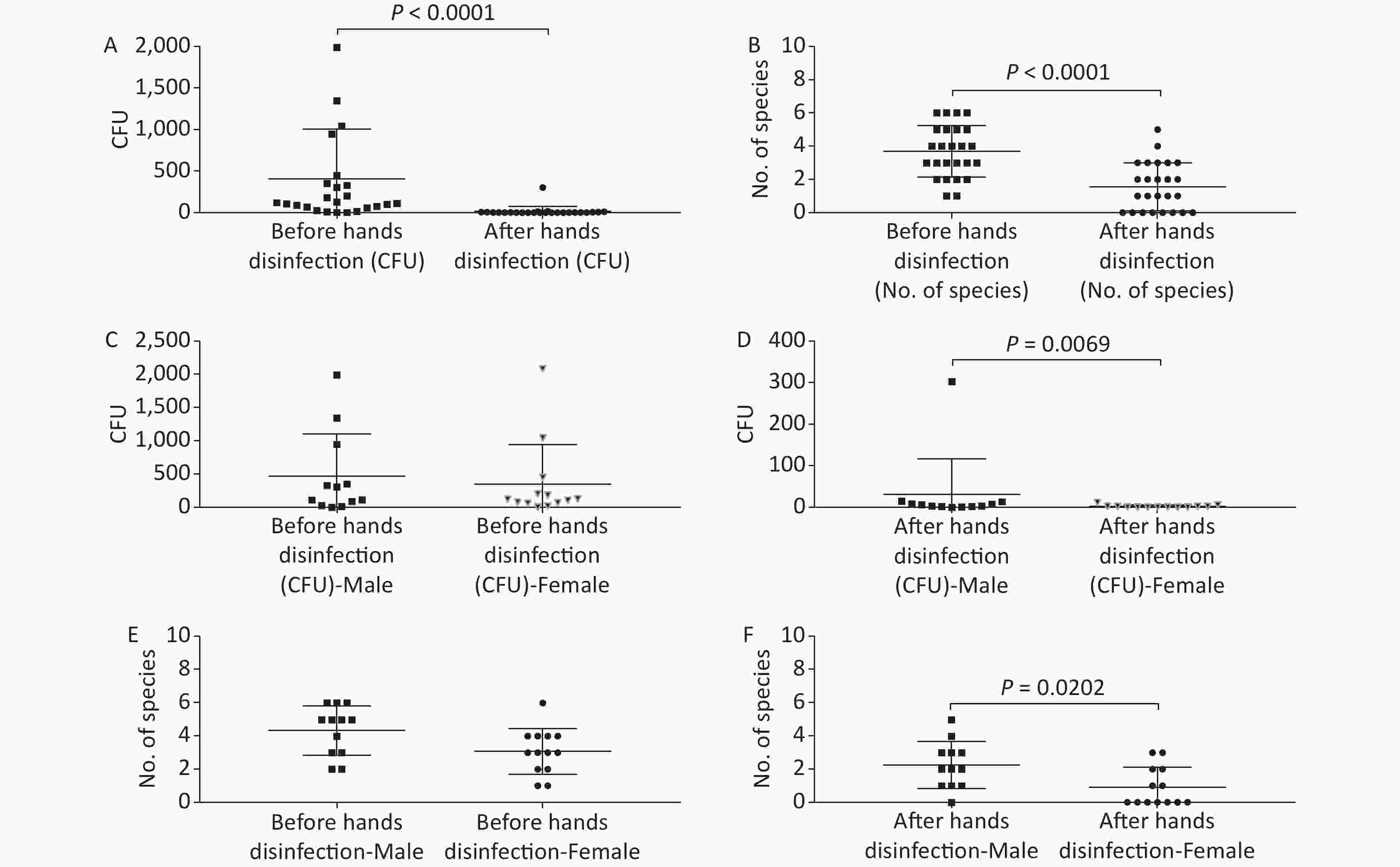
Figure 2. Baseline microorganisms' growth on the hands of anesthesiologists and the effectiveness of disinfection. (A) and (B) represent microorganisms' growth before and after hands disinfection in terms of the total CFUs and number of species, respectively. (C) and (D) indicate microorganisms' growth before and after hands disinfection between male and female anesthesiologists in terms of total CFUs, respectively. (E) and (F) represent microorganisms' growth before and after hands disinfection between male and female anesthesiologists in terms of number of species, respectively. CFU, colony-forming units. The two-tailed Mann-Whitney test was used in this section, and a P-value < 0.05 was considered significant.
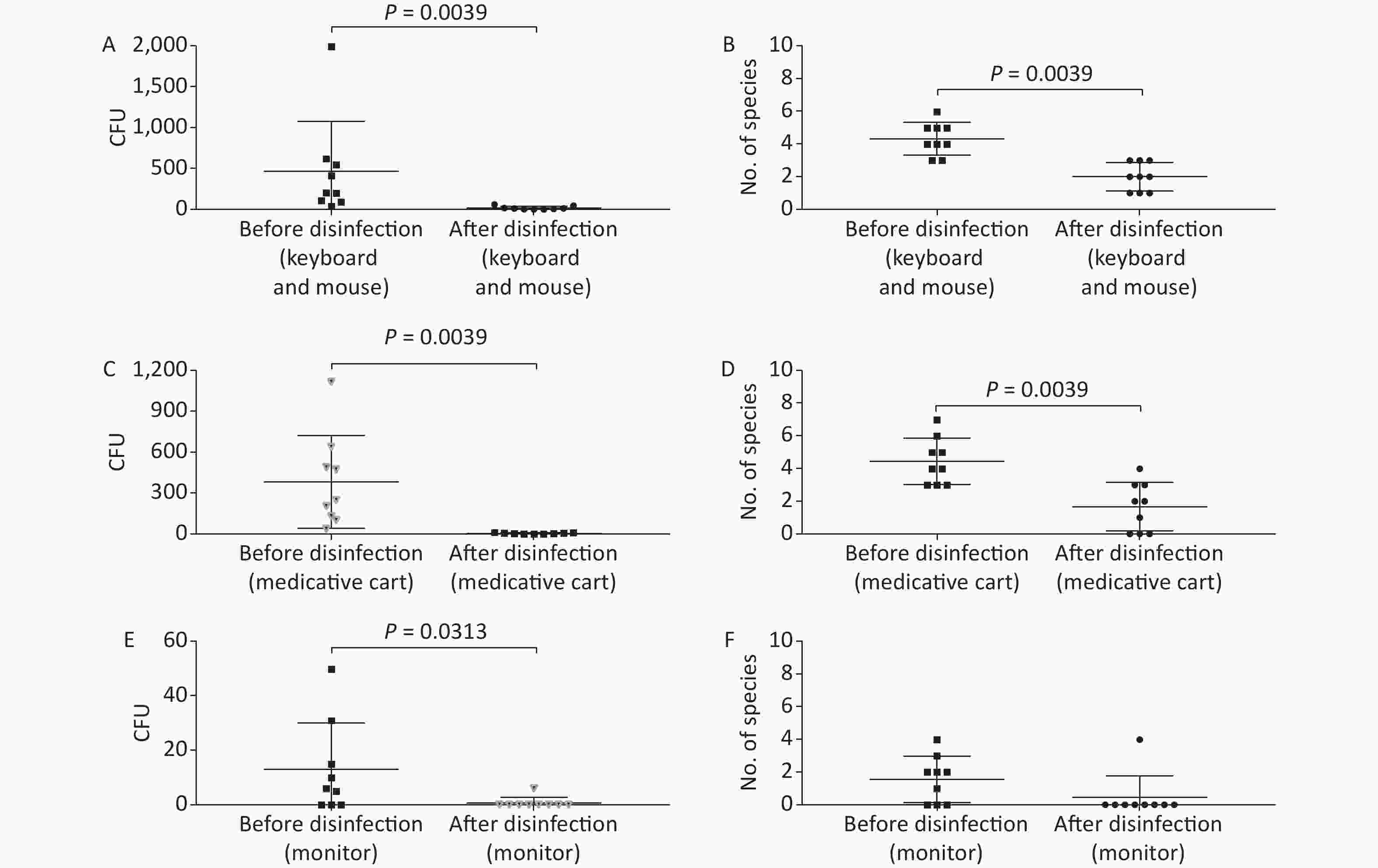
Figure 3. Baseline microorganisms' growth in the anesthesia environment and the effectiveness of disinfection. (A) and (B) represent microorganisms' growth before and after disinfection at the key board and mouse in terms of CFUs and number of species, respectively. (C) and (D) indicate microorganisms' growth before and after disinfection at the medicative cart in terms of CFUs and number of species, respectively. (E) and (F) represent microorganisms' growth before and after disinfection at the monitor in terms of CFUs and number of species, respectively. CFU, colony-forming units. The two-tailed Mann-Whitney test was used in this section, and a P-value < 0.05 was considered significant.
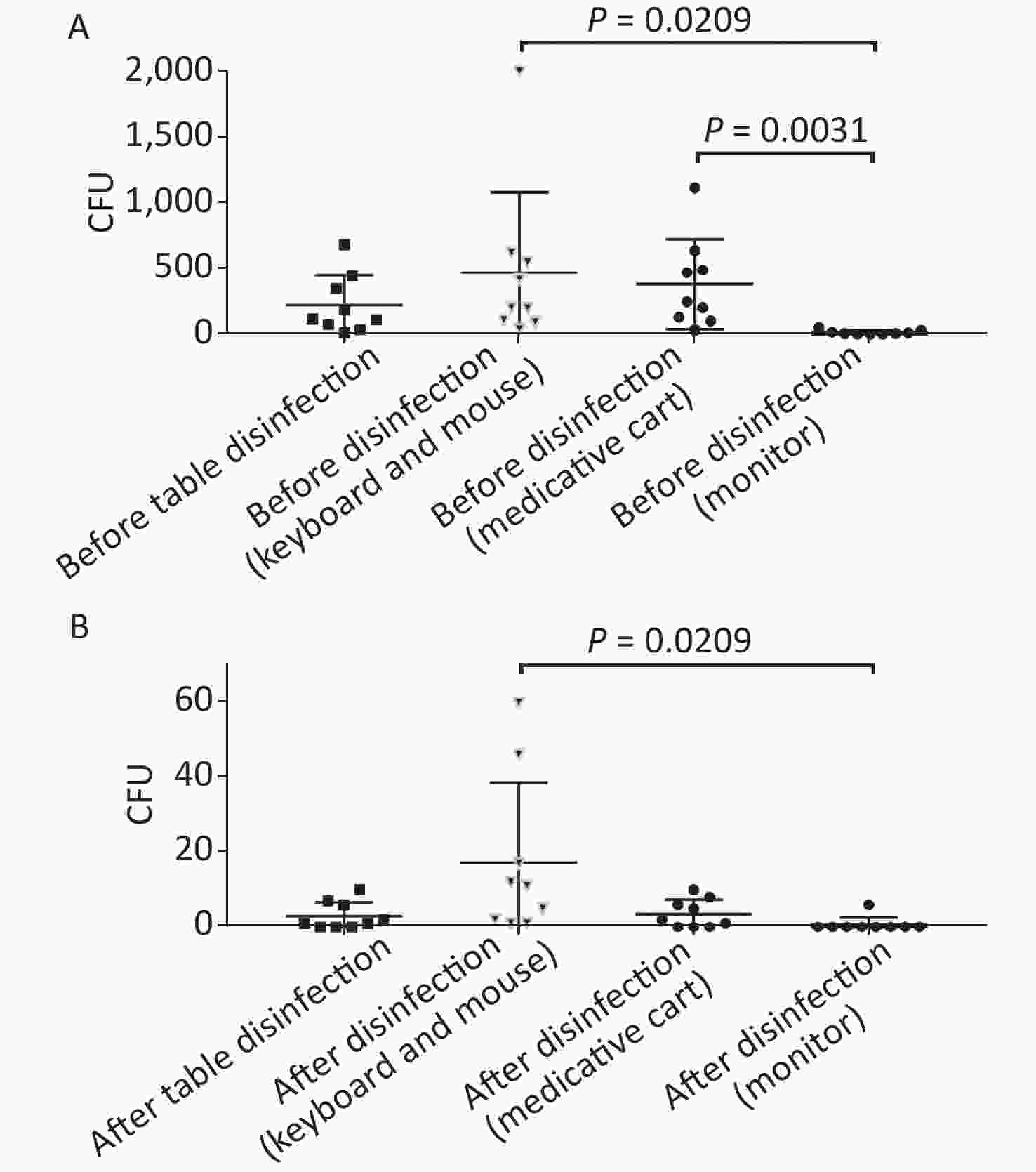
Figure 4. Comparison of microorganisms' growth among different parts of anesthesia environment and the effectiveness of disinfection. (A) and (B) represent microorganisms' growth before and after disinfection at different parts of anesthesia environment in terms of CFUs, respectively. CFU, colony-forming units. The one-way ANOVA test (multiple comparisons) was used in this section, and a P-value < 0.05 was considered significant.
-
The microorganisms' growth results from the hands of the 25 anesthesiologists sampled at a ready-to-use operating room before patient contact showed that the overall mean number of total CFUs and CFU/cm2 were 407 (range: 2–2091) and 7 (range: 0–35) (Figure 2, Supplementary Table S1, available in www.besjournal.com), respectively. Five anesthesiologists’ hands (Providers 3, 7, 10, 23, and 25) (5/25, 20%) carried microorganisms more than 10 CFU/cm2 (Supplementary Table S1, Figure 2). Only one major pathogen Staphylococcus aureus was isolated, from 5/25 anesthesiologists (20%; Providers 1, 2, 3, 4, and 13) (Supplementary Table S1, Figure 2). Considering the possible impact of gender, we compared the microorganisms' growth between male and female anesthesiologists before uniform disinfection, and found no differences (Figure 2). Not surprisingly, after disinfection, the microorganism growth significantly decreased regardless of the total CFUs (P < 0.0001) or the number of species (P < 0.0001), but obvious divergences were determined between male and female providers, notably that female anesthesiologists performed hand hygiene better than male anesthesiologists, resulting in fewer CFUs (P = 0.0069) and fewer species (P = 0.0202) (Figure 2).
Table S1. Bacterial growth on the hands of anesthesiologists before and after disinfection
Name Gender Before Disinfection of Hands After Disinfection of Hands CFU/total CFU/cm2 Bacteria CFU/total CFU/cm2 Bacteria Provider 1 M 114 2 Brevibacillus parabrevis 1 0 Bacillus cereus Kocuria marina Staphylococcus aureus Staphylococcus epidermidis Staphylococcus hominis Provider 2 M 305 5 Staphylococcus epidermidis 3 0 Bacillus megaterium Bacillus cereus Staphylococcus capitis Moraxella osloensis Kocuria rhizophila Staphylococcus aureus Staphylococcus hominis Provider 3 M 950 16 Staphylococcus aureus 304 5 Micrococcus luteus Staphylococcus haemolyticus Kocuria palustris Micrococcus luteus Staphylococcus aureus Staphylococcus capitis Staphylococcus warneri Provider 4 M 28 0 Bacillus firmus 6 0 Lactobacillus plantarum Bacillus megaterium Staphylococcus capitis Enterobacter cloacae Staphylococcus epidermidis Staphylococcus aureus Staphylococcus capitis Staphylococcus epidermidis Provider 5 M 89 1 Bacterium* 2 0 Corynebacterium tuberculostearicum Neisseria mucosa Staphylococcus epidermidis Staphylococcus capitis Staphylococcus epidermidis Staphylococcus hominis Staphylococcus pettenkoferi Provider 6 M 330 6 Corynebacterium tuberculostearicum 3 0 Bacterium* Micrococcus antarcticus Micrococcus luteus Staphylococcus epidermidis Bacterium* Provider 7 M 2000 33 Staphylococcus capitis 7 0 Staphylococcus epidermidis Kocuria marina Provider 8 M 10 0 Bacillus cereus 8 0 Bacillus cereus Bacillus megaterium Staphylococcus capitis Moraxella osloensis Bacterium* Provider 9 M 2 0 Aerococcus viridans 0 0 Lactobacillus fermentum Provider 10 M 1352 23 Acinetobacter baumannii 15 0 Bacillus flexus Acinetobacter nosocomialis Brevibacterium linens Brevibacterium linens Sphingomonas aerolata Staphylococcus epidermidis Sphingomonas paucimobilis Provider 11 M 352 6 Bacillus cereus 13 0 Bacillus infantis Bacillus flexus Bacillus megaterium Bacillus infantis Bacillus simplex Bacillus megaterium Staphylococcus cohnii Dermacoccus nishinomiyaensis Provider 12 M 109 2 Bacillus megaterium 2 0 Micrococcus luteus Micrococcus luteus Neisseria meningitidis Serratia rubidaea Staphylococcus sp Provider 13 F 120 2 Micrococcus luteus 2 0 Bacillus horikoshii Moraxella sp Staphylococcus capitis Staphylococcus hominis Staphylococcus aureus Provider 14 F 67 1 Micrococcus luteus 2 0 Bacillus pseudofirmus Ralstonia syzygii Staphylococcus capitis Provider 15 F 77 1 Micrococcus luteus 0 0 Staphylococcus epidermidis Staphylococcus hominis Provider 16 F 15 0 Bacterium* 0 0 Kocuria marina Staphylococcus capitis Staphylococcus hominis Provider 17 F 202 3 Bacillus mojavensis 11 0 Brevibacillus borstelensis Kocuria rhizophila Brevibacterium linens Staphylococcus sciuri Bacterium* Staphylococcus epidermidis Provider 18 F 2 0 Bacillus subtilis 0 0 Staphylococcus capitis Provider 19 F 450 8 Brevibacterium linens 0 Bacterium* Provider 20 F 127 2 Acinetobacter pittii 2 0 Bacillus idriensis Brevundimonas vesicularis Micrococcus antarcticus Yeast* Micrococcus luteus Ralstonia syzygii Staphylococcus caprae Provider 21 F 103 2 Pasteurella dagmatis 0 0 Brachybacterium conglomeratum Bacterium* Brachybacterium conglomeratum Provider 22 F 60 1 Bacillus cereus 5 0 Neisseria perflava Staphylococcus cohnii Staphylococcus epidermidis Provider 23 F 1047 17 Bacillus cereus 0 0 Bacillus megaterium Staphylococcus cohnii Staphylococcus saprophyticus Provider 24 F 183 3 Bacillus cereus 0 0 Provider 25 F 2091 35 Bacillus sp 2 0 Bacillus circulans Brevibacterium casei Neisseria meningitidis Note. *Indicates identification scores of ≤ 60, considered “not reliable”. CFU, colony-forming units. -
In this study, anesthesia environment samples were obtained from all nine operating rooms at the hospital. In each ready-to-use operating room we sampled the anesthesia cart, monitor screen, working table, and keyboard and mouse of the anesthesia machine (Figure 1). The microorganisms' growth results showed that the overall mean numbers of total CFUs found on the anesthesia cart, monitor screen, working table, and keyboard and mouse of the anesthesia machine were 376 (range: 36–1,114), 13 (range: 0–50), 223 (range: 13–684), and 469 (range: 38–2,000) (Supplementary Tables S2 and S3, available in www.besjournal.com), respectively. Based on CFU/cm2 results, five of the nine operating rooms (55.6%) carried microorganisms more than 5 CFU/cm2 on the anesthesia cart (Rooms 3 & 8), working table (Room 7), or the keyboard and mouse of anesthesia machine (Rooms 1 & 9) (Supplementary Tables S2 and S3). One major pathogen, Staphylococcus aureus, was isolated in three rooms (33.3%; Rooms 1, 3, and 7) (Supplementary Tables S2 and S3). Considering the importance of disinfection, we disinfected the working areas determined in this study with 5.5%–6.5% chlorine and then re-sampled after 30 minutes. Not surprisingly, after disinfection, microorganisms' growth significantly decreased regardless of the total CFUs (P < 0.05) or numbers of species (P < 0.05) (Figure 3). Furthermore, Figure 4 shows that the microorganisms' growth are very likely to be hand contact-related because the keyboard and mouse of the anesthesia machines, as the most frequently touched areas, carried the highest number of colonies, whereas the monitor screens carried the fewest colonies; these differences remained statistically significant even after disinfection. In this study, we also monitored the microorganisms' growth sampled from the working table of the anesthesia machines at different time points to determine how soon a repeat cleaning is needed. The results showed that microorganisms regrowth began 1 hour after disinfection, and increased gradually over time (Figure 5) until reaching an excessive amount (> 5 CFU/cm2) at 4 hours after disinfection (Supplementary Table S3).
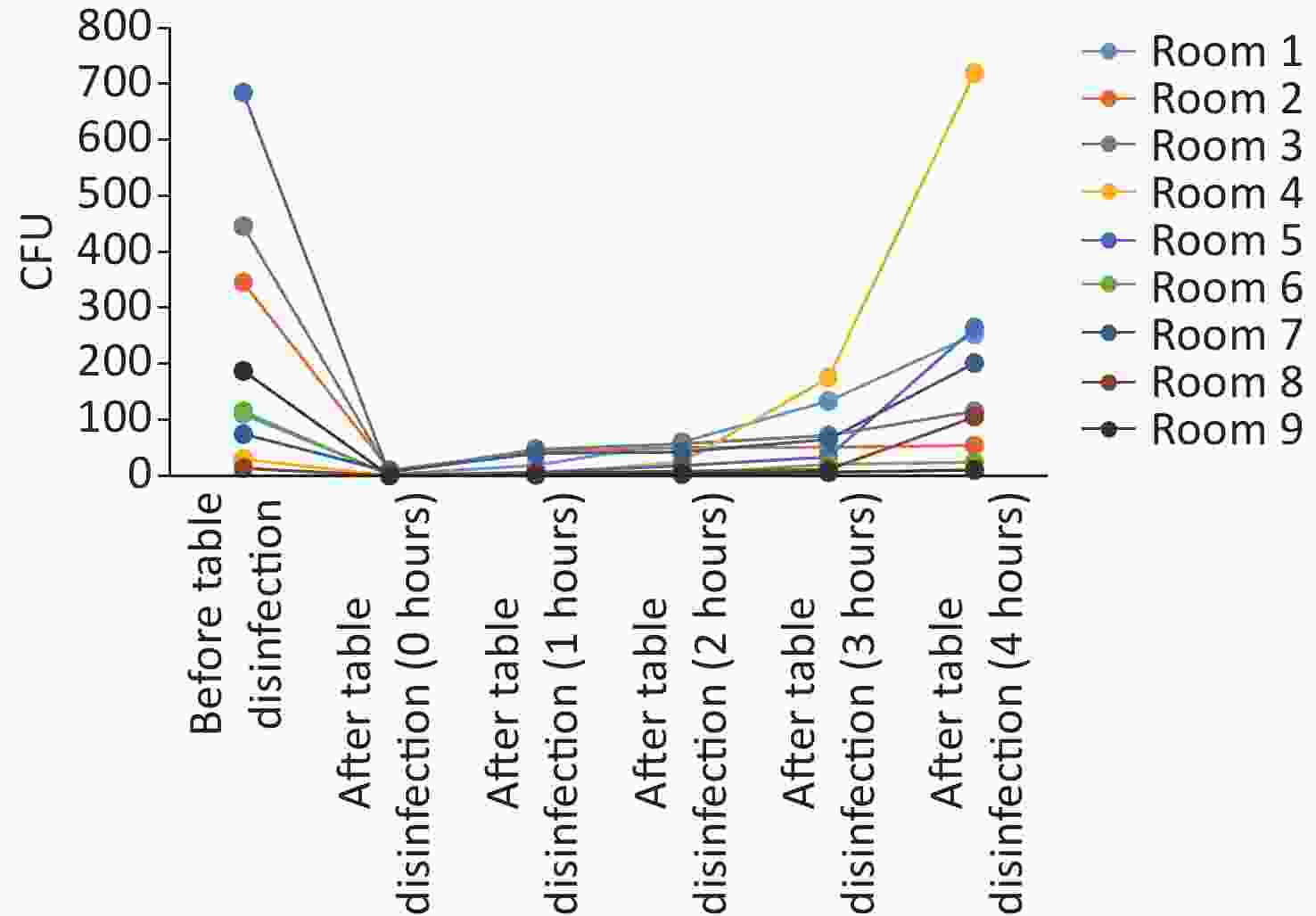
Figure 5. Microorganisms’ growth on anesthesia machines (working tables) and the effectiveness of disinfection. CFU, colony-forming units.
Table S2. Bacterial growth in the anesthesia environment before and after disinfection
No. of Operating Room Before disinfection
(Keyboard and Mouse)After disinfection
(Keyboard and Mouse)Before disinfection
(Medicative Cart)After disinfection
(Medicative Cart)Before disinfection
(Monitor)After disinfection
(Monitor)CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria 1 2,000; 20 Micrococcus luteus 1; 0 Moraxella sp 129; 1 Bacillus firmus 0; 0 0; 0 0; 0 Staphylococcus epidermidis Bacillus licheniformis Staphylococcus pasteuri Kocuria rosea Bacterium* Bacterium* Staphylococcus hominis Staphylococcus pasteuri 2 106; 1 Bacterium* 2; 0 Bacillus sp 203; 2 Bacillus megaterium 8; 0 Micrococcus luteus 0; 0 0; 0 Bacillus simplex Staphylococcus epidermidis Kocuria rhizophila Staphylococcus capitis Nocardia higoensis Staphylococcus cohnii Staphylococcus epidermidis Bacterium* Staphylococcus epidermidis Staphylococcus cohnii Staphylococcus hominis Staphylococcus haemolyticus 3 548; 5 Corynebacterium aurimucosum 60; 1 Corynebacterium tuberculostearicum 637; 6 Arthrobacter aurescens 10; 0 Yeast* 31; 0 Micrococcus luteus 0; 0 Micrococcus luteus Staphylococcus capitis Kocuria palustris Micrococcus antarcticus Moraxella osloensis Kocuria marina Micrococcus antarcticus Bacterium* Staphylococcus hominis Staphylococcus hominis Moraxella osloensis Staphylococcus haemolyticus Staphylococcus warneri Staphylococcus cohnii Staphylococcus hominis 4 38; 0 Kocuria rhizophila 17; 0 Staphylococcus hominis 100; 1 Bacillus cereus 1; 0 Paenibacillus lautus 50; 1 Bacillus cereus 0; 0 Staphylococcus epidermidis Micrococcus luteus Kocuria rhizophila Staphylococcus pettenkoferi Staphylococcus epidermidis Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus hominis Staphylococcus lugdunensis 5 91; 1 Bacterium* 1; 0 Paenibacillus glucanolyticus 208; 2 Corynebacterium aurimucosum 0; 0 6; 0 Moraxella osloensis 0; 0 Kocuria rosea Micrococcus antarcticus Staphylococcus warneri Staphylococcus epidermidis Staphylococcus warneri Staphylococcus hominis Bacterium* 6 416; 4 Micrococcus luteus 46; 0 Micrococcus luteus 488; 5 Acinetobacter lwoffii 0; 0 10; 0 Staphylococcus cohnii 0; 0 Staphylococcus epidermidis Pseudomonas luteola Micrococcus luteus Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus saprophyticus Staphylococcus cohnii Staphylococcus hominis 7 202; 2 Escherichia coli 5; 0 Actinomyces funkei 36; 0 Bacillus cereus 6; 0 Bacillus licheniformis 15; 0 Bacillus subtilis 6; 0 Bacillus firmus Moraxella osloensis Micrococcus antarcticus Micrococcus luteus Staphylococcus capitis Bacillus licheniformis Staphylococcus aureus Staphylococcus epidermidis Moraxella osloensis Staphylococcus epidermidis Lactobacillus paracasei Staphylococcus hominis Staphylococcus haemolyticus Lysinibacillus sphaericus 8 197; 2 Dietzia maris 12; 0 Enterococcus pallens 1114; 11 Yeast* 2; 0 Micrococcus luteus 0; 0 0; 0 Kocuria rhizophila Micrococcus luteus Pandoraea pulmonicola Staphylococcus capitis Staphylococcus epidermidis Staphylococcus hominis Staphylococcus haemolyticus Staphylococcus saprophyticus 9 621; 6 Bacillus idriensis 11; 0 Micrococcus antarcticus 471; 5 Yeast* 5; 0 Kocuria rosea 5; 0 Bacterium* 0; 0 Bacterium* Micrococcus luteus Staphylococcus epidermidis Bacillus sp Staphylococcus hominis Staphylococcus cohnii Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus warneri Note. *Indicates identification scores of ≤ 60, considered “not reliable”. CFU, colony-forming units. Table S3. Bacterial growth in the anesthesia environment (working table) before and after disinfection
No. of Operating Room Before Table Disinfection After Table Disinfection
(0 hours)After Table Disinfection
(1 hour)After Table Disinfection
(2 hours)After Table Disinfection
(3 hours)After Table Disinfection
(4 hours)CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria 1 110; 1 Micrococcus antarcticus 2; 0 Staphylococcus hominis 19; 0 Bacillus mycoides 60; 1 Kocuria rosea 133; 1 Pseudomonas avellanae 252; 3 Micrococcus luteus Micrococcus luteus Staphylococcus epidermidis Paenibacillus apiarius Staphylococcus capitis Staphylococcus capitis Staphylococcus aureus Staphylococcus hominis Staphylococcus cohnii Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus capitis Staphylococcus hominis Staphylococcus hominis Staphylococcus haemolyticus Staphylococcus cohnii Bacterium* Staphylococcus hominis 2 345; 3 Kocuria rosea 10; 0 Bacillus megaterium 43; 0 Bacillus megaterium 50; 1 Bacillus vallismortis 51; 1 Yeast* 54; 1 Bacillus subtilis Moraxella osloensis Bacillus sp Bacillus sp Kocuria rhizophila Kocuria rosea Bacterium* Staphylococcus capitis Kocuria rhizophila Bacterium* Neisseria perflava Staphylococcus capitis Moraxella osloensis Staphylococcus epidermidis Staphylococcus hominis Staphylococcus epidermidis Staphylococcus capitis Staphylococcus cohnii Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus epidermidis Staphylococcus kloosii Staphylococcus pettenkoferi Bacterium* 3 445; 4 Bacillus pseudofirmus 6; 0 Staphylococcus epidermidis 47; 0 Micrococcus luteus 57; 0 Candida parapsilosis 72; 1 Bacterium* 115; 1 Moraxella osloensis Kocuria rhizophila Staphylococcus hominis Staphylococcus aureus Staphylococcus hominis Brachybacterium conglomeratum Yeast* Moraxella osloensis Staphylococcus haemolyticus Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus aureus Staphylococcus sciuri Staphylococcus saprophyticus Staphylococcus capitis Staphylococcus epidermidis Staphylococcus hominis 4 30; 0 Kocuria marina 0; 0 3; 0 Bacterium* 25; 0 Bacterium* 175; 2 Yeast* 719; 7 Yeast* Paenibacillus chitinolyticus Staphylococcus capitis Enterobacter cloacae Micrococcus antarcticus Micrococcus luteus Bacterium* Staphylococcus epidermidis Micrococcus luteus Moraxella osloensis Staphylococcus hominis Staphylococcus capitis Staphylococcus capitis 5 684; 7 Bacterium* 1; 0 Bacterium* 6; 0 Staphylococcus capitis 18; 0 Bacillus cohnii 34; 0 Bacillus sp 264; 3 Bacterium* Neisseria subflava Yeast* Staphylococcus cohnii Staphylococcus warneri Staphylococcus hominis Staphylococcus lugdunensis Staphylococcus sciuri Bacterium* 6 115; 1 Bacillus cereus 1; 0 Staphylococcus hominis 3; 0 Bacterium* 3; 0 Brevibacterium iodinum 20; 0 Micrococcus luteus 24; 0 Bacillus megaterium Enterobacter cloacae Staphylococcus cohnii Bacterium* Paenibacillus massiliensis Moraxella sp Micrococcus antarcticus Staphylococcus epidermidis Kocuria marina Pseudomonas flavescens Staphylococcus cohnii Staphylococcus cohnii Staphylococcus epidermidis Staphylococcus hominis 7 74; 1 Kocuria marina 7; 0 Bacillus mannanilyticus 40; 0 Neisseria perflava 42; 0 Staphylococcus epidermidis 65; 1 Micrococcus luteus 201; 2 Bacillus megaterium Micrococcus antarcticus Lactobacillus paracasei Rothia mucilaginosa Staphylococcus haemolyticus Neisseria flavescens Neisseria flavescens Staphylococcus capitis Neisseria perflava Staphylococcus aureus Staphylococcus pettenkoferi Neisseria sicca Staphylococcus hominis Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus hominis Rothia mucilaginosa Staphylococcus haemolyticus 8 13; 0 Kocuria rhizophila 0; 0 5; 0 Arcanobacterium phocae 8; 0 Bacillus cereus 11; 0 Staphylococcus epidermidis 105; 1 Alkalibacillus haloalkaliphilus Kocuria rosea Cardiobacterium sp Corynebacterium sp Staphylococcus haemolyticus Micrococcus luteus Moraxella osloensis Staphylococcus hominis Kocuria palustris Staphylococcus capitis Staphylococcus capitis Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus hominis 9 187; 2 Bacillus simplex 0; 0 1; 0 Staphylococcus haemolyticus 3; 0 Bacillus smithii 6; 0 Micrococcus luteus 10; 0 Corynebacterium afermentans Staphylococcus epidermidis Micrococcus antarcticus Staphylococcus epidermidis Staphylococcus hominis Staphylococcus hominis Staphylococcus epidermidis Staphylococcus hominis Staphylococcus warneri Staphylococcus saprophyticus Note. *Indicates identification scores of ≤ 60, considered “not reliable”. CFU, colony-forming units. -
Hand-mediated transmission is a paramount factor causing infection associated with healthcare [10]. Effective and timely hand disinfection before patient contact will decrease the incidence of transfer of potential pathogens [11]. Anesthesiologists are usually a neglected population who may still lack consciousness in operating room infection control and hand hygiene [1-6]. In China, two regulatory instructions were issued on regulation of disinfection in healthcare settings (WS/T 367-2012) and health workers [7, 8], but whether the anesthesiologists and cleaning staff implemented properly was still unknown. In this study, microorganisms’ growth results showed that the hands of 20% (5/25) of anesthesiologists carried excessive bacteria or fungi, and significantly decreased after disinfection with fewer CFUs and species. It is well worth mentioning that the method of hand hygiene used in this study had already been standardized (WS/T 313-2019) [8], specifying that the whole hand and fingers (particularly the tips) should be exposed to the alcohol hand sanitizer after rubbing them for 10 to 15 seconds, and that alcohol hand sanitizer should be conveniently placed. We found that female anesthesiologists performed hand hygiene better than did their male counterparts, because men had a higher CFU count and number of species. Therefore, male anesthesiologists need to pay more attention to the standard operating procedures and effect evaluation of hand hygiene. However, whether the results mentioned above indicate that male anesthesiologists are more likely to cause hand-mediated transmission and higher incidence of subsequent hospital-acquired infections remains unknown and will require further study.
Surfaces in the anesthesia environment, especially the anesthesia cart and the anesthesia machine, which are used frequently during operations, are often neglected as important potential sources of bacterial transmission [12]. Munoz-Price and Birnbach [13] found that pathogenic organisms were present in 16.6% of ready-to-use operating room surfaces. Our study found that 55.6% of ready-to-use operating rooms carried excessive bacteria or fungi on the anesthesia cart, working table, or keyboard and mouse of anesthesia machine (Supplementary Tables S2 and S3). Disinfection can largely reduce microorganisms' growth with fewer CFUs (P < 0.05) and number of species (P < 0.05) (Figure 3), but the regrowth began quickly (1 hour) after disinfection, and increased gradually over time (Figure 5) until reaching excessive levels at 4 hours after disinfection (Supplementary Table S3). Jefferson et al. [14] evaluated 71 operating rooms across six acute care hospitals and found an average daily cleaning rate of 25% of the objects monitored. A similar study [13] also found a baseline daily cleaning rate of 47%. In this study, all operating rooms are cleaned daily. Yet the results of this study confirm that daily cleaning rate may be insufficient because unawareness of hand contact with excessively bacteria-colonized surfaces may increase the risk of subsequent hospital-acquired infections.
The hospital environment is a major reservoir of multidrug-resistant bacteria, including MRSA, vancomycin-resistant enterococci, C. difficile, and A. baumannii [15-21], even in areas such as operating rooms that were previously thought to be “sterile” [22]. Staphylococcus aureus, usually colonized on the skin of human beings and on environmental surfaces, is a common cause of healthcare-associated infections worldwide and has become a major screened and monitored pathogen on admission as a key infection prevention strategy [23-26]. In this study, the major pathogen Staphylococcus aureus was isolated from the hands of 20% of anesthesiologists and in 33.3% of operating rooms, but we did not determine the antimicrobial susceptibility of these isolates, and therefore whether they were MRSA or MSSA is unknown. Loftus et al [27] found that 7% (12/164) and 11% (18/164) of anesthesia providers’ hands were contaminated with MRSA and MSSA, respectively, and MRSA and MSSA can also be isolated from anesthesia machines.
A key point of the present study is giving us a specific name list of all possible pathogens in hands of anesthesiologists and in the anesthesia environment. Most of the detected species were not thought to be pathogenic, but commensal species have been confirmed to serve as reservoirs of antibiotic resistance and virulence genes for the pathogenic species [28,29], which may not take a toll on patients now but long-term colonization on surfaces in the anesthesia environment is still a potential risk because patients admitted in cancer hospitals are usually more vulnerable to microorganisms [30]. It is also worth mentioning that we found that the number of detected species seemed to change much less than the counts. This is likely to be expected, previous study [31] also reported that cleaning procedures were very effective in eliminating coliforms, in contrast, gram-positive bacteria were not totally eliminated, possibly due to the greater resistance of gram-positive bacteria (with their thicker peptidoglycan cell wall layer) to ethanol-based sanitizers and disinfectants.
There are some limitations in this study. Firstly, because this is a single-center study, the results obtained may not be applicable to other hospitals. Secondly, we sampled hands only in the short time period immediately before patient contact, which may underestimate the importance of hand hygiene throughout the entire process of patient care because Loftus and others [32-35] suggested that hand hygiene use of 4–8 times/hour reduced surgical site infections. Thirdly, the test procedures employed in the present study were dictated by the national standards, which probably limit the external validity.
Despite these limitations, we characterized the baseline levels of contamination on the hands of anesthesiologists and in the anesthesia environment. Our study indicates that male anesthesiologists need to pay more attention to the standard operating procedures and effect evaluation of hand hygiene, daily cleaning rate of the operating room may be insufficient, and we would suggest that there should be a repeat cleaning every four hours. These results of this study provide a theoretical basis for the formulation of future measures to control and prevent nosocomial infection.
-
LIU Hong Lei helped design the study, conduct the study, analyze the data, and write the manuscript. LIU Ya Li helped design the study, has seen the original study data, reviewed the analysis of the data, approved the final manuscript. LI Zong Chao has seen the original study data, reviewed the analysis of the data, approved the final manuscript. TAN Hong Yu, SUN Fang Yan and XU Ying Chun helped design the study, approved the final manuscript.
doi: 10.3967/bes2022.128
Hand Hygiene among Anesthesiologists and Microorganisms Contamination in Anesthesia Environments: A Single-Center Observational Study
-
Abstract:
Objective To investigate the baseline levels of microorganisms' growth on the hands of anesthesiologists and in the anesthesia environment at a cancer hospital. Methods This study performed in nine operating rooms and among 25 anesthesiologists at a cancer hospital. Sampling of the hands of anesthesiologists and the anesthesia environment was performed at a ready-to-use operating room before patient contact began and after decontamination. Results Microorganisms' growth results showed that 20% (5/25) of anesthesiologists’ hands carried microorganisms (> 10 CFU/cm2) before patient contact began. Female anesthesiologists performed hand hygiene better than did their male counterparts, with fewer CFUs (P = 0.0069) and fewer species (P = 0.0202). Our study also found that 55.6% (5/9) of ready-to-use operating rooms carried microorganisms (> 5 CFU/cm2). Microorganisms regrowth began quickly (1 hour) after disinfection, and increased gradually over time, reaching the threshold at 4 hours after disinfection. Staphylococcus aureus was isolated from the hands of 20% (5/25) of anesthesiologists and 33.3% (3/9) of operating rooms. Conclusion Our study indicates that male anesthesiologists need to pay more attention to the standard operating procedures and effect evaluation of hand hygiene, daily cleaning rate of the operating room may be insufficient, and we would suggest that there should be a repeat cleaning every four hours. -
Key words:
- Hand hygiene /
- Anesthesiologists /
- Contamination /
- Anesthesia environments
&These authors contributed equally to this work.
注释:1) CONFLICTS OF INTEREST: -
Figure 2. Baseline microorganisms' growth on the hands of anesthesiologists and the effectiveness of disinfection. (A) and (B) represent microorganisms' growth before and after hands disinfection in terms of the total CFUs and number of species, respectively. (C) and (D) indicate microorganisms' growth before and after hands disinfection between male and female anesthesiologists in terms of total CFUs, respectively. (E) and (F) represent microorganisms' growth before and after hands disinfection between male and female anesthesiologists in terms of number of species, respectively. CFU, colony-forming units. The two-tailed Mann-Whitney test was used in this section, and a P-value < 0.05 was considered significant.
Figure 3. Baseline microorganisms' growth in the anesthesia environment and the effectiveness of disinfection. (A) and (B) represent microorganisms' growth before and after disinfection at the key board and mouse in terms of CFUs and number of species, respectively. (C) and (D) indicate microorganisms' growth before and after disinfection at the medicative cart in terms of CFUs and number of species, respectively. (E) and (F) represent microorganisms' growth before and after disinfection at the monitor in terms of CFUs and number of species, respectively. CFU, colony-forming units. The two-tailed Mann-Whitney test was used in this section, and a P-value < 0.05 was considered significant.
Figure 4. Comparison of microorganisms' growth among different parts of anesthesia environment and the effectiveness of disinfection. (A) and (B) represent microorganisms' growth before and after disinfection at different parts of anesthesia environment in terms of CFUs, respectively. CFU, colony-forming units. The one-way ANOVA test (multiple comparisons) was used in this section, and a P-value < 0.05 was considered significant.
S1. Bacterial growth on the hands of anesthesiologists before and after disinfection
Name Gender Before Disinfection of Hands After Disinfection of Hands CFU/total CFU/cm2 Bacteria CFU/total CFU/cm2 Bacteria Provider 1 M 114 2 Brevibacillus parabrevis 1 0 Bacillus cereus Kocuria marina Staphylococcus aureus Staphylococcus epidermidis Staphylococcus hominis Provider 2 M 305 5 Staphylococcus epidermidis 3 0 Bacillus megaterium Bacillus cereus Staphylococcus capitis Moraxella osloensis Kocuria rhizophila Staphylococcus aureus Staphylococcus hominis Provider 3 M 950 16 Staphylococcus aureus 304 5 Micrococcus luteus Staphylococcus haemolyticus Kocuria palustris Micrococcus luteus Staphylococcus aureus Staphylococcus capitis Staphylococcus warneri Provider 4 M 28 0 Bacillus firmus 6 0 Lactobacillus plantarum Bacillus megaterium Staphylococcus capitis Enterobacter cloacae Staphylococcus epidermidis Staphylococcus aureus Staphylococcus capitis Staphylococcus epidermidis Provider 5 M 89 1 Bacterium* 2 0 Corynebacterium tuberculostearicum Neisseria mucosa Staphylococcus epidermidis Staphylococcus capitis Staphylococcus epidermidis Staphylococcus hominis Staphylococcus pettenkoferi Provider 6 M 330 6 Corynebacterium tuberculostearicum 3 0 Bacterium* Micrococcus antarcticus Micrococcus luteus Staphylococcus epidermidis Bacterium* Provider 7 M 2000 33 Staphylococcus capitis 7 0 Staphylococcus epidermidis Kocuria marina Provider 8 M 10 0 Bacillus cereus 8 0 Bacillus cereus Bacillus megaterium Staphylococcus capitis Moraxella osloensis Bacterium* Provider 9 M 2 0 Aerococcus viridans 0 0 Lactobacillus fermentum Provider 10 M 1352 23 Acinetobacter baumannii 15 0 Bacillus flexus Acinetobacter nosocomialis Brevibacterium linens Brevibacterium linens Sphingomonas aerolata Staphylococcus epidermidis Sphingomonas paucimobilis Provider 11 M 352 6 Bacillus cereus 13 0 Bacillus infantis Bacillus flexus Bacillus megaterium Bacillus infantis Bacillus simplex Bacillus megaterium Staphylococcus cohnii Dermacoccus nishinomiyaensis Provider 12 M 109 2 Bacillus megaterium 2 0 Micrococcus luteus Micrococcus luteus Neisseria meningitidis Serratia rubidaea Staphylococcus sp Provider 13 F 120 2 Micrococcus luteus 2 0 Bacillus horikoshii Moraxella sp Staphylococcus capitis Staphylococcus hominis Staphylococcus aureus Provider 14 F 67 1 Micrococcus luteus 2 0 Bacillus pseudofirmus Ralstonia syzygii Staphylococcus capitis Provider 15 F 77 1 Micrococcus luteus 0 0 Staphylococcus epidermidis Staphylococcus hominis Provider 16 F 15 0 Bacterium* 0 0 Kocuria marina Staphylococcus capitis Staphylococcus hominis Provider 17 F 202 3 Bacillus mojavensis 11 0 Brevibacillus borstelensis Kocuria rhizophila Brevibacterium linens Staphylococcus sciuri Bacterium* Staphylococcus epidermidis Provider 18 F 2 0 Bacillus subtilis 0 0 Staphylococcus capitis Provider 19 F 450 8 Brevibacterium linens 0 Bacterium* Provider 20 F 127 2 Acinetobacter pittii 2 0 Bacillus idriensis Brevundimonas vesicularis Micrococcus antarcticus Yeast* Micrococcus luteus Ralstonia syzygii Staphylococcus caprae Provider 21 F 103 2 Pasteurella dagmatis 0 0 Brachybacterium conglomeratum Bacterium* Brachybacterium conglomeratum Provider 22 F 60 1 Bacillus cereus 5 0 Neisseria perflava Staphylococcus cohnii Staphylococcus epidermidis Provider 23 F 1047 17 Bacillus cereus 0 0 Bacillus megaterium Staphylococcus cohnii Staphylococcus saprophyticus Provider 24 F 183 3 Bacillus cereus 0 0 Provider 25 F 2091 35 Bacillus sp 2 0 Bacillus circulans Brevibacterium casei Neisseria meningitidis Note. *Indicates identification scores of ≤ 60, considered “not reliable”. CFU, colony-forming units. S2. Bacterial growth in the anesthesia environment before and after disinfection
No. of Operating Room Before disinfection
(Keyboard and Mouse)After disinfection
(Keyboard and Mouse)Before disinfection
(Medicative Cart)After disinfection
(Medicative Cart)Before disinfection
(Monitor)After disinfection
(Monitor)CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria 1 2,000; 20 Micrococcus luteus 1; 0 Moraxella sp 129; 1 Bacillus firmus 0; 0 0; 0 0; 0 Staphylococcus epidermidis Bacillus licheniformis Staphylococcus pasteuri Kocuria rosea Bacterium* Bacterium* Staphylococcus hominis Staphylococcus pasteuri 2 106; 1 Bacterium* 2; 0 Bacillus sp 203; 2 Bacillus megaterium 8; 0 Micrococcus luteus 0; 0 0; 0 Bacillus simplex Staphylococcus epidermidis Kocuria rhizophila Staphylococcus capitis Nocardia higoensis Staphylococcus cohnii Staphylococcus epidermidis Bacterium* Staphylococcus epidermidis Staphylococcus cohnii Staphylococcus hominis Staphylococcus haemolyticus 3 548; 5 Corynebacterium aurimucosum 60; 1 Corynebacterium tuberculostearicum 637; 6 Arthrobacter aurescens 10; 0 Yeast* 31; 0 Micrococcus luteus 0; 0 Micrococcus luteus Staphylococcus capitis Kocuria palustris Micrococcus antarcticus Moraxella osloensis Kocuria marina Micrococcus antarcticus Bacterium* Staphylococcus hominis Staphylococcus hominis Moraxella osloensis Staphylococcus haemolyticus Staphylococcus warneri Staphylococcus cohnii Staphylococcus hominis 4 38; 0 Kocuria rhizophila 17; 0 Staphylococcus hominis 100; 1 Bacillus cereus 1; 0 Paenibacillus lautus 50; 1 Bacillus cereus 0; 0 Staphylococcus epidermidis Micrococcus luteus Kocuria rhizophila Staphylococcus pettenkoferi Staphylococcus epidermidis Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus hominis Staphylococcus lugdunensis 5 91; 1 Bacterium* 1; 0 Paenibacillus glucanolyticus 208; 2 Corynebacterium aurimucosum 0; 0 6; 0 Moraxella osloensis 0; 0 Kocuria rosea Micrococcus antarcticus Staphylococcus warneri Staphylococcus epidermidis Staphylococcus warneri Staphylococcus hominis Bacterium* 6 416; 4 Micrococcus luteus 46; 0 Micrococcus luteus 488; 5 Acinetobacter lwoffii 0; 0 10; 0 Staphylococcus cohnii 0; 0 Staphylococcus epidermidis Pseudomonas luteola Micrococcus luteus Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus saprophyticus Staphylococcus cohnii Staphylococcus hominis 7 202; 2 Escherichia coli 5; 0 Actinomyces funkei 36; 0 Bacillus cereus 6; 0 Bacillus licheniformis 15; 0 Bacillus subtilis 6; 0 Bacillus firmus Moraxella osloensis Micrococcus antarcticus Micrococcus luteus Staphylococcus capitis Bacillus licheniformis Staphylococcus aureus Staphylococcus epidermidis Moraxella osloensis Staphylococcus epidermidis Lactobacillus paracasei Staphylococcus hominis Staphylococcus haemolyticus Lysinibacillus sphaericus 8 197; 2 Dietzia maris 12; 0 Enterococcus pallens 1114; 11 Yeast* 2; 0 Micrococcus luteus 0; 0 0; 0 Kocuria rhizophila Micrococcus luteus Pandoraea pulmonicola Staphylococcus capitis Staphylococcus epidermidis Staphylococcus hominis Staphylococcus haemolyticus Staphylococcus saprophyticus 9 621; 6 Bacillus idriensis 11; 0 Micrococcus antarcticus 471; 5 Yeast* 5; 0 Kocuria rosea 5; 0 Bacterium* 0; 0 Bacterium* Micrococcus luteus Staphylococcus epidermidis Bacillus sp Staphylococcus hominis Staphylococcus cohnii Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus warneri Note. *Indicates identification scores of ≤ 60, considered “not reliable”. CFU, colony-forming units. S3. Bacterial growth in the anesthesia environment (working table) before and after disinfection
No. of Operating Room Before Table Disinfection After Table Disinfection
(0 hours)After Table Disinfection
(1 hour)After Table Disinfection
(2 hours)After Table Disinfection
(3 hours)After Table Disinfection
(4 hours)CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria CFU/total; CFU/cm2 Bacteria 1 110; 1 Micrococcus antarcticus 2; 0 Staphylococcus hominis 19; 0 Bacillus mycoides 60; 1 Kocuria rosea 133; 1 Pseudomonas avellanae 252; 3 Micrococcus luteus Micrococcus luteus Staphylococcus epidermidis Paenibacillus apiarius Staphylococcus capitis Staphylococcus capitis Staphylococcus aureus Staphylococcus hominis Staphylococcus cohnii Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus capitis Staphylococcus hominis Staphylococcus hominis Staphylococcus haemolyticus Staphylococcus cohnii Bacterium* Staphylococcus hominis 2 345; 3 Kocuria rosea 10; 0 Bacillus megaterium 43; 0 Bacillus megaterium 50; 1 Bacillus vallismortis 51; 1 Yeast* 54; 1 Bacillus subtilis Moraxella osloensis Bacillus sp Bacillus sp Kocuria rhizophila Kocuria rosea Bacterium* Staphylococcus capitis Kocuria rhizophila Bacterium* Neisseria perflava Staphylococcus capitis Moraxella osloensis Staphylococcus epidermidis Staphylococcus hominis Staphylococcus epidermidis Staphylococcus capitis Staphylococcus cohnii Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus epidermidis Staphylococcus kloosii Staphylococcus pettenkoferi Bacterium* 3 445; 4 Bacillus pseudofirmus 6; 0 Staphylococcus epidermidis 47; 0 Micrococcus luteus 57; 0 Candida parapsilosis 72; 1 Bacterium* 115; 1 Moraxella osloensis Kocuria rhizophila Staphylococcus hominis Staphylococcus aureus Staphylococcus hominis Brachybacterium conglomeratum Yeast* Moraxella osloensis Staphylococcus haemolyticus Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus aureus Staphylococcus sciuri Staphylococcus saprophyticus Staphylococcus capitis Staphylococcus epidermidis Staphylococcus hominis 4 30; 0 Kocuria marina 0; 0 3; 0 Bacterium* 25; 0 Bacterium* 175; 2 Yeast* 719; 7 Yeast* Paenibacillus chitinolyticus Staphylococcus capitis Enterobacter cloacae Micrococcus antarcticus Micrococcus luteus Bacterium* Staphylococcus epidermidis Micrococcus luteus Moraxella osloensis Staphylococcus hominis Staphylococcus capitis Staphylococcus capitis 5 684; 7 Bacterium* 1; 0 Bacterium* 6; 0 Staphylococcus capitis 18; 0 Bacillus cohnii 34; 0 Bacillus sp 264; 3 Bacterium* Neisseria subflava Yeast* Staphylococcus cohnii Staphylococcus warneri Staphylococcus hominis Staphylococcus lugdunensis Staphylococcus sciuri Bacterium* 6 115; 1 Bacillus cereus 1; 0 Staphylococcus hominis 3; 0 Bacterium* 3; 0 Brevibacterium iodinum 20; 0 Micrococcus luteus 24; 0 Bacillus megaterium Enterobacter cloacae Staphylococcus cohnii Bacterium* Paenibacillus massiliensis Moraxella sp Micrococcus antarcticus Staphylococcus epidermidis Kocuria marina Pseudomonas flavescens Staphylococcus cohnii Staphylococcus cohnii Staphylococcus epidermidis Staphylococcus hominis 7 74; 1 Kocuria marina 7; 0 Bacillus mannanilyticus 40; 0 Neisseria perflava 42; 0 Staphylococcus epidermidis 65; 1 Micrococcus luteus 201; 2 Bacillus megaterium Micrococcus antarcticus Lactobacillus paracasei Rothia mucilaginosa Staphylococcus haemolyticus Neisseria flavescens Neisseria flavescens Staphylococcus capitis Neisseria perflava Staphylococcus aureus Staphylococcus pettenkoferi Neisseria sicca Staphylococcus hominis Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus hominis Rothia mucilaginosa Staphylococcus haemolyticus 8 13; 0 Kocuria rhizophila 0; 0 5; 0 Arcanobacterium phocae 8; 0 Bacillus cereus 11; 0 Staphylococcus epidermidis 105; 1 Alkalibacillus haloalkaliphilus Kocuria rosea Cardiobacterium sp Corynebacterium sp Staphylococcus haemolyticus Micrococcus luteus Moraxella osloensis Staphylococcus hominis Kocuria palustris Staphylococcus capitis Staphylococcus capitis Staphylococcus haemolyticus Staphylococcus epidermidis Staphylococcus hominis 9 187; 2 Bacillus simplex 0; 0 1; 0 Staphylococcus haemolyticus 3; 0 Bacillus smithii 6; 0 Micrococcus luteus 10; 0 Corynebacterium afermentans Staphylococcus epidermidis Micrococcus antarcticus Staphylococcus epidermidis Staphylococcus hominis Staphylococcus hominis Staphylococcus epidermidis Staphylococcus hominis Staphylococcus warneri Staphylococcus saprophyticus Note. *Indicates identification scores of ≤ 60, considered “not reliable”. CFU, colony-forming units. -
[1] Biddle C, Shah J. Quantification of anesthesia providers' hand hygiene in a busy metropolitan operating room: what would Semmelweis think? Am J Infect Control, 2012; 40, 756−9. [2] Fernandez PG, Loftus RW, Dodds TM, et al. Hand hygiene knowledge and perceptions among anesthesia providers. Anesth Analg, 2015; 120, 837−43. doi: 10.1213/ANE.0000000000000408 [3] Paul ET, Kuszajewski M, Davenport A, et al. Sleep safe in clean hands: improving hand hygiene compliance in the operating room through education and increased access to hand hygiene products. Am J Infect Control, 2019; 47, 504−8. doi: 10.1016/j.ajic.2018.10.021 [4] Sahni N, Biswal M, Gandhi K, et al. Quantification of hand hygiene compliance in anesthesia providers at a tertiary care center in northern India. Am J Infect Control, 2015; 43, 1134−6. doi: 10.1016/j.ajic.2015.05.035 [5] Scheithauer S, Rosarius A, Rex S, et al. Improving hand hygiene compliance in the anesthesia working room work area: more than just more hand rubs. Am J Infect Control, 2013; 41, 1001−6. doi: 10.1016/j.ajic.2013.02.004 [6] Segal S, Harris HM, Gunawan A, et al. A simple method for estimating hand hygiene use among anesthesia personnel: development, validation, and use in a quality improvement project. Anesth Analg, 2019; 129, 1549−56. doi: 10.1213/ANE.0000000000004106 [7] Regulation of disinfection technique in healthcare settings (WS/T 367-2012) The Health Industry Standard of the People's Republic of China, 2012. [8] Specification of hand hygiene for healthcare workers (WS/T 313-2019). The Health Industry Standard of the People's Republic of China, 2019. [9] Ning YT, Yang WH, Zhang W, et al. Developing two rapid protein extraction methods using focused-Ultrasonication and zirconia-silica beads for filamentous fungi identification by MALDI-TOF MS. Front Cell Infect Microbiol, 2021; 11, 687240. doi: 10.3389/fcimb.2021.687240 [10] Stone S, Teare L, Cookson B. Guiding hands of our teachers. Lancet, 2001; 357, 479−80. doi: 10.1016/S0140-6736(05)71287-3 [11] Pittet D, Hugonnet S, Harbarth S, et al. Effectiveness of a hospital-wide programme to improve compliance with hand hygiene. Lancet, 2000; 356, 1307−12. doi: 10.1016/S0140-6736(00)02814-2 [12] Birnbach DJ, Rosen LF, Fitzpatrick M, et al. The use of a novel technology to study dynamics of pathogen transmission in the operating room. Anesth Analg, 2015; 120, 844−7. doi: 10.1213/ANE.0000000000000226 [13] Munoz-Price LS, Birnbach DJ. Hand hygiene and anesthesiology. Int Anesthesiol Clin, 2013; 51, 79−92. doi: 10.1097/AIA.0b013e31826f2db6 [14] Jefferson J, Whelan R, Dick B, et al. A novel technique for identifying opportunities to improve environmental hygiene in the operating room. AORN J, 2011; 93, 358−64. doi: 10.1016/j.aorn.2010.08.022 [15] Cole DC, Baslanti TO, Gravenstein NL, et al. Leaving more than your fingerprint on the intravenous line: a prospective study on propofol anesthesia and implications of stopcock contamination. Anesth Analg, 2015; 120, 861−7. doi: 10.1213/ANE.0b013e318292ed45 [16] DiDiodato G. Has improved hand hygiene compliance reduced the risk of hospital-acquired infections among hospitalized patients in Ontario? Analysis of publicly reported patient safety data from 2008 to 2011. Infect Control Hosp Epidemiol, 2013; 34, 605−10. doi: 10.1086/670637 [17] Weinstein RA, Hota B. Contamination, disinfection, and cross-colonization: are hospital surfaces reservoirs for nosocomial infection? Clin Infect Dis, 2004; 39, 1182−9. [18] Huang SS, Datta R, Platt R. Risk of acquiring antibiotic-resistant bacteria from prior room occupants. Arch Intern Med, 2006; 166, 1945−51. doi: 10.1001/archinte.166.18.1945 [19] Loftus RW, Koff MD, Brown JR, et al. The dynamics of Enterococcus transmission from bacterial reservoirs commonly encountered by anesthesia providers. Anesth Analg, 2015; 120, 827−36. doi: 10.1213/ANE.0000000000000123 [20] Munoz-Price LS, Weinstein RA. Acinetobacter infection. N Engl J Med, 2008; 358, 1271−81. doi: 10.1056/NEJMra070741 [21] Otter JA, Yezli S, French GL. The role played by contaminated surfaces in the transmission of nosocomial pathogens. Infect Control Hosp Epidemiol, 2011; 32, 687−99. doi: 10.1086/660363 [22] Vajravelu RK, Guerrero DM, Jury LA, et al. Evaluation of stethoscopes as vectors of Clostridium difficile and methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol, 2012; 33, 96−8. doi: 10.1086/663338 [23] Bode LGM, Kluytmans JAJW, Wertheim HFL, et al. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med, 2010; 362, 9−17. doi: 10.1056/NEJMoa0808939 [24] Hidron AI, Edwards JR, Patel J, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2006-2007. Infect Control Hosp Epidemiol, 2008; 29, 996−1011. doi: 10.1086/591861 [25] Huang SS, Yokoe DS, Hinrichsen VL, et al. Impact of routine intensive care unit surveillance cultures and resultant barrier precautions on hospital-wide methicillin-resistant Staphylococcus aureus bacteremia. Clin Infect Dis, 2006; 43, 971−8. doi: 10.1086/507636 [26] Jain R, Kralovic SM, Evans ME, et al. Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N Engl J Med, 2011; 364, 1419−30. doi: 10.1056/NEJMoa1007474 [27] Loftus RW, Muffly MK, Brown JR, et al. Hand contamination of anesthesia providers is an important risk factor for intraoperative bacterial transmission. Anesth Analg, 2011; 112, 98−105. doi: 10.1213/ANE.0b013e3181e7ce18 [28] Krawczyk B, Wityk P, Gałęcka M, et al. The many faces of Enterococcus spp.-Commensal, probiotic and opportunistic pathogen. Microorganisms, 2021; 9, 1900. doi: 10.3390/microorganisms9091900 [29] Orazi G, Collins AJ, Whitaker RJ. Prediction of prophages and their host ranges in pathogenic and commensal Neisseria species. mSystems, 2022; 7, e0008322. doi: 10.1128/msystems.00083-22 [30] Ramos-Zayas A, López-Medrano F, Urquiza-Fornovi I, et al. The impact of healthcare-associated infections in patients undergoing oncological microvascular head and neck reconstruction: a prospective Multicentre study. Cancers, 2021; 13, 2109. doi: 10.3390/cancers13092109 [31] Joga M, Palombo EA. Removal of contaminating bacteria from computers by disinfection and hand sanitation. Am J Infect Control, 2012; 40, 389−90. doi: 10.1016/j.ajic.2011.11.018 [32] Koff MD, Brown JR, Marshall EJ, et al. Frequency of hand decontamination of intraoperative providers and reduction of postoperative healthcare-associated infections: a randomized clinical trial of a novel hand hygiene system. Infect Control Hosp Epidemiol, 2016; 37, 888−95. doi: 10.1017/ice.2016.106 [33] Noritomi DT, Chierego M, Byl B, et al. Is compliance with hand disinfection in the intensive care unit related to work experience? Infect Control Hosp Epidemiol, 2007; 28, 362-4. [34] Pittet D, Stéphan F, Hugonnet S, et al. Hand-cleansing during postanesthesia care. Anesthesiology, 2003; 99, 530−5. doi: 10.1097/00000542-200309000-00006 [35] Larson EL, Albrecht S, O’keefe M. Hand hygiene behavior in a pediatric emergency department and a pediatric intensive care unit: comparison of use of 2 dispenser systems. Am J Crit Care, 2005; 14, 304−11. doi: 10.4037/ajcc2005.14.4.304 -
 22123Supplementary Materials.pdf
22123Supplementary Materials.pdf

-




 下载:
下载:





 Quick Links
Quick Links