-
Salidroside (SAL) is a glucoside of tyrosol and a key constituent of Rhodiola rosea L. The diverse pharmacological activities of SAL include anti-inflammatory, cardioprotective, and neuroprotective effects[1]. Our previous investigation showed that the anti-aging effects of SAL involve reductions of the levels of advanced glycation end products in a D-galactose induced mouse model[2]. Additionally, in 2BS human embryonic lung diploid fibroblasts in which cell senescence was induced using hydrogen peroxide, the protective potential of SAL against senescence was also proven by determination of senescence associated proteins, including P21 and P16[3]. However, the associated gene biomarkers responsible for the anti-senescence property of SAL remain unclear. In this study, we used the 2BS cell premature senescence model to detect the protective potential of SAL against senescence using whole genome mRNA microarray analysis and to reveal the possible molecular mechanisms.
As we demonstrated previously[2], the cells are considered to be young when the population doubling (PD) number is ≤ 30 and near replicative senescence (which can be induced by telomere shortening) at PD 50. Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 mg/L) at 37 ℃ in a humidified atmosphere containing 5% CO2. SAL was dissolved in dimethyl sulfoxide (DMSO) as a 10 mmol/L stock solution. Aliquots from the resulting culture containing 2 × 106 cells were individually dispensed in a 10 cm dish and cultured for 24 h. Cells at PD 30 were treated with 10 μmol/L SAL or 0.1% DMSO (control) for 24 h. Total RNA was isolated from the collected cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. The extracted RNA was reverse-transcribed to cDNA using total mRNA as a template. Then, 4 × 180 k human gene microarrays analysis of 2BS cells was conducted by Shanghai Biotechnology Corporation (China). Slides were scanned using a G2565CA microarray scanner (Agilent Technologies, Santa Clara, CA, USA). Raw data were extracted using Feature Extraction Software (Agilent Technologies. Quantitative RT-PCR was performed to detect the expression of mRNA according to the SYBR green method to confirm the microarray analysis results. The data are expressed as mean ± SD. One-way ANOVA analysis was done using SPSS 19.0 software (SPSS Inc., Chicago, IL, USA) to compare differences between groups. P-values < 0.05 were considered statistically significant.
Based on the results of the Agilent chip experiment, a gene ontology (GO) enrichment analysis of differentially expressed mRNAs was performed to identify GO categories with higher confidence. Enrichment provides a measure of the significance of the function. As GO enrichment increases, the corresponding function is more specific, which helps to conclusively identify GO-related functions[4]. Presently, the two most enriched GO categories associated with differentially expressed transcripts were 'response to cGMP' (GO: 0070305; biological process; P = 0.001) and 'cellular response to cGMP' (GO: 0071321; biological process; P = 0.001) (Supplementary Figure S1 available in www.besjournal.com).
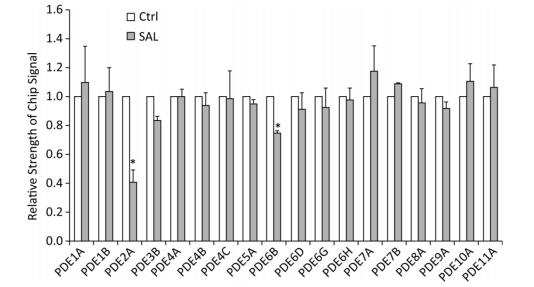
GO analysis of these target genes revealed the close relationship of 3′, 5'-cyclic nucleotide phosphodiesterases (PDEs) genes with 'response to cGMP', which was significantly expressed induced by SAL in old (PD 50) 2BS cells (Figure 1). Guanosine 3', 5'-cyclic monophosphate (cGMP) is an intracellular second messenger molecule that is critical in intracellular signaling during the development and function of the brain. PDEs comprise a family of enzymes (PDE1-11) that hydrolyze cGMP and the associated compound cyclic adenosine-3-5- monophosphate (cAMP). The 11 PDEs differ in their substrate specificity, kinetic properties, mode of regulation, intracellular localization, and patterns of tissue expression. Each PDE could be a unique target molecule in modulating separate physiological processes[5]. Thus, selectively targeting PDEs with small molecules or biologics may be one way to modulate cGMP in different biological processes. PDE isozymes 1, 2, 4, and 9 are believed to have the most influential effects on cGMP signaling in various diseases[6]. Presently, the gene for PDE2A was the most differentially expressed gene induced by SAL in 2BS cells (Figures 1 and 2).
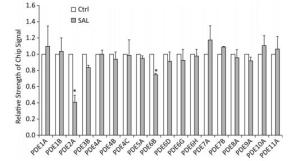
Figure 1. The effect of salidroside (SAL) on the expression of PDE members in 2BS human fibroblasts. The histogram was based on the microarray data (*P < 0.05). Ctrl: control.
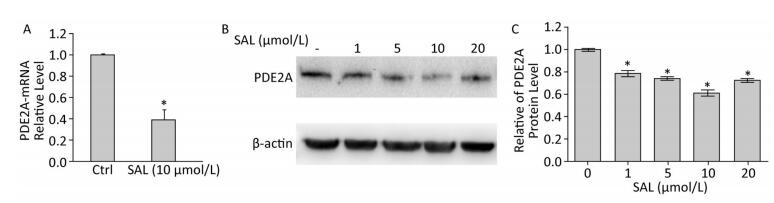
Figure 2. Salidroside (SAL) inhibits PDE2A expression at the mRNA (A) and protein (B, C) levels in 2BS human fibroblasts. (A) The relative expression levels of PDE2A mRNAs induced by SAL in 2BS cells. Each value represents the mean ± SD of three repeat experiments, *P < 0.05. (B, C) Western blot demonstrating the effect of SAL concentration on the expression of PDE2A protein in 2BS human fibroblast cells. Each value represents the mean ± SD of three repeat experiments, *P < 0.05. The 2BS cells recovered at PD 50 were incubated with SAL (1, 5, 10, 20 μmol/L) or dimethyl sulfoxide (control, Ctrl) for 24 h and analyzed for protein or mRNA levels.
PDE2 is a dual-substrate PDE that is widely expressed in different tissues and organs. The highest concentrations are produced in the striatum, cortex, and hippocampus[7]. PDE2A is activated by binding of cGMP to an allosteric site in the N-terminal domain, which enhances the rate of cAMP and cGMP hydrolysis. Thus, inhibition of PDE2A would result in elevation of both cAMP and cGMP levels, which may be one of its mechanisms in different biological processes. This and other aspects of PDE2A in cell senescence require further study.
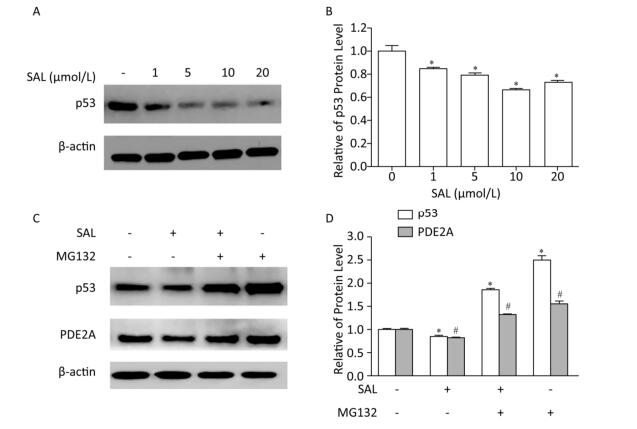
We previously described that SAL induces the significant reduction of p53, p21, p16, as well as their downstream targets CDK4 and CDK6 in PD 50 2BS cells[3, 8]. Another study described a series of potential candidates, including PDE2A, GAS6, HRAS, KITLG, and TGFA, which could interact with p53 through p53 binding sites[9]. Similar results was also predicted by using the MAPPERI Database (see Supplementary Table S1, available in www.besjournal.com). In the present study, SAL suppressed PDE2A and p53 protein expression in a dose-dependent manner (Figure 2B and Figure 3A).
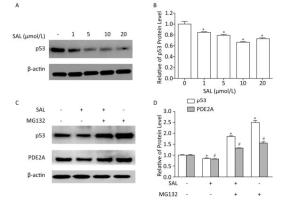
Figure 3. Salidroside (SAL) inhibition of PDE2A expression is associated with p53 down-regulation in 2BS human fibroblasts. (A, B) Western blot assessment of the effect of SAL on p53 protein expression in 2BS cells. Each value represents the mean ± SD of three repeat experiments, *P < 0.05. 2BS cells obtained at PD 50 were incubated in fresh medium alone or medium supplemented with different concentrations of SAL for 24 h before harvest. (C, D) Western blot assessment of the effect of SAL on p53 and PDE2A protein expression in 2BS cells. Each value represents the mean ± SD of three repeat experiments, *P < 0.05, #P < 0.05. 2BS cells recovered at PD 50 were dispensed (2 × 106 cells) in a 10-cm diameter dish and incubated for 24 h. The cells were then incubated in fresh medium supplemented with 10 μmol/L SAL for 24 h, and then treated with MG132 (10 μmol/L) for 4 h before harvest for the detection of the expression of p53 and PDE2A proteins.
To further investigate the p53 dependence of SAL on PDE2A inhibition, a proteasome inhibitor (MG132), which causes the accumulation of p53 protein[10], was used with the SAL treatment of PD 50 2BS cells. The down-regulation of PDE2A by SAL was blocked by MG132, which induced an incremental change in p53 protein level (Figure 3C). The findings indicated that SAL can reduce PDE2A expression in a p53-dependent manner.
In conclusion, the findings provide new insights regarding the multifunctional mode of action of SAL. SAL could be a potential PDE2A inhibitor that exerts its activity in a p53-dependent manner. Furthermore, the findings indicate a new interventional strategy against age-related diseases, in particular neurodegenerative disorders in humans, using SAL supplementation.
Salidroside Reduces PDE2A Expression byDown-regulating p53 in Human Embryonic Lung Fibroblasts
doi: 10.3967/bes2019.020
Chinese traditional medicine science and technology projects of Zhejiang Province 2015ZA002
the Health Bureau of Zhejiang Province 2017KY189
the Science Technology Department of Zhejiang Province 2016C34002
the National Natural Science Foundation of China 81501113
the National Natural Science Foundation of China 81771520
the Health Bureau of Zhejiang Province 2019RC091
the Health Bureau of Zhejiang Province 2015KYA001
the Health Bureau of Zhejiang Province 2019KY257
the Health Bureau of Zhejiang Province 2015DTA001
Chinese traditional medicine science and technology projects of Zhejiang Province 2017ZB002
the Health Bureau of Zhejiang Province 2017KY188
Chinese traditional medicine science and technology projects of Zhejiang Province 2014ZB001
the Natural Science Foundation of Zhejiang province LQ16H020006
- Received Date: 2018-11-20
- Accepted Date: 2019-01-10
| Citation: | XING Wen Min, CHEN Sha Sha, WANG San Ying, GAO Wen Yan, WAN Xiao Qing, SU Hui Li, YANG Yi, ZHANG Jing, YAN Jing, MAO Gen Xiang. Salidroside Reduces PDE2A Expression byDown-regulating p53 in Human Embryonic Lung Fibroblasts[J]. Biomedical and Environmental Sciences, 2019, 32(2): 140-143. doi: 10.3967/bes2019.020 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: