-
Enteroviruses (EVs) are members of the genus Enterovirus within the order Picornavirales, family Picornaviridae, and consist of 12 species, including EV-A, EV-B, EV-C, and EV-D, which are associated with human infections. Coxsackievirus B6 (CV-B6) belongs to the species EV-B, which currently consists of 63 serotypes, including echovirus (serotypes 1–7, 9, 11–21, 24–27, 29–33), coxsackievirus group A (CV-A9), coxsackievirus group B (CV-B, serotypes 1–6), the newly identified EVs (serotypes EV-B69, B73–75, B77–88, B93, B97–98, B100–101, B106–107, and B110–113), and the simian enterovirus SA5.
EVs are small non-enveloped viruses containing 60 copies of each of the capsid proteins (VP4, VP2, VP3, and VP1) and enclose a single-stranded, positive-sense RNA genome. The viral RNA (approximately 7.4–7.5 kb) contains a long open reading frame flanked by a 5′-untranslated region (UTR) and a 3′-UTR. The 5′-UTR is about 740–750 bp in length and contains a cloverleaf structure and a secondary structure called the internal ribosome entry site (IRES) involved in the replication and internal initiation of translation of genomic RNA[1]. A single polyprotein translated from the RNA strand is first cleaved into 3 polyprotein precursors: P1, P2, and P3. P1 is processed to yield 4 structural proteins (VP1–VP4), whereas P2 and P3 are the precursors of nonstructural proteins 2A–2C and 3A–3D, respectively.
CV-B is classified into six serotypes (CV-B1–CV-B6), and can cause a variety of serious human diseases including acute myocarditis, acute aseptic meningitis, encephalitis, and acute flaccid paralysis. CV-B6 is one member of the CV-B viruses and the prototype strain CV-B6 (strain Schmitt/PHI/1953) was isolated in the Philippines in 1953[2]. Unlike other serotypes of CV-B, which are viruses with high detection rates and wide distributions, CV-B6 was rarely reported[3] and only a few nucleotide sequences were available in the GenBank; only three complete genome sequences (Strains Schmitt, DUCT, and LEV15) have been published[2,4]. The three CV-B6 complete genome sequences had very high nucleotide identity with each other, with only 10–18 nucleotide substitutions (4–10 amino acid substitutions), indicating that the strains DUCT and LEV15 were more like variants of the CV-B6 strain Schmitt than the epidemic strains. Hence, in the present study, the first two full-length genome sequences of Chinese CV-B6 strains were determined and compared with those of other EV-B viruses.
The two Chinese CV-B6 strains, 99148/XZ/CHN/1999 and X0012/XZ/CHN/2000 (hereafter referred to as 99148 and X0012), were isolated from two healthy children in the Xigazê Prefecture and Lhasa City of the Tibet Autonomous Region of China in 1999 and 2000, respectively. Stool samples from the 2 children were collected and sent to National Polio Laboratory in China supporting the Global Poliovirus Eradication Initiative. To maintain the high level of sensitivity of acute flaccid paralysis cases surveillance system in Tibet, 150 stool samples were collected from healthy children under 5 years old since 1996 due to the number of acute flaccid paralysis cases is too little (less than ten cases each year).
The stool sampls were processed according to the standard procedures recommended by the World Health Organization[5]. The samples were then inoculated into the following 2 cell lines: human rhabdomyosarcoma (RD) and human laryngeal epidermoid carcinoma (HEp-2). An EV-like cytopathic effect (CPE) was observed in both RD and HEp-2 cells. Viral RNA was extracted from infected cell culture supernatants that produced typical CPE in RD or HEp-2 cells using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, USA). The complete VP1 region of each virus was amplified using the serotype specific primers (upstream primer, CVB6Y: GCCACTGGGAGCTCAATCTA, and downstream primer, CVB6Z: TGCTCACAAGGAGGTCCCTA), and RT-PCR was performed with an Access RT-PCR Kit (Promega, Madison, Wisconsin, USA) according to the manufacturer’s instructions. The PCR products obtained were purified using a QIAquick Gel Extraction Kit (Qiagen), and the amplicons were bi-directionally sequenced using an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems, Hitachi, Japan). The two viral isolates were identified as CV-B6 based on a molecular typing method for enteroviruses by using the online Enterovirus Genotyping Tool, which revealed 74.5%–75.3% nucleotide sequence similarity and 95.7%–96.4% amino acid sequence similarity with the VP1 sequence of the prototype CV-B6 strain (strain Schmitt). In contrast, the two isolates showed less than 70% nucleotide similarity and less than 85% amino acid similarity to the other EVs serotypes.
The two CV-B6 isolates were further characterised by full-length genome sequencing. Primers were designed to anneal to highly conserved sites among the CV-B6 sequences available in the GenBank, and full-length genome sequencing was achieved using the 'primer-walking' strategy[6]. Viral RNA extraction, RT-PCR amplification, and PCR product purification were performed using standard methods, as described previously[7]. Sequencing was performed in both directions on an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems), and every nucleotide position from each strand was sequenced at least once. The 5′ segment sequences were determined using a 5′ rapid amplification of cDNA ends (RACE) core set (Takara Biomedicals), according to the manufacturer’s instructions.
Phylogenetic analyses were performed based on the entire VP1 region of the two Chinese CV-B6 strains and the other CV-B6 strains available in the GenBank, and based on the P1, P2, and P3 regions of the two Chinese CV-B6 strains and all the EV-B prototype strains. Phylogenetic trees were constructed using the neighbour-joining method using the MEGA program (version 7.0; Sudhir Kumar, Arizona State University, Tempe, AZ, USA). The topology of the trees was obtained by majority-rule consensus among 1,000 bootstrap replicates shown as percentages, and bootstrap values greater than 80% were considered statistically significant for grouping. The full-length genome sequences of the two Chinese CV-B6 strains described in this study were deposited in the GenBank under the accession numbers KY262575 and KY262576.
The two Chinese CV-B6 strains (99148 and X0012) were 7,399 and 7,398 nucleotides (nt) in length, respectively, encoding a polypeptide of 2,184 amino acids. The coding sequences were flanked by a non-coding 5′-UTR of 742 nt in both, a non-coding 3′-UTR of 102 and 101 nt, respectively, and a poly(A) tail composed of a long sequence of adenine nucleotides to the 3′ end of the genome. Analysis of the full-length genomic sequences of the two Chinese CV-B6 strains showed that both their genomes were collinear with that of the prototype strain (Schmitt, GenBank No. AF105342), and both strains have nucleotide deletions at position 374 in the 5′-UTR and nucleotide insertions at position 7308 in the 3′-UTR, and that strain 99148 has one more nucleotide insertion at position 7298 in the 3′-UTR. The overall base compositions of strain 99148 and X0012 were 28.03% and 27.98% A, 24.44% and 24.52% G, 24.00% and 24.07% C, and 23.53% and 23.43% U, respectively. The polypeptide cleavage sites were predicted using the complete genome sequence of the CV-B6 prototype strain. Table 1 shows the nucleotide and amino acid sequence identities between the Chinese CV-B6 strains, the prototype strain, and other EV-B prototype strains.
Region Nucleotide identity (%)/Amino acid identity (%) 99148/XZ/CHN/1999 X0012/XZ/CHN/2000 Prototype of CV-B6 Prototype of other EV-B Prototype of CV-B6 Prototype of other EV-B 5′-UTR 85.5 79.9–90.3 86.6 80.7–90.9 VP4 78.2 (95.6) 70.0–82.6 (76.8–100.0) 77.2 (95.6) 69.5–82.6 (76.8–100.0) VP2 78.4 (95.0) 65.9–72.7 (74.0–85.0) 77.9 (95.0) 66.4–73.8 (74.0–84.2) VP3 78.1 (97.0) 62.8–72.2 (67.7–84.0) 77.8 (97.0) 62.7–72.6 (67.3–83.6) VP1 79.1 (96.4) 56.0–67.3 (56.2–75.6) 78.6 (95.7) 55.5–67.9 (56.9–74.9) 2A 80.0 (93.3) 75.7–90.2 (84.0–96.6) 80.0 (93.3) 76.0–86.0 (85.3–96.6) 2B 79.1 (95.9) 75.4–90.5 (92.9–98.9) 79.4 (96.9) 76.7–88.2 (92.9–97.9) 2C 80.3 (97.8) 79.0–86.1 (96.3–99.0) 81.8 (97.8) 79.9–86.2 (96.9–99.0) 3A 84.6 (100.0) 76.0–88.3 (92.1–100.0) 80.8 (100.0) 74.1–86.5 (92.1–100.0) 3B 78.7 (100.0) 72.7–86.3 (90.9–100.0) 83.3 (100.0) 71.2–84.8 (90.9–100.0) 3C 76.3 (96.1) 75.7–87.4 (93.4–99.4) 78.3 (96.1) 74.8–85.9 (93.4–98.9) 3D 79.5 (96.7) 76.8–88.3 (95.0–98.7) 78.6 (96.7) 77.2–86.7 (94.3–98.9) 3′-UTR 85.5 76.9–95.1 84.6 75.0–93.2 Table 1. Pair-wise nucleotide and amino acid sequence identities between Chinese CV-B6 isolates, the CV-B6 prototype strain, and other prototype strains of the EV-B species
The nucleotide sequences available for the distinct CV-B6 strains differ considerably. The sequence of the VP1 coding region of the two Chinese CV-B6 strains displayed a genetic distance of 0.203-0.264 with the prototype strain (Schmitt), strain LEV15/RUS/2011 (GenBank No. JQ041368) and strain VMC-1/YN/CHN/1991 (GenBank No. AF225469). The strain LEV15 showed 99.4% nucleotide similarity with strain Schmitt whereas strain VMC-1 had a high genetic distance of 0.257–0.264 with the above two CV-B6 strains.
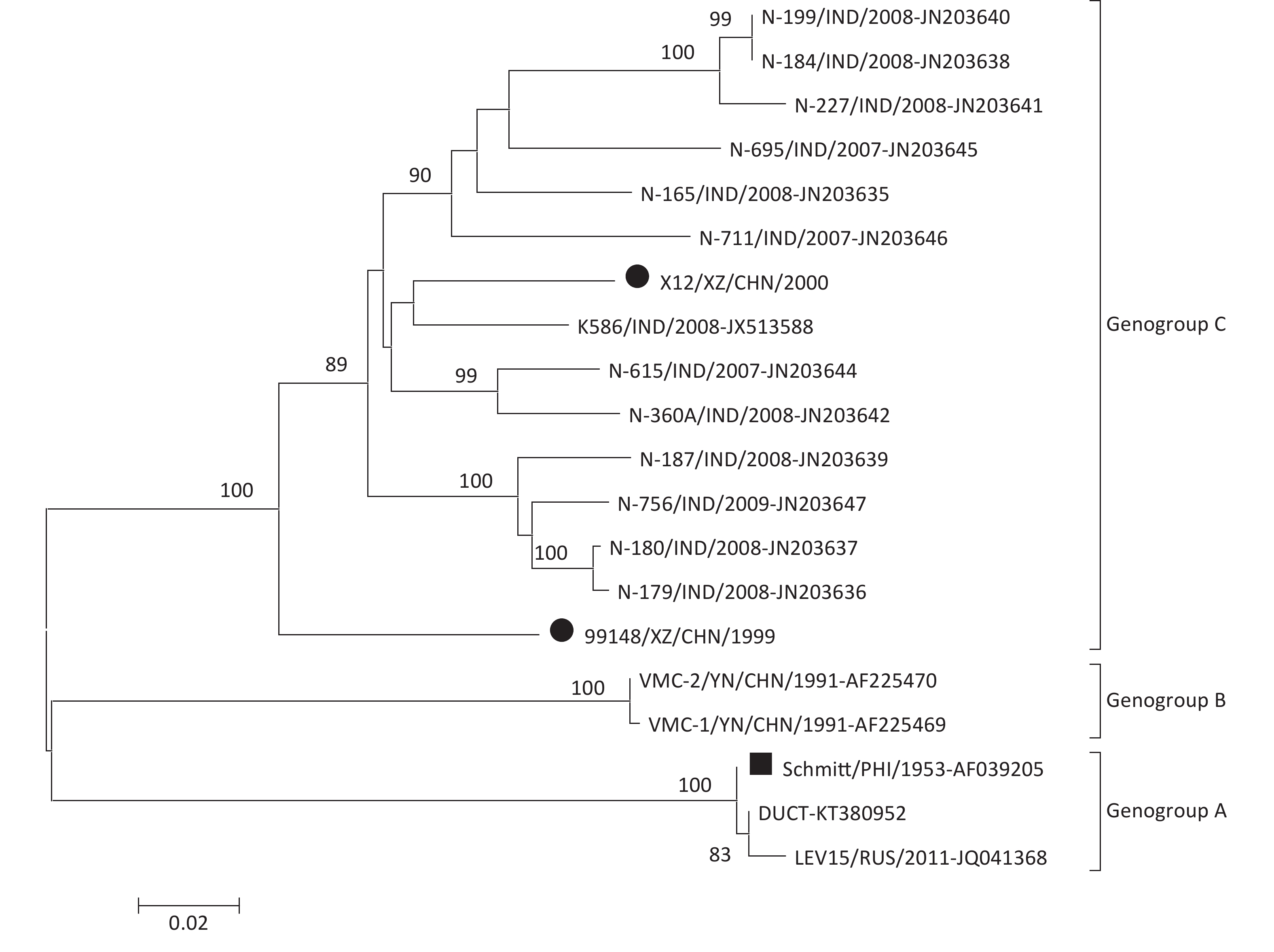
To determine the molecular epidemiology of the CV-B6 strains, a phylogenetic dendrogram based on the entire VP1 sequence was constructed with the two Chinese CV-B6 strains isolated in this study, together with all 18 CV-B6 sequences available in the GenBank database (Figure 1). The phylogenetic analysis based on the VP1 sequence, which was supported by a high bootstrap value, indicated that all the CV-B6 strains clustered into genogroups A, B, and C. The CV-B6 prototype strain (Strain Schmitt) together with the other two Schmitt-like strains (Strains DUCT and LEV15) form genogroup A, whereas genogroup B contains two CV-B6 strains that were isolated from patients with heart muscle disease from a selenium-deficient area of China. The two CV-B6 strains isolated in this study clustered with the CV-B6 strains isolated from 2007 to 2009 in India, and belong to genogroup C with a genetic distance 0.082–0.148.

Figure 1. Phylogenetic analyses of two Chinese CV-B6 isolates and other CV-B6 strains from GenBank database based on the entire VP1 nucleotide sequence (846 bp).
In the phylogenetic tree based on the P1 sequences, the Chinese CV-B6 strains clustered along with the CV-B6 prototype strain resulting in a bootstrap value of 100% (Figure 2A), which confirms the preliminary molecular typing results, but not in the P2 and P3 nonstructural regions (Figure 2B and 2C). Interestingly, in the nonstructural P2 region, both strain 99148 and strain X0012 had highest nucleotide identities (87.5% and 86.5%, respectively) with the EV-B86 prototype strain, and in the nonstructural P3 region, strain 99148 had the highest nucleotide identity (86.9%) with EV-B85 prototype strain, while strain X0012 had the highest nucleotide identity (85.2%) with EV-B75 prototype strain. Based on the genetic characterisations of these two Chinese CV-B6 isolates, it can be concluded that recombination events occurred in the P2 and P3 nonstructural region of these two Chinese CV-B6 strains, and that they had co-circulated with different EV-B viruses for a relatively long term in Tibet.

Figure 2. Phylogenetic relationship of CV-B6 strains and other EV-B strains generated from nucleotide sequence alignment using the neighbour-joining algorithm in MEGA 5.0.
In recent years, only a small number of CV-B6 were identified and could only be identified in Bangladesh and India, during acute flaccid paralysis (AFP) case surveillance activities in support of global polio eradication; however, it was not detected in some provinces of China that have already performed a detailed analysis of non-polio enteroviruses isolated from AFP patients[8]. China has also carried out large-scale enterovirus surveillance in sewage and disease surveillance related to enteroviruses such as hand, foot, and mouth disease case surveillance[9,10], but unfortunately, there is no CV-B6 identified until now.
Unlike other CV-B viruses, CV-B6 are seldom identified from patients with disease outbreaks; hence, the epidemiology of CV-B6 is different since its first isolation in the Philippines in 1953. The reasons for this special epidemiological characteristic remain unclear; it could be that CV-B6 is less infectious than other CV-B viruses, and the virulence of CV-B6 may also weak, resulting in very few cases (most CV-B6 were isolated from asymptomatic healthy individuals including the prototype strain and the two Chinese strains in this study)[2]. To better understand the circulation pattern of CV-B6, a prevalent CV-B6 antibody seroepidemiology study using clinical isolates needs to be conducted, and the enterovirus surveillance should be strengthened and expanded in order to monitor the trends of CV-B6 transmission.
Acknowledgments We would like to acknowledge the staffs of the national polio eradication program in the Tibet Autonomous Region Center for Disease Control and Prevention for the stool specimens of the healthy children used in this study.
Compliance with Ethical Standards The authors declare that they have no conflicts of interest. This study did not involve human participants or human experimentation; the only human materials used were stool samples collected from two healthy children at the instigation of the Ministry of Health P. R. of China for public health purposes. Informed consent was obtained from all individual participants included in the study. This study was approved by the Ethics Review Committee of the National Institute for Viral Disease Control and Prevention (NIVDC), Chinese Center for Disease Control and Prevention.
Disclosure of Potential Conflicts of Interest The authors declare that no competing interests exist.
Molecular Characterisation of Two Coxsackievirus B6 Strains from the Tibet Autonomous Region of China
doi: 10.3967/bes2019.088
- Received Date: 2019-07-15
- Accepted Date: 2019-08-30
| Citation: | SUN Qiang, HONG Mei, FENG Ning, ZHANG Yong. Molecular Characterisation of Two Coxsackievirus B6 Strains from the Tibet Autonomous Region of China[J]. Biomedical and Environmental Sciences, 2019, 32(9): 699-703. doi: 10.3967/bes2019.088 |







 Quick Links
Quick Links
 DownLoad:
DownLoad:
