-
Epstein-Barr virus (EBV) and cytomegalovirus (CMV) are double-stranded DNA viruses belonging to the human herpesvirus family[1], which causes primary and recurrent diseases and seriously endanger human life and health, especially in patients with low immunity[1-3,5]. EBV infection is the main cause of glandular fever and infectious mononucleosis (IM). CMV infection is highly related to human congenital infectious diseases[2]. Both EBV and CMV can cause IM, nervous system infections, unexplained fever, and other clinical disorders[1-5]. The clinical symptoms of the diseases caused by these two pathogens are very similar; therefore, a simple, rapid, convenient, and applicable nucleic acid detection method, especially for grassroots applications, is needed.
Recently, a newly developed recombinase-aided amplification (RAA) technique has become broadly used for the rapid detection of pathogens[6-8]. RAA can achieve exponential amplification of target genes by using enzymes and proteins in a reaction system under an optimal temperature of 37–42 °C in vitro within 30 min. In this study, we developed a real-time internal control reference (ICR) recombinase-aided amplification assay (ICR-RAA) for the detection of EBV and CMV. In parallel, we compared the specificity and sensitivity of the two assay with those of standard quantitative polymerase chain reaction (qPCR) methods. We also attempted detection without nucleic acid extraction to further simplify the novel RAA assays, and carried out preliminary clinical validation.
From March to August 2020, we collected 250 clinical samples from patients with suspected EBV or CMV infections; these were divided into two groups (Supplementary Table S1, available in www.besjournal.com). Group A comprised 125 samples from patients (73 male, 52 female) aged from 0 to 58 years with suspected EBV infection, of which 75 (61 peripheral blood and 14 sera) and 50 serum samples were collected at the Hebei General Hospital in Hebei Province and the Capital Institute of Pediatrics in Beijing, respectively. Group B comprised 125 samples from patients (65 male, 60 female) aged from 0 to 36 years with suspected CMV infection, of which 25 (11 peripheral blood and 14 sera) and 100 serum samples were collected at the Hebei General Hospital and Capital Institute of Pediatrics, respectively. The study was approved by the ethics review committee of the Hebei General Hospital, Capital Institute of Pediatrics, and the Institute of virus Diseases, Chinese Center for Disease Control and Prevention. Prior to the collection of the clinical samples, written informed consent was obtained per routine from all the subjects in the above indicated institutions. Written consent from parents or guardians was required if the subjects were under 18 years old.
Group Sample source
(number and type)Patients information
(number)Reference method Result judgment Internal
controlPositive Negative A Hebei (61 peripheral blood and
14 serum) Beijing (50 serum)Sex: 73 female, 52 male;
Age: 0–58 yearsEBV quantitative PCR kit
(DAAN GENE, Guangzhou, China)CT ≤ 30 CT > 30 NAa B Hebei (11 peripheral blood and
14 serum) Beijing (100 serum)Sex: 65 female, 60 male;
Age: 0–35 yearsCMV quantitative PCR kit
(DAAN GENE, Guangzhou, China)CT ≤ 30 CT > 30 NAa Note. aNA:Not Applied. Table S1. The sample information and the reference methods used in each group
The specificity panel included the stock strains EBV (B95-8 strain; GenBank:M80517. 1), West Nile virus (XJ11129 strain; GenBank: JX442279), nucleic acids of CMV, varicella-zoster virus (VZV), human herpesvirus type 1, 2, and 6 (HHV-1/2/6), hepatitis B virus, and influenza A virus, which were obtained from samples previously preserved in our laboratory[4,6,7].
For the samples from Hebei General Hospital and the stock strains, total DNA was extracted using the QIAamp Blood DNA Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. For the samples from the Capital Institute of Pediatrics, total DNA was extracted using the DAAN3200 automatic nucleic acid extractor (DAAN GENE, Guangzhou, China), according to the manufacturer’s protocol. A total of 48 randomly selected samples from the A and B groups were treated using thermal lysis. Briefly, 50 μL of each sample and 50 μL of DNA extraction buffer (DAAN GENE) were mixed and heated at 99 °C for 10 min, then centrifuged for 5 min at 13,800 ×g. The DNA pellet was stored at 4 °C until use. For whole blood samples, we first used an erythrocyte lysis buffer, then centrifuged twice for 5 min at 13,800 ×g and removed the supernatant, and then applied the above described lysis steps.
The available genome sequences of EBV and CMV were obtained from the National Center for Biotechnology Information database, and the highly conserved sequences were selected and aligned using Vector NTI v11.1 software (Invitrogen, Waltham, MA, USA). According to the principle of RAA primer and probe design, the primers and probes of EBV and CMV were manually designed, and an ICR probe was designed to contain 52 bp of the rosette sequence commonly used in our laboratory (Table 1)[6]. Oligo Primer Analysis Software v.7 (OLIGO, Colorado Springs, CO, USA) and NCBI Primer-Blast were used to analyze and evaluate the specificity of the primers and probes. All primers and probes were synthesized by Shanghai Sangon Biological Co. (Shanghai, China). The EBV (nucleotides: 177631–177771; GenBank accession: M80571.1), CMV (nucleotides: 172141–172304; GenBank accession: KC407666.1), and internal control targeted sequences were cloned into the pUC57 vector by Tsingke Biotechnology Corp (Beijing, China). Qubit dsDNA HS Analysis Kit and Qubit 2.0 fluorometer (Life Technologies, Waltham, MA, USA) were used to quantify the recombinant plasmids.
Assay Primers and probes Sequence 5’–3’ Primer
length (bp)Product
size (bp)EBV dual real-time
ICR-RAAForward Primer CTACCTGTGCCGCATGAAACTGGGCGAGACCGA 33 135 Reverse Primer CATGTCACAGTAAGGACAGAGAAGTCTGGG 30 Probea GAACACCTGAGCGTGGTGAAGCCTCTAACGC-[FAMdT]-[THF]-[BHQ1-dT]-CTGTCCACTCCGAA-C3-spacer 48 ICR Probea GTAAGGTGCTAGACTAAAATTGTTGGGACTT-[HEXdT]-G[THF]-A-[BHQ1-dT]-CTCTGAAGTAAAAGG-C3-spacer 51 CMV dual real-time
ICR-RAAForward Primer CAGATGAGGAAGAKGCTATTGYAGCCTACAC 31 163 Reverse Primer ATCACTCTGYTCACTTTCTTCCTGATCACT 30 Probea CYCCAGAGTCCCCTGTACCCGCGACTA-[FAMdT]-CC-[THF]-[BHQ1-dT]-CTGTCCTCAGTRATT-C3-spacer 47 ICR Probea GTAAGGTGCTAGACTAAAATTGTTGGGACTT-[HEXdT]-G[THF]-A-[BHQ1-dT]-CTCTGAAGTAAAAGG-C3-spacer 51 Note. aProbe modifications: BHQ, black hole quencher; C3-Spacer, 3′ phosphate blocker; FAM, 6-carboxyfuorescein; HEX, 5-hexachlorofuorescein; THF, tetrahydrofuran. Abbreviations: CMV, cytomegalovirus; EBV, Epstein-Barr virus; ICR-RAA, internal control reference recombinase-aided amplification. Table 1. List of primers and probes used in the study
According to the instructions of the fluorescence RAA reaction kit (Qitian Biological Co., Jiangsu, China), the RAA reaction was carried out in a 50 μL system containing 25 μL of buffer, 2.1 μL of each forward and reverse primer (10 μmol/L), 0.6 μL of the target gene and ICR probes (10 μmol/L), and 1 μL of the ICR plasmid; the volume of nuclease free water and template was adjusted to obtain the final reaction volume. The reaction system was placed into pre-freeze-dried reaction tubes (SSB800 30 ng/μL, uvsX120 30 ng/μL, and DNA polymerase 30 ng/μL). Lastly, 2.5 μL of magnesium acetate (280 mmol/L) was added to the cap and the unit tube was transferred to the QT-RAA-B6100 constant-temperature oscillating mixing instrument (Jiangsu Qitian biological Co.) at 39.0 °C for 4 min. Later, the tubes were transferred to the QT-RAA-F1620 real-time fluorescence detector (Jiangsu Qitian biological Co.) set at 39.0 °C for 30 min. A negative control (nuclease free water) was set in each round. The positive curve was determined by QT-RAA-F1620 by setting the slope K value ≥ 25.
Serially diluted plasmids of EBV and CMV (104 to 101, 5 and 1 copies/test) were used to determine the sensitivity of the EBV ICR-RAA and CMV ICR-RAA with eight replicates. To ensure that the amplification of ICR did not affect the normal amplification of the target gene, we optimized the concentration of the ICR template by detecting standard plasmids in the presence of different ICR template concentrations (500, 200, 100, and 50 copies/μL). We used a specific panel of pathogens to explore the specificity of EBV ICR-RAA and CMV ICR-RAA assays. Furthermore, clinical samples in groups A and B were detected by EBV ICR-RAA and CMV ICR-RAA, respectively. A total of 48 randomly selected clinical samples were also tested by real-time EBV ICR-RAA and CMV ICR-RAA using the obtained DNA by thermal lysis. In parallel, two commercial real-time qPCR assays for EBV and CMV detection were performed using 2 μL of extracted nucleic acid. The parameters of the two commercial qPCR kits are listed in Supplementary Table S1.
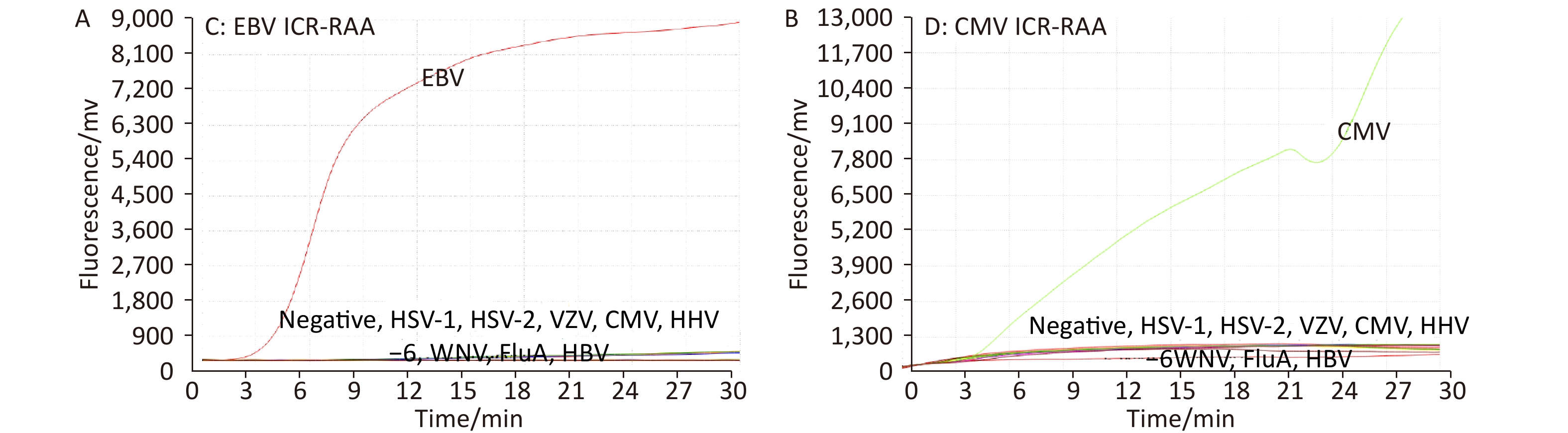
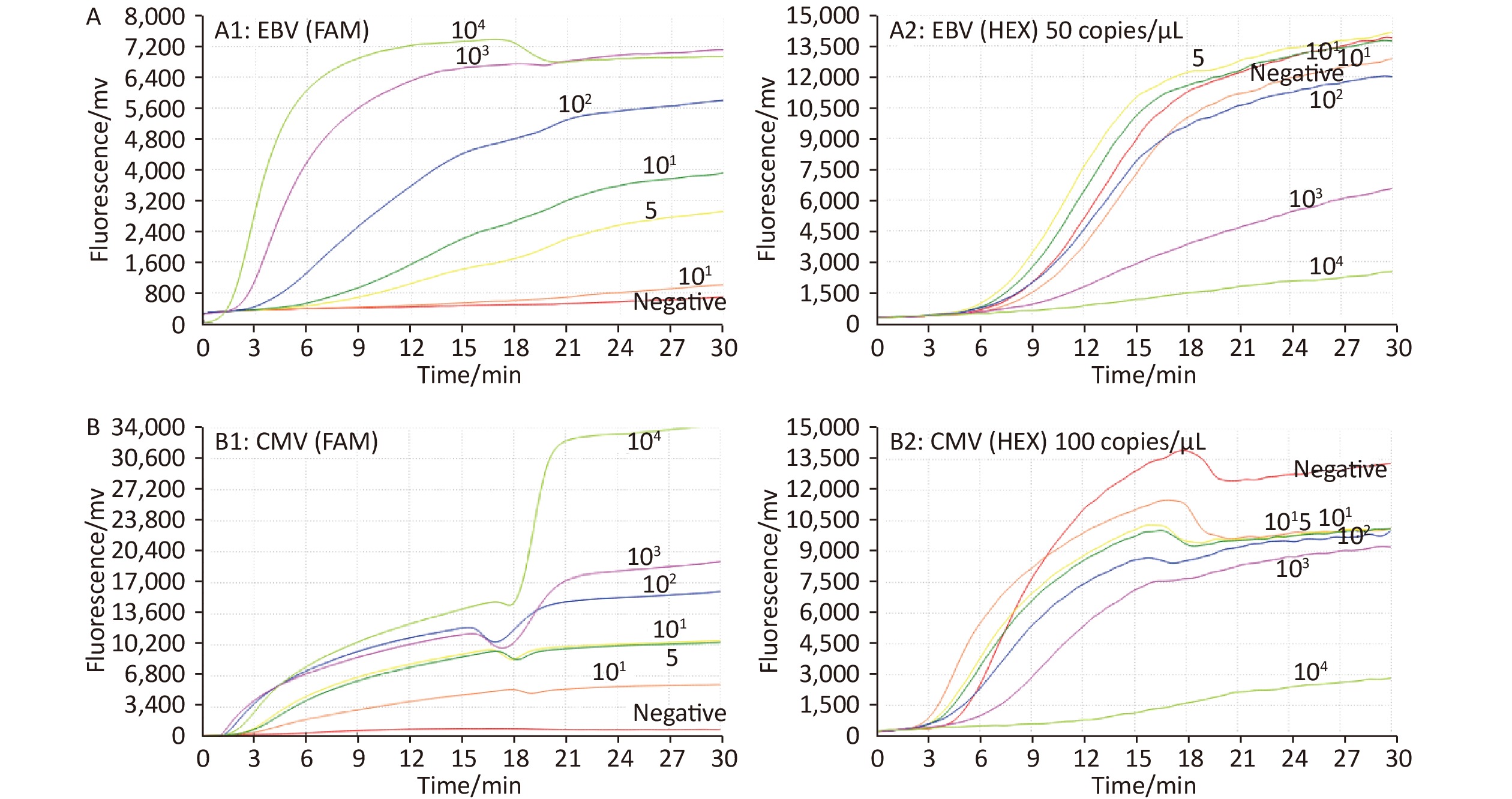
At an ICR concentration of 50 copies/μL, a panel of serially diluted standard plasmid (104 to 101, and 5 copies/test) were positive by EBV ICR-RAA (Figure 1). In the presence of 100 copies/μL of ICR, serially diluted standard plasmids (104 to 101, 5 and 1 copies/test) were positive by CMV ICR-RAA. The low detection limits (LOD) of EBV ICR-RAA and CMV ICR-RAA were calculated to be 5 and 1 copies/reaction, respectively, using probit probability regression analysis. No cross reaction with other human herpesvirus (HHV-1, HHV-2, VZV, or HHV-6) or other pathogens was observed (Figure 2).
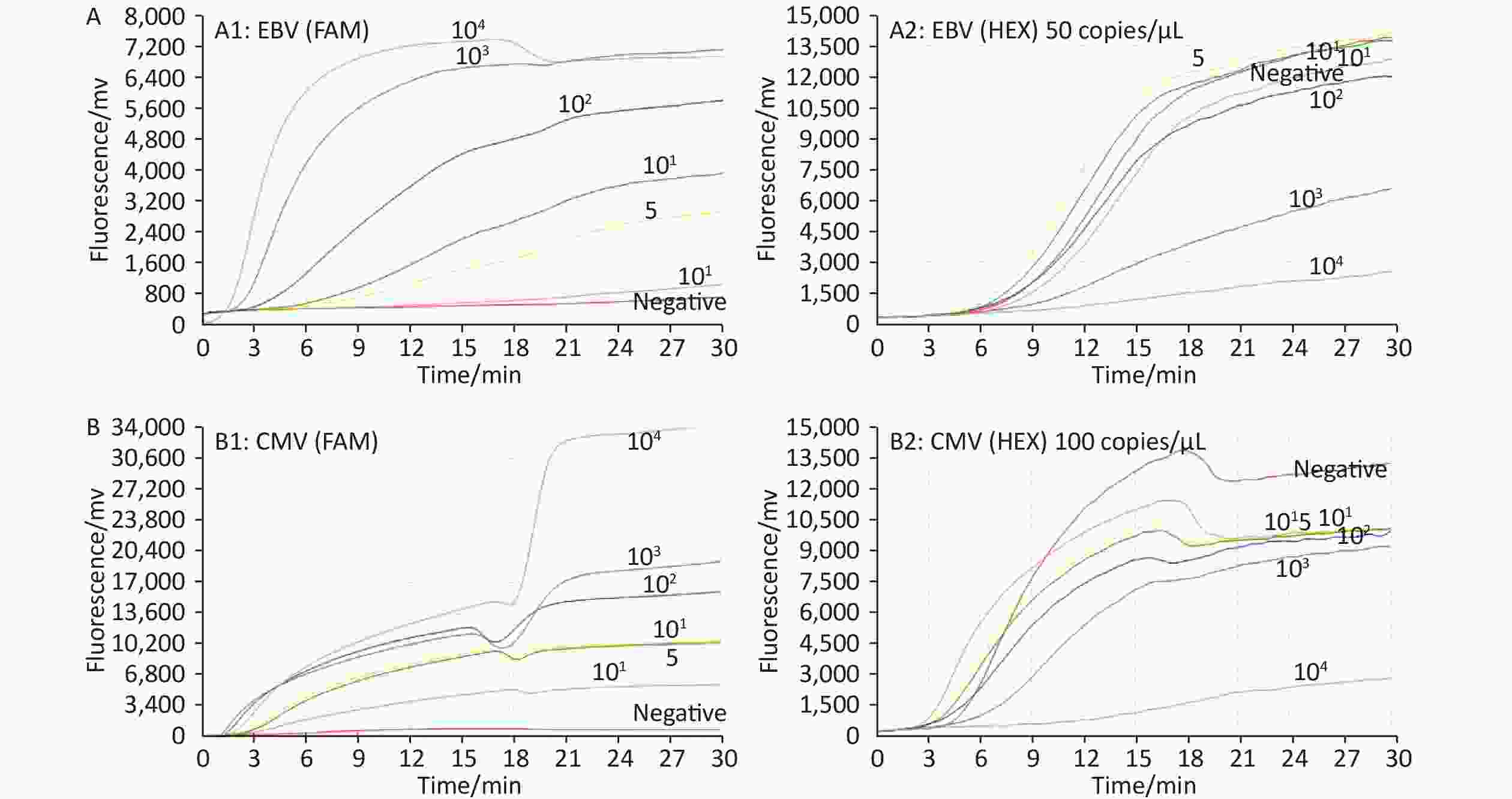
Figure 1. A series of diluted standard plasmid (104 to 101; 5 and 1 copies/reaction) and negative control were used to determine the sensitivity of the Epstein-Barr virus (EBV) and cytomegalovirus (CMV) internal control reference recombinase-aided amplification (ICR-RAA) assays. (A) Sensitivity of the EBV ICR-RAA assay with 50 copies/μL of ICR. A1 and A2 show the amplification curves of the FAM (target gene) and HEX (ICR) channels, respectively. (B) Sensitivity of the CMV ICR-RAA method with 100 copies/μL of ICR. B1 and B2 show the amplification curves of the FAM (target gene) and HEX (ICR) channels, respectively. Abbreviations: CMV, cytomegalovirus; EBV, Epstein-Barr virus; ICR-RAA, internal control reference recombinase-aided amplification; FAM, 6-carboxyfuorescein; HEX, 5-hexachlorofuorescein.
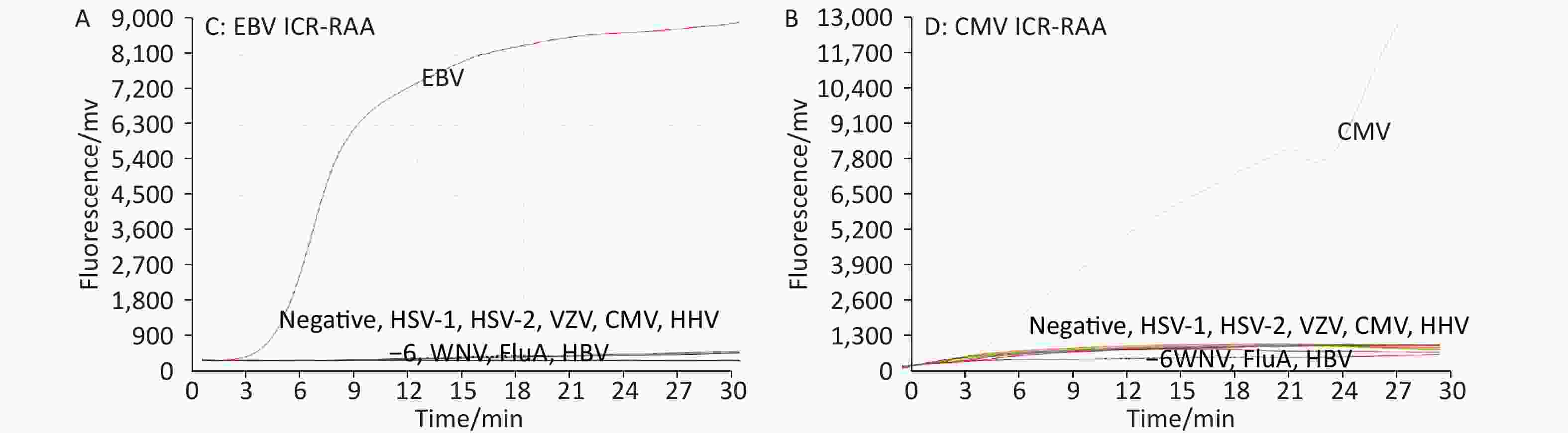
Figure 2. Pathogens in a specificity panel and negative control were used to explore the specificity of the Epstein-Barr virus (EBV) and cytomegalovirus (CMV) internal control reference recombinase-aided amplification (ICR-RAA) assays. (A) RAA channel for the EBV strain B95-8 showed a positive amplification curve, with the RAA results for other pathogens and negative control being judged as negative. (B) The RAA results of CMV DNA were positive, whereas for the remaining pathogens and negative control, the results were determined as negative. Abbreviations: CMV, cytomegalovirus; EBV, Epstein-Barr virus; ICR-RAA, internal control reference recombinase-aided amplification. WNV, West Nile virus; VZV, varicella-zoster virus; HSV-1, human herpesvirus type 1; HSV-2, human herpesvirus type 2: HSV-6, human herpesvirus type 6; human herpesvirus type 1, 2, and 6 (HSV-1/2/6), HBV, hepatitis B virus; FluA, influenza A virus; FAM, 6-carboxyfuorescein; HEX, 5-hexachlorofuorescein.
In group A, 45 samples tested positive by EBV qPCR, with the viral load ranging from 2.94 × 102 to 1.17 × 105 copies/mL [cycle threshold (Ct): 19.05 to 29.86]. Three samples with viral loads ranging from 5.49 × 101 to 6.28 × 102 copies/mL were positive by qPCR but negative by EBV ICR-RAA assay. In addition, one sample was positive by EBV ICR-RAA but negative by qPCR. In group B, 33 samples were tested positive by CMV qPCR with viral loads ranging from 8.27 × 102 to 1.06 × 108 copies/mL (Ct: 13.16 to 29.89). Five samples with viral load ranging from 1.20 × 103 to 7.02 × 103 copies/mL tested positive by qPCR but negative by ICR-RAA assay. Samples with discrepant results were sequenced according to a previously described method[4], and positive results were obtained in all cases. Compared with qPCR, the sensitivity of the EBV and CMV ICR-RAAs was 93.33% and 84.84%, respectively, and the specificity was 98.75% and 100.00%, respectively (Table 2).
Variables qPCR Performance of clinical samples Positive Negative Total Sensitivity (%) Specificity (%) Kappa (P < 0.05) EBV dual real-time
ICR-RAAPositive 42 1 43 93.33 98.75 0.930 Negative 3 79 82 Total 45 80 125 CMV dual real-time
ICR-RAAPositive 28 0 28 84.84 100.00 0.892 Negative 5 92 97 Total 33 92 125 Note. CMV, cytomegalovirus; EBV, Epstein-Barr virus; ICR-RAA, internal control reference recombinase-aided amplification; qPCR, quantitative polymerase chain reaction. Table 2. Clinical evaluation of EBV and CMV RAA assays using extracted DNA and qPCR
Previously, loop-mediated isothermal amplification (LAMP) was reported for the specific detection of human herpesvirus within 60 min[9,10], which overcomes some of the limitations of the standard method, such as the complexity of PCR dependent high-precision instruments and the difficulty of application in grassroots units. However, LAMP did not introduce ICR to monitor the reaction process. As illustrated in our previous reports[6–8], the proposed EBV and CMV ICR-RAA assays integrated ICR to monitor the whole RAA reaction system, avoiding false results caused by experimental operation and systematic errors. The fact that the ICR amplification curve of all the clinical samples tested positive in the present study confirmed that all the results were reliable.
In this study, the analytical sensitivity and LOD of ICR-RAA assays for EBV and CMV were comparable to those reported for LAMP[9,10]. However, the clinical sensitivity of both ICR-RAA assays was slightly lower than that of commercial qPCR. All false negatives with moderate Ct values (about 30) were obtained for serum samples for which automatic nucleic acid extraction was performed, suggesting that the ICR-RAA assay may not be fully compatible with this extraction method. LAMP methods normally require at least 1 h for detection[9,10], whereas ICR-RAA methods can be completed within 30 min. Except nucleic acid extraction, the amplification curves of most samples can be observed in 25 and 10 min for EBV and CMV, respectively. In addition, the RAA operation steps and personnel training procedures are relatively simple, detector and vortex mixer are portable, and the kits are transported and stored at a constant temperature, which is convenient for field tests and screening of large numbers of samples[6].
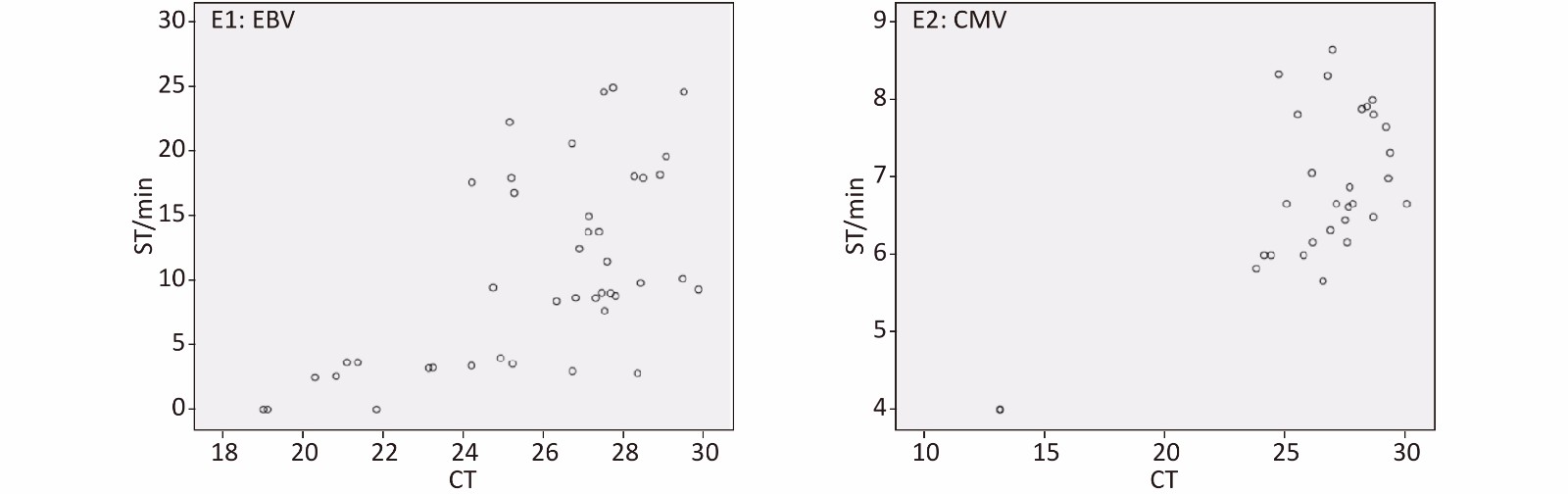
Quantitative detection of EBV and CMV is helpful for the treatment and prevention of diseases. In this study, we preliminarily explored the relationship between the time distribution of all EBV- and CMV-positive specimens by qPCR and ICR-RAA to explore the possibility of quantitative detection of ICR-RAA (Supplementary Figure S1, available in www.besjournal.com). The regression equation was y = 1.528x − 28.889 (gradient: 1.528, 95% CI: 0.881 to 2.176; intercept: −28.889, 95% CI: −45.731 to −12.047, P < 0.0001) for EBV and y = 0.205x + 1.423 (gradient: 0.205, 95% CI: 0.126 to 0.285; intercept: 1.423, 95% CI: −0.677 to 3.524, P < 0 005) for CMV. These results showed a weak (R2 = 0.362) and moderate (R2 = 0.498) correlation between the slope time of EBV ICR-RAA and CMV ICR-RAA, respectively, and the CT value by qPCR. These observations indicated that ICR-RAA is a more rapid and qualitative assay than a quantitative tool in the detection of EBV, with Ct values ranging from 19.05 to 29.86; for CMV, the Ct values range from 13.16 to 29.89.
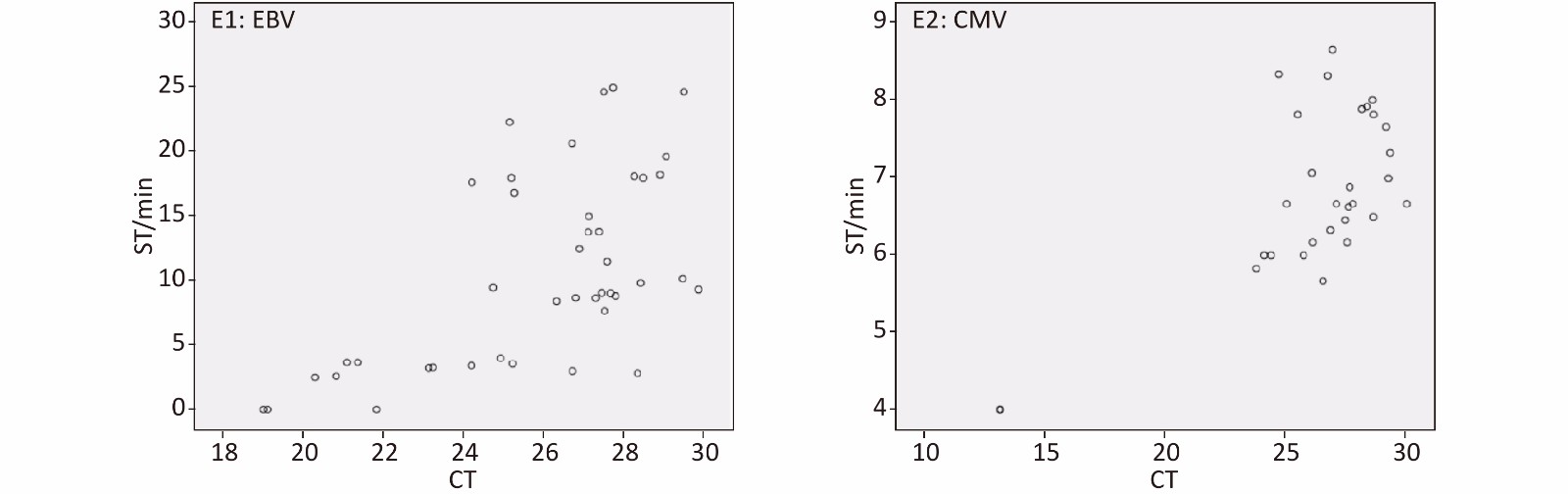
Figure S1. Scatter of ICR-RAA ST (y-axis) and q-PCR cycle threshold values (CT) (x-axis). Data determined by SPSS 21.0 software. q-PCR, fluorescence quantitative real-time PCR; ICR-RAA, internal control reference recombinase–aided amplification; ST, slope time. E1 shows time distribution of EBV ICR-RAA. E2 shows time distribution of CMV ICR-RAA.
The use of simplified nucleic acid extraction methods, such as thermal decomposition and solution lysis, may greatly reduce the turnover time of nucleic acid detection[6,8]. In this study, we adopted the thermal lysis method such that the nucleic acid extraction and result interpretation could be completed within 40 min. However, the sensitivity of direct EBV ICR-RAA reduced to 72.22% and that of CMV ICR-RAA to 80.00%; nonetheless, their specificities remained 100.00% (Supplementary Table S2, available in www.besjournal.com). These findings indicated that the presence of inhibitors in the raw blood or serum samples might have a significant adverse effect on ICR-RAA reaction. Alternative sample pretreatment or optimal thermal lysis conditions and assessment of large sample sizes are needed to address this challenge in the future.
Item q-PCR Performance of clinical samples Positive Negative Total Sensitivities (%) Specificities (%) Kappa (P < 0.05) EBV ICR-RAA positive 13 0 13 72.22 100.00 0.764 negative 5 30 35 total 18 30 48 CMV ICR-RAA positive 4 0 4 80.00 100.00 0.878 negative 1 43 44 total 5 43 48 Table S2. Comparison of clinical evaluation of EBV and CMV RAA assays using DNA obtained by thermal lysis and q-PCR assay
In conclusion, the proposed ICR-RAA assays offer a simple and rapid tool for the detection of EBV and CMV in blood samples.
Declarations All the authors approved the final manuscript and have no conflicts of interest to declare.
Author Contributions MA Xue Jun, FENG Zhi Shan, and SHEN Xin Xin designed the study. GAO Yuan, TIE Yan Qing, ZHAO Lin Qing, TAN He, DING Nan, DING Ya Xin, and GUO Qi collected all clinical samples. GAO Yuan, ZHANG Rui Qing, WANG Jin Rong, CHEN Zi Wei, and FAN Guo Hao performed the experiments. GAO Yuan, TIE Yan Qing, ZHAO Lin Qing, and ZHANG Rui Qing analyzed and interpreted the data. MA Xue Jun, FENG Zhi Shan, SHEN Xin, and GAO Yuan wrote the paper.
Acknowledgements We gratefully acknowledge the assistance from the National Institute for Viral Disease Control and Prevention (IVDC), the Chinese Center for Disease Control and Prevention (CCDC), the Hebei General Hospital, the Capital Institute of Pediatrics, and the Institute of Virus Diseases.
Rapid Internal Control Reference Recombinase-Aided Amplification Assays for EBV and CMV Detection
doi: 10.3967/bes2021.091
- Received Date: 2020-10-20
- Accepted Date: 2021-01-22
-
Key words:
- Recombinase-aided amplification /
- Isothermal nucleic acid detection /
- Epstein-Barr virus /
- Cytomegalovirus
Abstract: Epstein-Barr virus (EBV) and cytomegalovirus (CMV), two of the most prevalent human herpesviruses, cause a wide spectrum of diseases and symptoms and are associated with serious health problem. In this study, we developed an internal control reference recombinase-aided amplification (ICR-RAA) assay for the rapid detection of EBV and CMV within 30 min. The assay had a sensitivity of 5 and 1 copies/test for EBV and CMV, respectively, with no cross reaction with other pathogens. In comparison with those of the commercial quantitative polymerase chain reaction (qPCR), the sensitivity of the EBV and CMV ICR-RAAs using extracted DNA was 93.33% and 84.84%, respectively; the specificity was 98.75% and 100.00%, respectively; and the Kappa values were 0.930 and 0.892 (P < 0.05), respectively. In comparison with those of qPCR, the sensitivity of the EBV and CMV ICR-RAAs using the DNA by thermal lysis was 72.22% and 80.00%, respectively; the specificity was 100.00%; and the Kappa values were 0.764 and 0.878 (P < 0. 05), respectively. Thus, rapid and specific detection of EBV and CMV is possible using ICR-RAA assays.
&These authors contributed equally to this work.| Citation: | GAO Yuan, TIE Yan Qing, ZHAO Lin Qing, TAN He, DING Nan, DING Ya Xin, GUO Qi, ZHANG Rui Qing, WANG Jin Rong, CHEN Zi Wei, FAN Guo Hao, SHEN Xin Xin, FENG Zhi Shan, MA Xue Jun. Rapid Internal Control Reference Recombinase-Aided Amplification Assays for EBV and CMV Detection[J]. Biomedical and Environmental Sciences, 2021, 34(8): 650-655. doi: 10.3967/bes2021.091 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: