-
Type-VI secretion system (T6SS) is a widespread bacteriophage-like complex in bacteria that participates in multiple physiological processes, including metal ion uptake, bacterial competition, and biofilm formation [1]. Yersinia pestis is the causative agent of plague. There are five T6SS gene clusters in Y. pestis CO92, among which only the YPO0499-YPO0516 locus was well investigated [1, 2]. The expression of YPO0499-YPO0516 is induced at 26 ℃, suggesting that it plays a possible role in the natural cycle of Y. pestis [2]. Deletion of YPO0499-YPO0516 alters the uptake and intracellular growth of Y. pestis in macrophages, which is dependent on the pre-grown temperature [2]. However, it has no effect on the virulence in murine and on the ability of the pathogen to infect fleas [2]. The Hcp-like protein encoded by YPO0502 was shown to be an autoagglutination factor involved in the bacterial interaction [3].
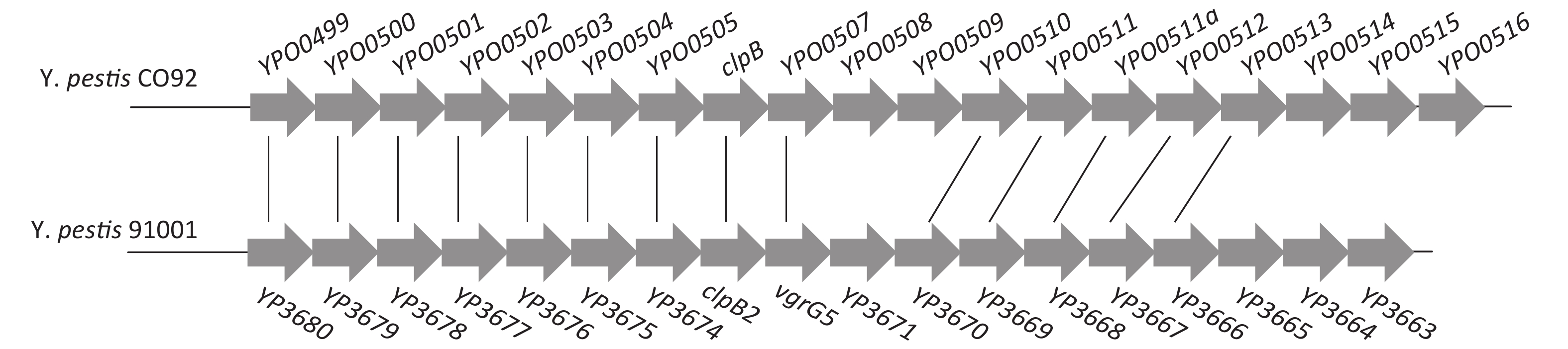
The transcriptional regulator RovA is required for the full virulence of Y. pestis through its regulatory activities on the virulence genes such as psaEFABC and CUS-2 loci [4]. In addition, RovA negatively regulates the biofilm formation by Y. pestis via repression of hmsT transcription and c-di-GMP production [5]. Moreover, a study showed that RovA positively regulates the transcription of YPO0499-YPO0516 genes but lacks detailed regulatory mechanisms [6]. The YP3680-YP3663 locus in Y. pestis Microtus strain 91001 is highly homologous to the YPO0499-YPO0516 locus in Y. pestis CO92 (Supplementary Figure S1 available in www.besjournal.com). Therefore, in the present study, Y. pestis 91001 (wild type, WT) and its non-polar rovA mutant (designated ΔrovA [5]) were employed to investigate the regulatory mechanisms of RovA on YP3680 transcription.
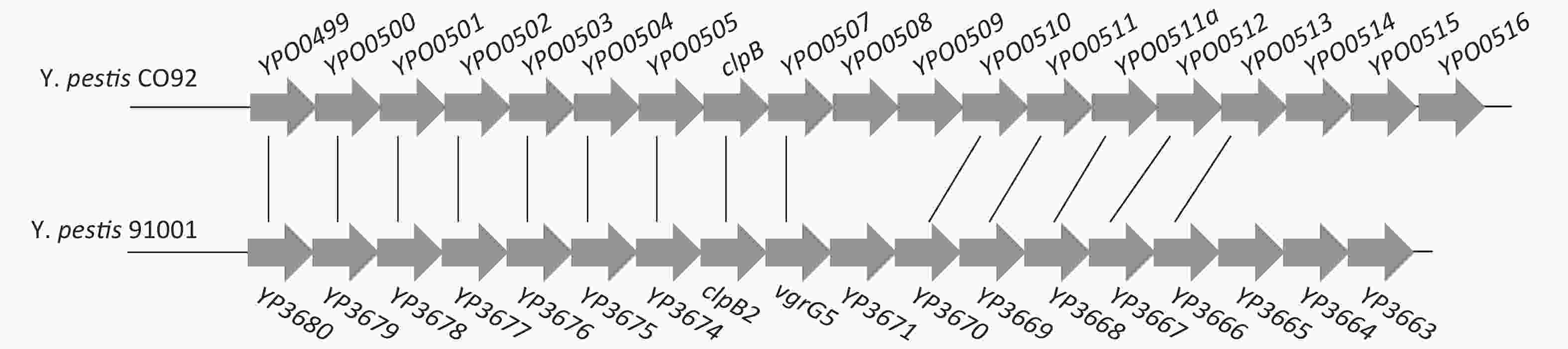
Figure S1. Organization of the T6SS gene cluster in Y. pestis. Genes connected by the lines indicate alleles in the two Yersinia strains.
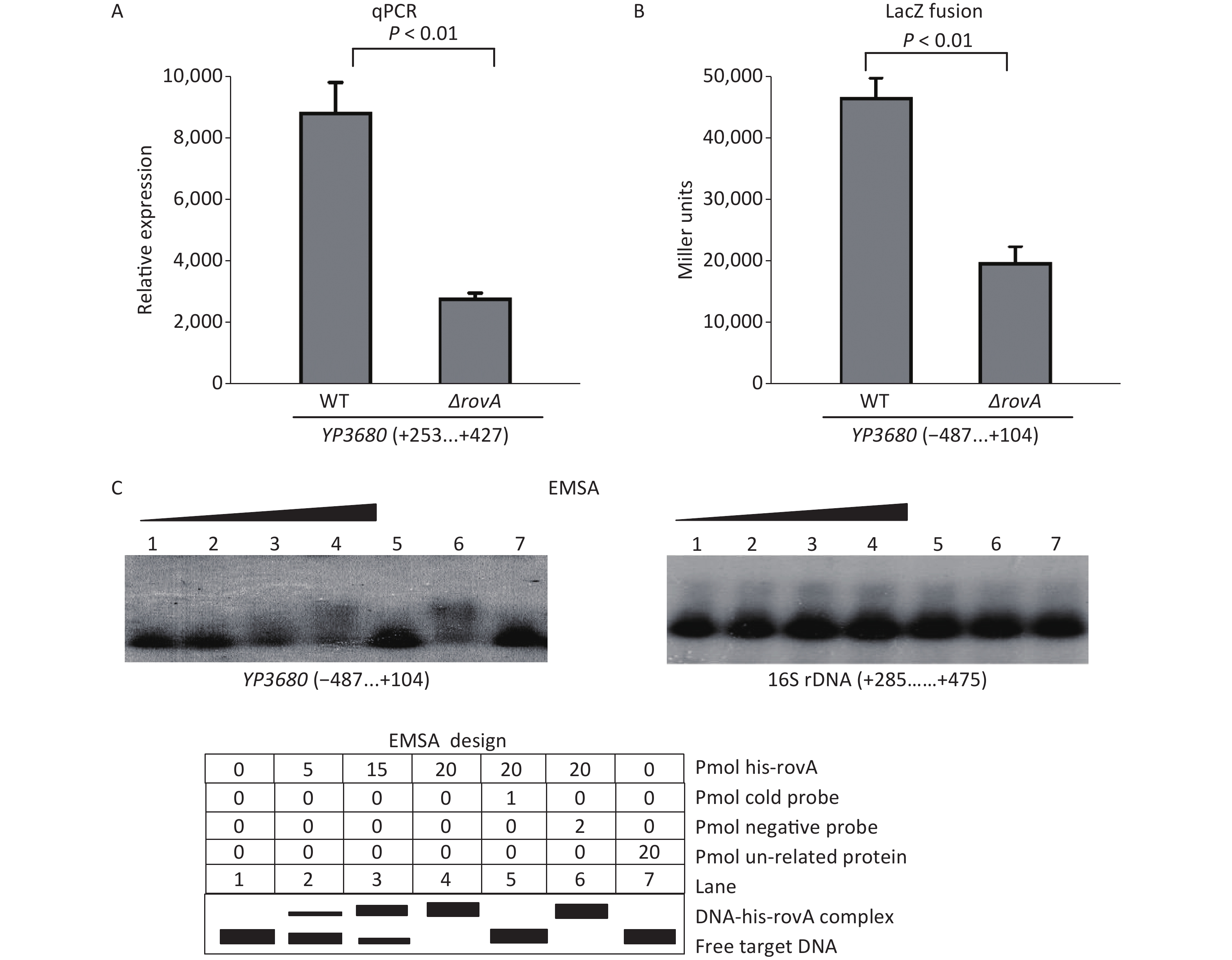
The overnight cell cultures of WT and ΔrovA were diluted 20-fold into 18 mL of fresh LB broth [1% tryptone (Oxoid), 0.5% yeast extract (Oxoid), and 1% NaCl (Merck Millipore)], and then cultured at 26 °C by shaking at 230 rpm to reach an optical density at 620 nm (OD620) value of approximately 1.0. The bacterial cells were harvested, and total RNAs were extracted using TRIzol Reagent (Invitrogen, USA). The qPCR assay was applied to detect the mRNA levels of YP3680 in WT and ΔrovA using a Light-Cycler system (Roche, Switzerland) together with the SYBR Green master mix (the primers are listed in Supplementary Table S1 available in www.besjournal.com). As shown in Figure 1A, the mRNA level of YP3680 significantly decreased in ΔrovA compared to that in WT, indicating that transcription of YP3680 was activated by RovA. To detect whether RovA has regulatory action on the YP3680 promoter activity, the promoter-proximal DNA region of YP3680 was cloned into the pRW50 plasmid harboring a promoterless lacZ reporter gene. The recombinant pRW50 was then transferred into WT and ΔrovA to determine the β-galactosidase activities in each strain using a β-Galactosidase Enzyme Assay System (Promega, USA) according to the manufacturer’s instructions. As shown in Figure 1B, the promoter activity of YP3680 significantly decreased in ΔrovA relative to that in WT, suggesting that RovA acted as a positive regulator of YP3680 expression. In addition, a RovA box-like sequence, TCATCGTGCTAA, was identified in the promoter-proximal DNA region of YP3680 (Supplementary Figure S2 available in www.besjournal.com)[7], indicating that YP3680 transcription was probably under the direct control of RovA in Y. pesitis. Thus, the 487 bp upstream of YP3680 was amplified and then labeled with [γ-32P]-ATP at the 5′- ends used for the electrophoresis mobility shift assay (EMSA) DNA probe [5]. EMSA was performed as previously described [5], and the results showed that His-RovA was able to dose-dependently bind to the regulatory DNA region of YP3680, but it could not bind to the DNA fragment of 16S rDNA as the negative control (Figure 1C). Taken together, these results suggested that transcription of YP3680 was directly activated by RovA in Y. pestis biovar Microtus.
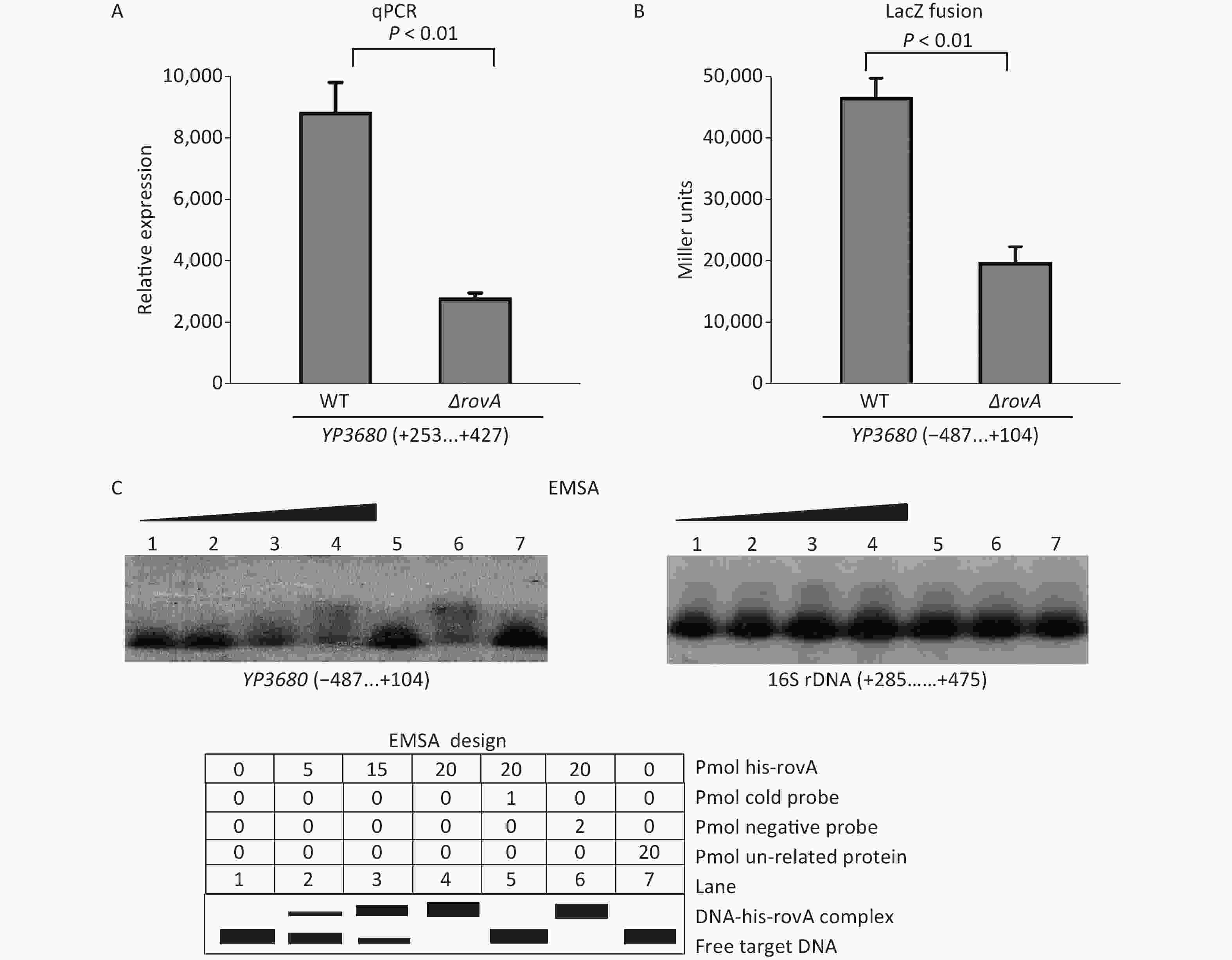
Figure 1. Positive regulation of YP3680 by RovA. The minus and positive numbers in the brackets indicated nucleotide positions upstream and downstream of YP3680. (A) qPCR. The relative mRNA level of YP3680 was compared between ΔrovA and WT. (B) LacZ fusion. The YP3680: lacZ fusion vector was transferred into WT and ΔrovA to determine the β-galactosidase activity (miller units) in the cellular extracts. (C) EMSA. The entire promoter-proximal region of YP3680 was incubated with increasing amounts of purified His-RovA, and then subjected to 4% (w/v) polyacrylamide gel electrophoresis. Shown below the binding was the schematic representation of the EMSA design.
Target Primers (forward/reverse, 5′–3′) qPCR YP3680 ATGACGCCTACCGGCTCAG/GATGCAGAGTGTTGATCAGGT 16S rDNA CACACTGGAACTGAGACAC/TGCTTCTTCTGCGAGTAAC LacZ fusion YP3680 GCAGGATCCGCATTCAAATCCAGCTCTGATT/GGTAAGCTTGAGAGGATGGACTCAATCT Primer extension YP3680 /TCCTGATTTACATACCTGTTATA EMSA YP3680 GCATTCAAATCCAGCTCTGATT/GAGAGGATGGACTCAATCT Table S1. Oligonucleotide primers designed in this study
The Rcs phosphorelay is a complex two-component system generally composed of three proteins, RcsB, RcsC, and RcsD [8]. The membrane protein RcsC is a sensor kinase catalyzing the phosphorylation of RcsD, which then transfers the phosphate group to RcsB [8]. Phosphorylated RcsB acts either alone or in combination with the RcsA to regulate the gene expression [8]. The gene encoding RcsA is nonfunctional in Y. pestis [9]. However, replacement of the rcsA pseudogene with the functional rcsA allele from Y. pseudotuberculosis to yield the RcsAB complex may strongly inhibit the Y. pestis biofilm formation, which is accomplished by directly repressing the transcription of hmsCDE, hmsT, and hmsHFRS [9]. A recent study showed that expression of T6SS in pathogenic Escherichia coli was strongly induced by the RcsB [10]. In this work, we detected whether the RcsAB complex has a regulatory activity on the transcription of YP3680 in Y. pestis.
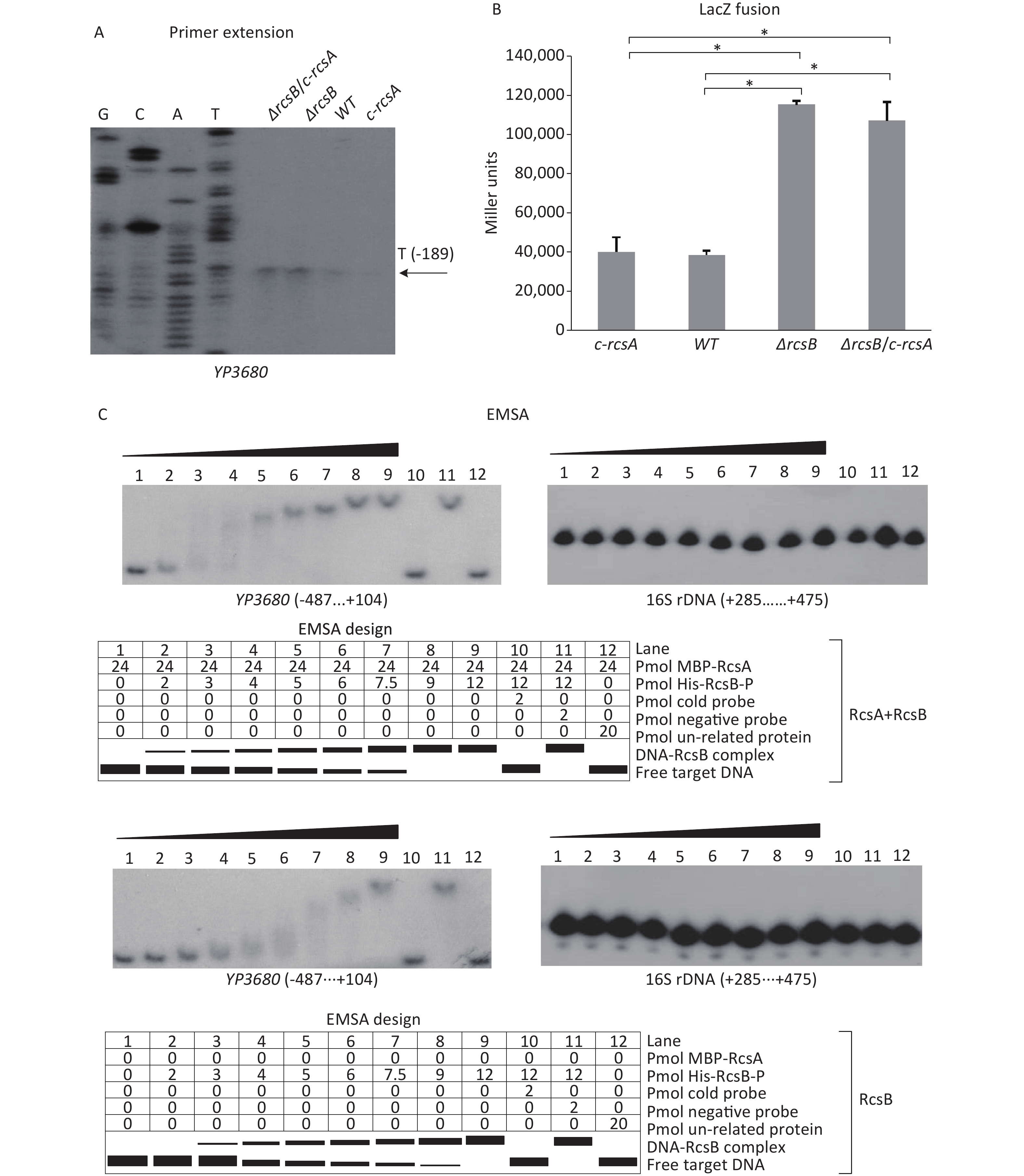
The mid-log phase (OD600≈1.0) bacterial cells of WT, ΔrcsB (rcsB mutant), c-rcsA (the WT strain transformed with the recombinant pACYC184 expressing RcsA of Y. pseudotuberculosis), and ΔrcsB/c-rcsA (the ΔrcsB strain transformed with the recombinant pACYC184 plasmid) [9] were harvested, and then total RNAs were extracted. The primer extension assay, which was performed as described in the previous studies [5, 9], was employed to map the transcription start sites of YP3680 and to investigate the relationship between RcsB regulation and YP3680 transcription. As shown in Figure 2A, the assay detected a single transcription start site for YP3680 located at 189 bp upstream of the translation start site, but both the −10 and −35 elements are a bad match with the consensus prokaryotic sequence (Supplementary Figure S2), suggesting that the promoter is relatively weak. The results of primer extension also showed that the mRNA levels of YP3680 significantly increased in the ΔrcsB and ΔrcsB/c-rscA than those in WT and c-rcsA, whereas no significant differences were observed between the ΔrcsB and ΔrcsB/c-rscA, but it seemed to be enhanced in WT relative to the c-rcsA (Figure 2A). In addition, the results of LacZ fusion assay showed that the promoter activities of YP3680 were much higher in ΔrcsB and ΔrcsB/c-rscA than those in WT and c-rcsA, whereas no significant differences were observed between ΔrcsB and ΔrcsB/c-rscA or WT and c-rcsA (Figure 2B). A RcsAB box-like sequence, TAAGTTTATCCCTA, was identified in the promoter-proximal DNA region of YP3680 [9], suggesting that transcription of YP3680 may be directly regulated by RcsAB. Indeed, the EMSA results showed that His-RcsB-P alone or mixed with the excess MBP-RcsA was able to specifically bind to the regulatory DNA region of YP3680 in a dose-dependent manner in-vitro (Figure 2C). The addition of excess MBP-RcsA improved the DNA-binding activity of His-RcsB-P (Figure 2C). Taken together, these results suggested that RcsB alone or upon binding with RcsA allele was able to specifically bind to the regulatory DNA region of YP3680 to repress its transcription.

Figure 2. Negative regulation of YP3680 by RcsAB. The minus and positive numbers in the brackets indicated nucleotide positions upstream and downstream of YP3680. (A) Primer extension. Lanes C, T, A, and G represented Sanger sequencing reactions. An oligonucleotide primer was designed to be complementary to the RNA transcript of YP3680. The primer extension product was analyzed with an 8 M urea −6% acrylamide sequencing gel. The transcription start site was indicated by the arrow with nucleotide and position. The LacZ fusion (B) and EMSA (C) were performed as given in Figure 1. *Asterisks indicate statistically significant differences (P < 0.01).
Both RovA and RcsAB are involved in repressing the biofilm formation by Y. pestis [5, 9]. Biofilms formed by Y. pestis in flea gut are essential for the transmission of plague [9]. Moreover, RovA has been demonstrated to be a protein thermometer that senses the temperature shift to change its conformation, thereby reducing its DNA-binding activity and enhancing its susceptibility to proteolytic degradation [11]. In addition, the temperature shift from 26 °C to 37 °C also triggered the down-regulation of rovA at the transcriptional level [4]. According to the observations in Y. pestis CO92, the YP3680-YP3663 locus may play key roles in the flea cycle and bacterial interaction, and its expression also probably manifests in a temperature-dependent manner [2, 3]. Thus, it is worth investigating the roles of the YP3680-YP3663 locus in the flea cycle and the regulation of RovA and RcsAB in this process in the future.
Taken together, the data disclosed that RovA bound to the promoter-proximal DNA region of YP3680 on the YP3680-YP3663 cluster to activate its transcription, whereas the RcsAB complex directly repressed the transcription of YP3680. These results provided us with a deeper understanding of T6SS regulation in Y. pestis biovar Microtus.
HTML
 21328Supplementary Materials.pdf
21328Supplementary Materials.pdf
|

|








 Quick Links
Quick Links
 DownLoad:
DownLoad: