-
Epstein-Barr virus (EBV), a ubiquitous human herpesvirus, was first reported as the etiology of Burkitt’s lymphoma, in 1964 [1-2]. More than 50% of children and 90% of adults worldwide were found to be infected with EBV, usually without causing long-term health problems [3]. EBV infects B lymphocytes (B cells) to cause long-term latent infections, and also induces memory B cells in a quiescent state [4, 5]. EBV also infects T lymphocytes (T cells) and natural killer cells (NK cells), resulting in hemophagocytic lymph histiocytosis and chronic active EBV infection [6-8]. EBV has also been linked to T-cell lymphoma, Hodgkin’s lymphoma, and NK leukemia or large granular lymphocyte leukemia [9, 10]. In immunodeficient patients, especially those treated with hematopoietic stem cell transplantation (HSCT), infection of EBV may lead to disordered lymphocyte proliferation [11]. EBV-associated post-transplant lymphoproliferative disorder (PTLD) has been recognized as a significant cause of morbidity and mortality in patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HSCT). The number of patients at risk of developing PTLD is increasing, partly as a result of highly immunosuppressive regimens [12].
EBV infection increases the risk of HSCT recipients to chronic graft versus host disease (GVHD) [13], which results in the failure of the transplant. Previous studies reported that the EBV infection rate was 8.8%–58.8% in children following HSCT [14-17]. Because there is no effective antiviral agent against EBV infection, prevention or preemptive therapy is vital in reducing morbidity/mortality or failure of transplantation caused by EBV. Recent evidence-based guidelines from the European Conference on Infections in Leukemia have recommended a weekly screening of EBV DNA for at least 3 months in high-risk allogeneic HSCT recipients [3].
Therefore, for treatment intervention during the early stages, it is important to quantitatively detect the EBV viral load and to investigate EBV infection as early as possible.
-
The BALF5 gene of EBV was cloned into the pUC57 plasmid to yield pUC57-BALF5. The plasmid was serially diluted by 3.0 × 106, 3.0 × 105, 3.0 × 104, 3.0 × 103, 3.0 × 102, and 3.0 × 10 copies/mL. The plasmid was also doubly-diluted 1:2, 1:4, 1:8, 1:16, 1:32, and 1:64 by 3.0 × 102 copies/mL. The EBV B95-8 strain was 10-fold serially diluted by 10−1, 10−2, 10−3, 10−4, 10−5, 10−6, 10−7, and 10−8. Herpes simplex virus 1 (HSV-1, KOS strain), herpes simplex virus 2 (HSV-2, G strain), varicella zoster virus (VZV, Ellen strain), human cytomegalovirus (HCMV, AD169 strain), human herpesvirus 6A (HHV-6A, GS strain) (HHV-6 Foundation, Santa Barbara, CA, USA), human herpesvirus 6B (HHV-6B, Z29 strain) (HHV-6 Foundation), and human herpesvirus 7 (HHV-7, JI strain) (HHV-6 Foundation) were stored at −80 °C. Each experiment had negative and positive controls, and the experiments were repeated three times.
The patients were treated according to the ethical guidelines of the Helsinki Declaration, which was approved by the Ethics Committee of the National Institute for Viral Disease Control and Prevention (Beijing, China) (IVDC2020-014). All methods were conducted in accordance with relevant guidelines and regulations.
-
From May 2019 to September 2020, 64 serum samples of children following HSCT were collected at the Beijing Capital Institute of Pediatrics Children’s Hospital (Beijing, China). Forty-one males and 23 females were enrolled, and the median age was 7.5 years. All cases were diagnosed using clinical and molecular biological methods. The project was approved by the Ethics Committee of the National Institute for Viral Disease Control and Prevention.
-
The primers and probe for qPCR and cdPCR, designed according to the EBV-BALF5 gene, were the following: EBV-F 5’-CGGAAGCCCTCTGGACTTC-3’; EBV-R 5’-CCCTGTTTATCCGATGGAATG-3’; and EBV-Probe 5’-FAM-TATACACGCACGAGAAATGCGCC-BHQ-3’.
-
A total of 140 μL EBV supernatant or clinical serum samples were extracted according to the QIAamp Virus DNA Blood Mini Kit (Qiagen, Hilden, Germany). The DNA was eluted in a volume of 60 μL doubly-distilled H2O and stored at −80 °C for future use.
-
Eight microliters of DNA of each sample was added to 25 μL of qPCR reaction system containing 12.5 μL of Premix Ex TaqTM (TaKaRa, Shiga, Japan), 0.5 μL of primers (10 μmol/L), 0.5 μL probe (10 μmol/L), and 3 μL doubly-distilled H2O. The qPCR procedure was the following: one cycle at 95 °C for 10 min; 40 cycles at 95 °C for 5 s, and 61 °C for 30 s using the qPCR machine (CFX96; Bio-Rad, Hercules, CA, USA).
-
The 25 μL cdPCR mixture contained 5 μL ToughMix buffer (Stilla, Villejuif, France), 2.5 μL fluorescein (1 μmol/L) (PEXBIO, Beijing, China), 0.5 μL primer (10 μmol/L), 0.5 μL probe (10 μmol/L) and 8 μL DNA template. Cycling conditions were the following: 95 °C for 10 min; 94 °C for 5 s, 61 °C for 30 s, and a total of 40 cycles in the cdPCR adopted Naica™ Crystal Digital PCR system (Stilla). Data were analyzed using CrystalMiner.
-
The data were processed using SPSS statistical software for Windows, version 20.0 (SPSS, Chicago, IL, USA). Data were compared using Student's t-test, and counting data were compared using the χ2-test.
-
Eight microliters of serially diluted plasmid was detected by qPCR and cdPCR. A total of 3.0 × 106, 3.0 × 105, 3.0 × 104, 3.0 × 103, and 3.0 × 102 copies/mL diluted plasmids were detected by qPCR and cdPCR. Thirty copies/milliliter of diluted plasmid was not detected by qPCR, but cdPCR showed that 30 copies/mL of the plasmid could be detected. The plasmid was doubly-diluted 1:2, 1:4, 1:8, 1:16, 1:32, and 1:64 using 3.0 × 102 copies/mL. The limit of detection (LOD) of EBV cdPCR was up to 110 copies/mL (2.1 copies/reaction) and the LOD of qPCR was 327 copies/mL for the plasmid dilutions. The results confirmed that the sensitivity of cdPCR was higher than that of qPCR. The consistency between cdPCR and the expected values of the diluted plasmid was r2 = 0.996, P = 0.84. The consistency between qPCR and the expected value of the diluted plasmid was r2 = 0.994, P = 0.89. Good consistency was observed between the diluted plasmid copies and those measured by cdPCR and qPCR.
-
After EBV was 10-fold serially diluted from 10−1, 10−2, 10−3, 10 −4, 10 −5, 10−6, 10−7 to 10−8, 8 μL DNA of each dilution was used for detection by qPCR and cdPCR. A total of 10−1, 10−2, 10−3, 10−4, and 10−5 dilutions of the EBV B95-8 strain were detected by qPCR, and 10−1, 10−2, 10−3, 10−4, 10−5, and 10−6 dilutions were detected by cdPCR. The 10−6, 10−7, and 10−8 dilutions were not detected by qPCR. The EBV cdPCR did not detect the 10−7 or 10−8 dilutions. Together, the results showed that the sensitivity of EBV cdPCR was 10-fold higher than that of qPCR, and the LOD of EBV cdPCR was up to 121 copies/mL (2.3 copies/reaction).
To determine the specificity of cdPCR, EBV cdPCR was used to detect the other eight herpesviruses, including herpes simplex virus 1, herpes simplex virus 2, varicella zoster virus, human cytomegalovirus, human herpesvirus 6A, human herpesvirus 6B, and human herpesvirus 7. The results showed that EBV cdPCR did not react with seven herpesviruses except EBV. The results also showed that EBV cdPCR had no cross-reaction with the other herpesviruses.
-
We then analyzed EBV infections in children after HSCT, based on the results of cdPCR. There were 41 males and 23 females, and 62 patients were allogeneic HSCT and two patients were autologous HSCT among the 64 children from May 2019 to September 2020. Among 64 children following HSCT, there were 22 cases of acute myelocytic leukemia (AML), 11 cases of acute lymphocytic leukemia (ALL), 7 cases of aplastic anemia (AA), 6 cases of myelodysplastic syndrome (MDS), 3 cases of lymphoma, 9 cases of Wiskott-Aldrich syndrome (WAS), and 6 cases of mucopolysaccharidosis (MPS) (Table 1). There were 4 cases of EBV infection among the 22 AML cases, with a viral load from 643 copies/mL to 1,530 copies/mL. There were 3 cases of EBV infection in the 11 ALL cases, and the viral load was from 397 to 3,209 copies/mL. There were 2 EBV-infected cases among the 6 MDS cases, with EBV viral loads of 617 and 797 copies/mL, respectively. Two cases of EBV infection were found in the nine WAS cases, and the viral loads were 140 and 1,551 copies/mL, respectively. There was only one EBV-infected among three lymphoma cases, with a viral load of 154 copies/mL. No EBV infections were found in both seven AA and six MPS cases. Complications and sequelae of 64 HSCT children included bacterial and fungal infections, rash, fever, agranulocytosis, GVHD, pneumonia, gastroenteritis, liver damage, secondary diabetes, hypertension, conjunctivitis, and sepsis.
Children characteristics No. of children (No. of EBV positive) Age (years) 0−6 39 (4) 7−12 17 (7) > 12 8 (1) Sex Male 41 (7) Female 23 (5) Disease AML 22 (4) ALL 11 (3) AA 7 (0) MDS 6 (2) Lymphoma 3 (1) WAS 3 (2) MPS 6 (0) HSCT Allogeneic 62 (12) Autologous 2 (0) Source of HSCT BM + PBSCT 58 (10) PBSCT 6 (2) Note. HSCT, hematopoietic stem cell transplantation; EBV, Epstein-Barr virus; AML, acute myelocytic leukemia; ALL, acute lymphocytic leukemia; AA, aplastic anemia; MDS, myelodysplastic syndrome; WAS, Wiskott-Aldrich syndrome; MPS, mucopolysaccharidosis; BM, bone narrow; PBHSCT, peripheral blood stem cell transplantation. Table 1. Characteristics of 64 children after hematopoietic stem cell transplantation
The incidence of EBV infection was 18.75% (12/64) in 64 children after HSCT, and the infection in males was 17.07% (7/41), and 21.74% (5/23) in females. The incidence of EBV infection was 10.26% (4/39) in the 0−6 years old age group, and the incidence of EBV infection was 41.18% (7/17) in the 7−12 years old age group. In the 0−6 years old age group, the incidence of males was 9.09% (2/22), and that of females was 11.76% (2/17). In the 7−12 years old age group, the incidence of males was 33.33% (5/15), and the incidence of females was 100% (2/2). For those over 12 years of age, the incidence of EBV was 12.50% (1/8), with only one female child infected (Table 2).
Age (years) No. EBV positive Positive rate
(%)Male Female Positive Negative Positive rate
(%)Positive Negative Positive rate
(%)0–6 39 4 10.26 2 20 9.09 2 15 11.76 7–12 17 7 41.18 5 10 33.33 2 0 100 > 12 8 1 12.50 0 4 0 1 3 25.00 Total 64 12 18.75 7 34 17.07 5 18 21.74 Table 2. Epstein-Barr virus infection in children after hematopoietic stem cell transplantation
The length of hospital stays were analyzed for 64 children after HSCT. The average hospital stay of children with EBV infection (184 ± 91 days) was longer than that of children without EBV infection (125 ± 79 days), P = 0.026 (Figure 1A).
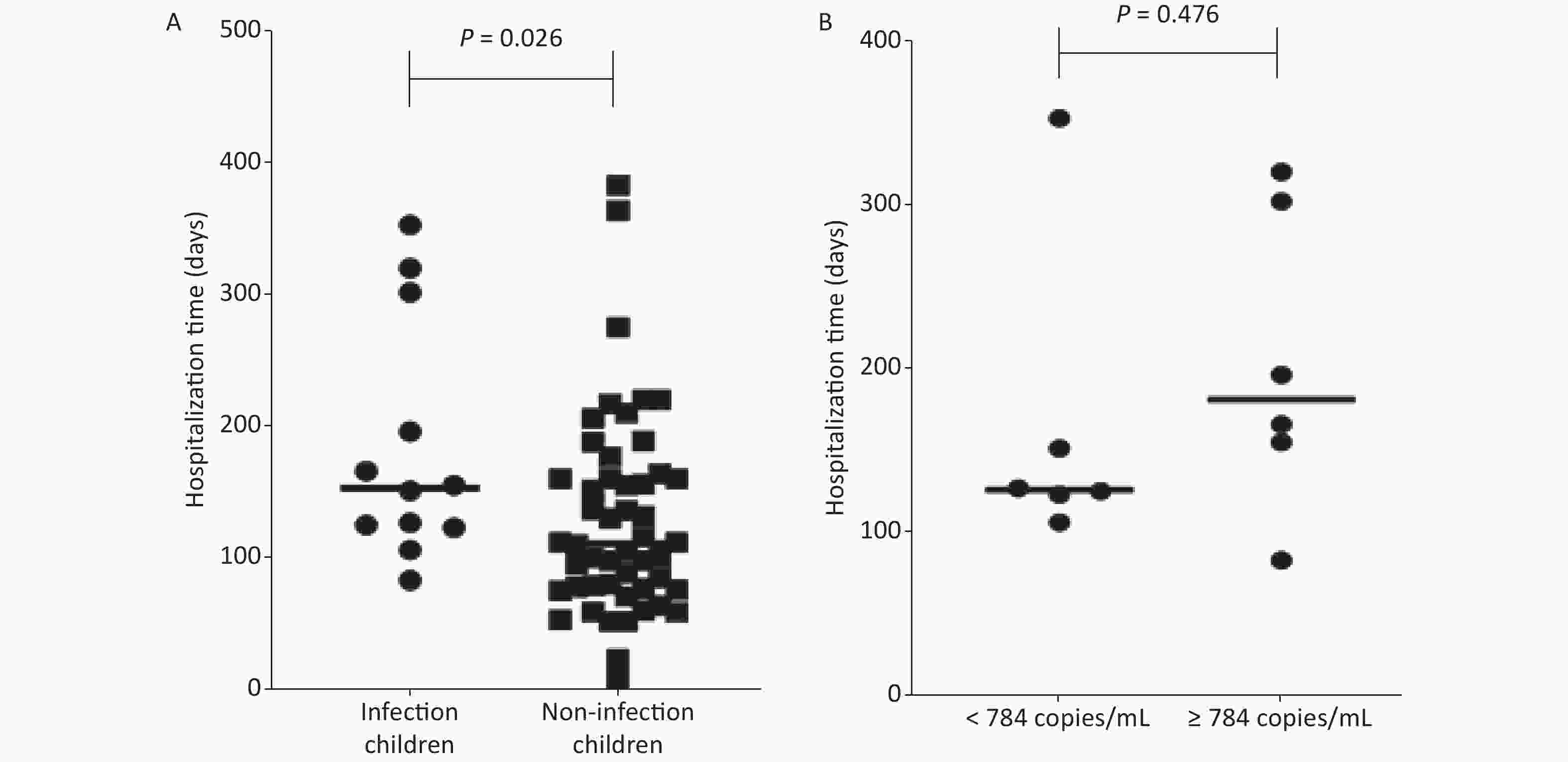
Figure 1. The length of hospital stays of children after hematopoietic stem cell transplantation. (A) Comparison of hospital stays between infected and non-infected children after hematopoietic stem cell transplantation. (B) The comparison of hospital stays between low Epstein-Barr virus (EBV) load (< 784 copies/mL) group and high EBV viral load (≥ 784 copies/mL) group.
-
For 12 positive children, the range of viral loads was from 140 copies/mL to 3,209 copies/ mL (median EBV viral load: 784 copies/mL). The viral loads of three children were less than 500 copies/mL, those of five children were between 500 copies/mL to 1,000 copies/mL, those of three children were between 1,000 copies/mL to 2,000 copies/ mL, and that of one patient was more than 3,000 copies/mL. Among 12 EBV positive children, except for patient 1, all other patients had complications and sequelae including GVHD (eight cases) and fever (two cases). The median viral load of EBV was 820 copies/mL for GVHD patients. Three children died after HSCT, with EBV viral loads of 820, 1,530 and 3,209 copies/mL. Co-infection with HCMV occurred in two of the three deaths (Table 3). Together, the results indicated that the high viral load of EBV may have caused more serious disease and even death for HSTC patients.
Cases Sex Age
(years)Disease Copies/mL The length of hospital
stay (days)Complications and sequelae Prognosis 1 F 5 AML 643 125 None 2 F 10 MDS 797 83 Secondary diabetes, GVHD 3 M 10 ALL 397 353 Secondary hypertension, GVHD 4 M 8 AML 820 196 Fungal pneumonia, acute upper respiratory tract infection, GVHD dead 5* M 7 ALL 3,209 320 Septic shock, pancreatitis, cholangitis, GVHD dead 6 M 5 Lymphoma 154 127 Liver damage 7* F 5 AML 1,530 302 Septicemia, type II respiratory failure, GVHD dead 8 M 10 WAS 140 106 Septicemia, liver damage, GVHD 9 F 15 MDS 617 123 Fever 10 M 7 ALL 771 151 Acute gastroenteritis, fever 11 M 2 WAS 1,551 166 Fungal pneumonia, enteritis, GVHD 12 F 9 AML 1,008 155 Thrombotic Microangiopathes, GVHD Note. F, female; M, male; GVHD, graft versus host disease. *Case 5 and case 7 involved co-infection with human cytomegalovirus. Table 3. Characteristics of 12 Epstein-Barr virus positive children after hematopoietic stem cell transplantation
The children following HSCT were divided into two groups according to the median EBV viral load of 784 copies/mL. The length of hospital stay of the low EBV viral load (< 784 copies/mL) group was 164 ± 94 days, and the length of hospital stay of the high EBV viral load (≥ 784 copies/mL) group was 203 ± 91 days. There was no significant difference between the two groups (P = 0.476) (Figure 1B).
-
EBV infection has been linked to a wide range of malignancies including Hodgkin lymphoma, Burkitt lymphoma, nasopharyngeal carcinoma, gastric carcinomas, and HIV-associated smooth muscle neoplasms. EBV infection is also associated with non-malignant disease, for example, lymphoproliferative disorder in immunocompromised individuals, and inflammatory pseudo-tumors of the liver and spleen [18-23]. Lymphoproliferative disorder significantly increases the mortality and morbidity of post allo-HSCT, and PTLD can lead to a mortality as high as 85% [24]. Major risk factors of EBV-related PTLD include HLA mismatch, graft T-cell depletion, and GVHD [25-28]. PTLD usually occurs before the recovery of the EBV-specific cytotoxic T-lymphocyte response after allo-HSCT transplantation [28]. To prevent the occurrence of PTLD, a specific threshold value of EBV viremia is needed for initiating pre-emptive therapy. However, because of no universal standard for PCR assays, it is difficult to obtain consistent results from different assays using serum, whole blood, or peripheral blood mononuclear cells.
In this study, we established a sensitive cdPCR method to detect the viral load of EBV infection. In contrast to qPCR, cdPCR is an absolute quantitative method, which does not depend on the standard curve, and can directly detect the copy number of the target sequence. The cdPCR is conducted using an advanced cutting-edge microfluidic chip (Sapphire chip) two-dimensional array microchamber to complete the PCR reaction. Like other chip-based biosensors used to detect EBV infection [29-31], cdPCR is time-saving and simple to conduct. Moreover, cdPCR technology has a stronger tolerance for the presence of inhibitors in the PCR reaction. The results of the present study showed that the minimum viral load was 140 copies/mL, and the maximum viral load was 3,209 copies/mL using cdPCR of our collected cases. Thus, the cdPCR method may be used to evaluate the viral load of HSCT children. Some studies have also reported an EBV threshold of 1,000, 10,000, or 40,000 copies/mL when detected in plasma, whole blood, or serum, respectively [3,32].
Studies has shown that low EBV viral load could be a predictor of poor survival, and that low EBV infection may result in an unbalanced control between EBV infection and the host immune system [33]. Because of its high sensitivity, cdPCR may become a new method to predict the prognoses of patients after HSCT. Thus, cdPCR is a simple, sensitive, and specific technique. The viral load of case 5 (ALL-L2) was higher than 3,000 copies/mL (3,209 copies/mL) in serum, and the child subsequently died during this study. However, our study found that two AML patients (case 4 and case 7) relapsed and died after transplantation, but the serum viral load was less than 2,000 copies/mL (820 copies/mL and 1,530 copies/mL, respectively).
We found that the two patients (case 5 and case 7) were co-infected with HCMV. Therefore, co-infection may be an important cause of transplantation failure. Our results also indicated that the threshold value of preemptive therapy should be judged together with the patient’s condition and co-infection. In our study, the average hospital stay was longer in patients with EBV infection, which may have been caused by EBV infection or the occurrence of complications and sequelae, leading to a prolonged course of the disease. However, due to the small sample size and short duration of this study, there was no significant difference of hospitalization time between low and high EBV viral loads.
-
In conclusion, a cdPCR method was established for detection of the EBV viral load. The incidence of EBV infection was 18.75% in 64 children after HSCT, when using cdPCR. The range of EBV viral load was from 140 copies/mL to 3,209 copies/mL in HSCT children. It is well-known that EBV is considered to be the major risk factor for transplantation. Thus, to decrease the mortality and recurrence of children after HSCT from EBV infection, continuous monitoring of the viral load and preemptive therapy should be conducted to prevent postoperative viremia.
-
The authors declare that they have no competing interests.
-
We thank the HHV-6 Foundation for providing human herpesvirus 6A (GS strain), human herpesvirus 6B (Z29 strain), and human herpesvirus 7 (JI strain). We gratefully acknowledge the teachers at the Virus Resource Center, without whom this study would not have been possible.
-
HAN Jun, YAO Hai Lan, LIU Rong conceived and designed the experiments. WANG Wen Jun, FENG Shun Qiao, HE Feng, FENG Miao, WANG Rui Fang, and LIU Mi performed the experiments. WANG Wen Jun, FENG Shun Qiao, MEI Guo Yong, and DU Hai Jun analyzed the data. WANG Wen Jun wrote the manuscript. HAN Jun revised the manuscript. All authors read and approved the final manuscript.
-
All children were confirmed according to diagnostic criteria. All patients were approved by the Ethics Committee of the National Institute for Viral Disease Control and Prevention. The protocol was approved by the Ethics Committee of the National Institute for Viral Disease Control and Prevention (Reference code: IVDC2020-014).
The Viral Load of Epstein-Barr Virus in Blood of Children after Hematopoietic Stem Cell Transplantation
doi: 10.3967/bes2022.052
- Received Date: 2021-12-27
- Accepted Date: 2022-03-31
-
Key words:
- Chip digital PCR /
- Epstein-Barr virus /
- Hematopoietic stem cell transplantation /
- Quantitative PCR
Abstract:
| Citation: | WANG Wen Jun, FENG Shun Qiao, HE Feng, DU Hai Jun, FENG Miao, WANG Rui Fang, MEI Guo Yong, LIU Mi, LIU Rong, YAO Hai Lan, HAN Jun. The Viral Load of Epstein-Barr Virus in Blood of Children after Hematopoietic Stem Cell Transplantation[J]. Biomedical and Environmental Sciences, 2022, 35(9): 804-810. doi: 10.3967/bes2022.052 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: