-
Tuberculosis (TB), caused by the Mycobacterium tuberculosis complex (MTBC), is the 13th leading cause of death and the second leading cause of infectious killer after COVID-19 (above HIV and AIDS), according to the World Health Organization’s 2022 Global TB Report. In 2021, 10.6 million people were diagnosed with a new episode of TB and 1.6 million people died as a result. Multidrug-resistant tuberculosis (MDR-TB) and rifampin-resistant tuberculosis (RR-TB) accounted for 0.437 million of these cases, a 3.0% increase from 2020. China reported 7.4% of TB and 7.3% of MDR-TB (RR-TB) globally, and is among 30 countries with a high MDR-TB burden[1].
MDR-TB strains are resistant to at least the two most potent anti-TB drugs: isoniazid (INH) and rifampin (RIF)[2]. Hangzhou is the capital city of the Zhejiang Province, with a population of 10 million. We evaluated the prevention and treatment of TB in the suburban districts of Hangzhou, comprised of seven counties that have poor medical care, including a lack of MDR-TB-designated hospitals. As a result, we found that the rate of MDR-TB showed a significant downward trend in 2018–2019 (2.60%) compared to 2011 (11.6%) and 2015 (8.0%)[3]. Molecular rapid diagnosis GeneXpert MTB/RIF (Cepheid Inc.) has been used in the suburban districts of Hangzhou since 2016, which significantly shortens the time of MDR-TB diagnosis, an important reason for the decline in the multidrug-resistance rate. However, the prevalence and characteristics of drug-resistance mutations (DRMs) in Hangzhou MDR-TB strains have not been well studied. To address this, we sequenced 62 MDR-TB strains isolated from patients in the suburban districts of Hangzhou from July 2018 to June 2022 using whole-genome sequencing to study the molecular characteristics of INH-resistant and RIF-resistant Mycobacterium tuberculosis (M.tb). Gene sequencing provided a distinct blueprint for RIF resistance, which was used to ascertain the accuracy of GeneXpert MTB/RIF.
Among the strains used, all were culture-positive specimens of suspected TB on Lowenstein–Jensen (LJ) medium from ten designated hospitals in the suburban districts in Hangzhou from July 2018 to June 2022. Species identification using p-nitrobenzoic acid and 2-thiophenecarboxylic acid hydrazide and phenotypic drug susceptibility tests (DST) using Lowenstein-Jensen culture (L-J) medium were conducted for 3 to 4 weeks at 37 °C, with cultures checked weekly for visible colony growth. The drug concentrations of DST were as follows: 0.2 mg/L INH, 40 mg/L RIF, 4.0 mg/L streptomycin (SM), 2.0 mg/L ethambutol (EMB), 4.0 mg/L ofloxacin (OFX), and 30 mg/L kanamycin (KM). The standard isolate H37Rv (M. tuberculosis ssp. tuberculosis ATCC 27294) and sterile deionized water (ddH2O) were used as quality control and negative controls, respectively, in all experiments[4]. GeneXpert MTB/RIF was performed on patients’ sputum after visiting the designated tuberculosis hospital following standard procedures, and the GeneXpert results were processed on the same day. The resulting bacterial colonies were scraped from L-J cultures, added 400 μL of TE buffer, and incubated for 30 min at 80 °C. Genomic DNA was extracted using the OMEGA Bacterial DNA Kit according to the manufacturer’s instructions. Next, genomes were sequenced using an Illumina HiSeq platform. Genome assembly was conducted using SOAPdenovoV2.04 and the genome gap was filled using Gapcloser (version 1.12). The assembled genome sequences were submitted to the NCBI for Biotechnology Information database for annotation. Finally, drug resistance was analyzed using TB-Profiler (version 4.3.0).
A total of 3,096 strains were identified as M.tb based on acid-fast staining, culture and GeneXpert, of which 72 were MDR-TB strains according to DST. All 72 strains were RR-TB strains based on GeneXpert. A total of 62 MDR-TB strains were whole-genome sequenced, and 10 strains were excluded, of which 6 strains were difficult to re-culture and 4 strains failed to be sequenced.
RIF is one of the most potent first-line anti-TB drugs, and RIF resistance is mostly associated with ropB, which encodes the DNA-dependent RNA polymerase β-subunit. A 96% RIF mutation was found in the 81-bp fragment of the RIF resistance-determining region (RRDR, 426–452), which was detected using GeneXpert[4]. In our study, all 62 strains harbored mutations in the RRDR according to whole-genome sequencing, which was consistent with the GeneXpert results (Table 1). According to the mutation characteristics of RIF resistance, GeneXpert plays an important role in the diagnosis of RR-TB in the suburban districts of Hangzhou. Patients were diagnosed with RR-TB on their first visit to a locally designated tuberculosis hospital in a suburban district. Diagnosed patients were immediately transferred to an MDR-TB-designated tuberculosis hospital in the urban area of Hangzhou for treatment. Rapid diagnosis reduces tuberculosis transmission and drug resistance mutations due to the misuse of antibiotics. Seventeen strains harbored two different mutation loci. The most common mutation was in codon 450 (n = 40, 64.5%), which had three types of amino acid substitutions: S450L (n = 37, 59.7%), S450P (n = 2, 3.2%), and S450T (n = 1, 1.6%). Previous research on the proportion of mutations at codon 450 found a rate of 37.8% in Vietnam, 59.0% in India, 58.7% in Nepal, 41.0% in Beijing, 62.7% in Ningbo Zhejiang Province, and 58.3-63.3% in other parts of China, all of which were lower than our data[2,5]. The second most common mutation was at codon 445 (n = 12, 19.4%), and the different amino acid substitutions were H445N (n = 6, 9.7%), H445Y (n = 3, 4.8%), H445A (n = 1, 1.6%), H445R (n = 1, 1.6%), and H445D (n = 1, 1.6%). Other mutation codons were followed by 430 (L430P, 8), 452 (L452P, 3; L452V, 1), 435 (D435G, 1; D435F, 1; D435V, 1), 434 (M434V, 2), and 432 (Q432P, 1) (Table 1). The most common rpoB gene mutations were observed at codons 450 (64.5%), 445 (19.4%), 430 (12.9%), 452 (6.5%), 435 (4.8%) while the order in Beijing was 450 (41.6%), 445 (20.2%), 435 (9.3%), 430 (6.4%), and 452 (3.5%)[2]. The order of our previous studies on 103 MDR strains isolated from the same area from January 1, 2016 to December 31, 2017 using Sanger sequencing of the rpoB fragment (3,519 bp) was 450 (61.2%), 445 (30.1%), 435 (2.9%), and 441 (1.9%), respectively[4]. We found that codons 450 and 445 were the most common and second most common mutation sites, respectively, in Hangzhou and Beijing. Compared to our previous studies from January 1, 2016 to December 31, 2017, the rpoB mutation sites were approximately the same, except for two new sites that were found at 430 and 452, while site 441 was missing from July 2018 to June 2022.
Amino acid changes in rpoB gene Other mutation Frequency
(no. of isolates)Relative frequency (%) rpoB L430P 3 4.8 rpoB L430P rpoB M434V 1 1.6 rpoB Q432P 1 1.6 rpoB D435G rpoB M434V 1 1.6 rpoB D435F 1 1.6 rpoB H445A rpoB L430P 1 1.6 rpoB H445R 1 1.6 rpoB H445N 2 3.2 rpoB H445N rpoB L430P 3 4.8 rpoB H445N rpoB L452V 1 1.6 rpoB H445D 1 1.6 rpoB H445Y rpoB D435V 1 1.6 rpoB H445Y rpoB E460G 1 1.6 rpoB H445Y 1 1.6 rpoB S450L 29 46.8 rpoB S450L rpoC G332R 4 6.5 rpoB S450L rpoC F452S 1 1.6 rpoB S450L rpoB A286V 3 4.8 rpoB S450F 2 3.2 rpoB S450W 1 1.6 rpoB L452P 3 4.8 Table 1. Amino acid changes of RIF resistance genes in 62 MDR-TB strains
We also discovered two strains harboring ropB mutations, E460G and A286V, outside the conventional 81-bp hotspot (RRDR, 426–452) and both strains had additional RRDR mutations. Five strains harbored rpoC mutations, including four at G332R and one at F452S (Table 1). rpoC mutations may compensate for fitness defects in RIF-resistant M. tuberculosis by altering gene expression in response to rifampin exposure[6]. Previous studies have shown that rpoC F452L alleles restore the transcriptional efficiency of RNA polymerase bearing the rpoB S450L mutation[6]. Similarly, all five strains in our study harbored the rpoC mutation together with the rpoB S450L mutation (Table 1). Among all 21 mutant genotype patterns considering all the RIF resistance genes, the most frequently changed codons were ropB S450L (29/62, 46.8%) (Supplementary Table S1, available in www.besjournal.com) which was higher than that reported in previous research in Beijing (38.2%), while mutations ropB S450L and rpoC G332R (4/62, 6.5%) reported similar rates[2].
Drug(s) Locus Mutation Other mutation Frequency (no. of isolates) Relative frequency (%) RIF rpoB S450L 29 46.8 S450L rpoC G332R 4 6.5 S450L rpoB A286V 3 4.8 L452P 3 4.8 L430P 3 4.8 H445N rpoB L430P 3 4.8 INH katG S315T 42 67.7 fabG1 -15C > T 3 4.8 RIF and INH katG and rpoB katG S315T, rpoB S450L 19 30.7 katG S315T, rpoB A286V, rpoB S450L 3 4.8 katG S315T, rpoB H445N, rpoB L430P 3 4.8 katG S315T, rpoB L430P 3 4.8 katG S315T, rpoB L452P 3 4.8 Table S1. Most frequently identified mutations among the 62 MDR-TB strains
Isoniazid (INH) is an effective drug that has been commonly used to treat tuberculosis since 1952. The emergence of INH-resistant TB continues to increase the utility of INH. INH resistance appears to be more complex and has been reported with katG, which encodes catalase-peroxidase and transforms INH into its active form; the upstream region of the fabG1-inhA operon encodes a putative mycolic acid synthesis enzyme involved in cell wall formation, while ahpC encodes alkyl hydroxyperoxidase, which acts as a component of antioxidant reductase. Previous studies have shown that mutations conferring INH resistance are most frequently detected in the katG gene, especially in codon 315[2,7]. In our study, 85.5% (53/62) of the 62 MDR showed mutations in the katG gene. We found that 17.7% (11/62) of patients possessed mutations in the upstream region of fabG1-inhA operon, whereas 4.8% (3/62) had mutations in the upstream region of ahpC. Of these strains, 11.3% (7/62) harbored mutations in katG and fabG1. One strain showed nucleotide substitutions in katG and ahpC. Among these, 85.5% (53/62) of katG mutations possessed 8 forms, including S315T (71.0%, 44/62), S315I (1.6%, 1/62), Q127P (3.2%, 2/62), N138H (1.6%, 1/62), Y155S (1.6%, 1/62), W191R (1.6%, 1/62), T251M (1.6%, 1/62), and W300G (1.6%, 1/62) and −10A > C (1.6%, 1/62). It had been previously reported that 10%–28% INH-resistant TB strains have a −15C > T in the upstream region of fabG1-inhA operon[2]. The fabG1 promoter mutations were fabG1 −15C > T (14.5%, 9/62) and fabG1 −8T > C (1.6%, 1/62), whereas the mutation of inhA was a G-to-A replacement 154-bp upstream of inhA. ahpC mutations had ahpC_c. −74G > A (1.6%, 1/62) and ahpC −52C > T (1.6%, 1/62) (Table 2). The most frequent mutation was a single gene mutation at the katG315 codon as a serine to threonine substitution (S315T) at a rate of 67.7% (42/62), which was approximate to the data (60.4%) from the Chinese national TB drug-resistance surveillance program[7] (Supplementary Table S1). The second most frequent mutation was fabG1 −15C > T (4.8%, 3/62) (Supplementary Table S1). Mutations in the katG and fabG1-inhA operons were discovered in 96.6% (57/59) of INH genotype-resistant isolates in our study, which is consistent with another study conducted in Zhejiang Province[5]. As expected, the most common combination of mutations was rpoB (S450L) + katG (S315T) (19/62, 30.7%) in the 62 MDR-TB cases (Supplementary Table S1).
Gene mutation and locus Other mutation Frequency (no. of isolates) Relative frequency (%) katG (53) katG S315T 42 67.7 katG S315T fabG1 -8T > C 1 1.6 katG S315T fabG1 -15C > T 1 1.6 katG S315Ile katG N138H 1 1.6 katG Q127P fabG1 -15C > T 2 3.2 katG Y155S 1 1.6 katG W191R fabG1 -15C > T 1 1.6 katG T251M fabG1 -15C > T 1 1.6 katG W300G fabG1 -15C > T 1 1.6 katG -10A > C ahpC -52C > T 1 1.6 *katG -667_*26327del 1 1.6 inhA (1) inhA -154G > A 1 1.6 fabG (10) fabG1 -15C > T 3 4.8 fabG1 -15C > T katG Q127P 2 3.2 fabG1 -15C > T katG W300G 1 1.6 fabG1 -15C > T katG W191R 1 1.6 fabG1 -15C > T katG T251M 1 1.6 fabG1 -15C > T katG S315T 1 1.6 fabG1 -8T > C katG S315T 1 1.6 ahpC (3) ahpC -74G > A 1 1.6 ahpC -52C > T katG -10A > C 1 1.6 ahpC -52C > T 1 1.6 No mutation 3 4.8 Table 2. Nucleotide and amino acid changes of INH resistance genes in 62 MDR-TB strains
Three strains (3/62, 4.84%) showed phenotypic INH resistance, but lacked any INH resistance genes. We scanned the whole-genome drug resistance of these three strains and did not find any other drug resistance genes, except for a ropB mutation. Phenotype DST revealed that the three strains were sensitive to streptomycin, ethambutol, ofloxacin, and kanamycin. These results highlight the need for further research to determine whether novel INH resistance genes exist.
One MDR-TB strain, Y210107, had a large genomic deletion (LGD) (26,994 bp) of 30 genes and two pseudogenes compared to H37Rv (Figure 1). Deletions were performed from Rv1978 to Rv1909c, including katG (Supplementary Table S2, available in www.besjournal.com). The gene category included nine genes related to intermediary metabolism and respiration (red), six genes related to cell wall and cell processes (green), one of lipid metabolism (blue), two of virulence and detoxification, including katG-conferred isoniazid resistance (orange), one regulatory protein (yellow), and 11 hypothetical proteins (indigo). A genome-wide scan of the strain showed no other resistance genes except for His445Ala and Leu430Pro in ropB. It conferred resistance to INH (0.2 mg/L) and RIF (40 mg/L) on L-J culture. The growth rate of the L-J culture was consistent with that of H37RV. LGDs of katG operon have rarely been reported, with a single report in China identifying strain W146, a clinical Beijing/W genotype MDR isolated from Wuxi, Jiangsu Province, which harbored a 15,925-bp furA-katG operon that included 16 genes from Rv1900c to Rv1915[8]. Vilchèze et al. reported LGDs (6–63 kbp) lacking katG in isoniazid-resistant M.tb mutants derived from four M. tuberculosis strains[9]. Research in Italy found that large fragment deletions had no effect on growth, but increased the MIC of INH. They found that the katG deletion M.tb strain conferred high-level resistance to INH (MIC > 25.6 μg/mL)[10]. In future, we aim to study the MIC of our strain and the whole-genome sequence to further examine the effects of the 26,994-bp deletion on genotype and transmission.
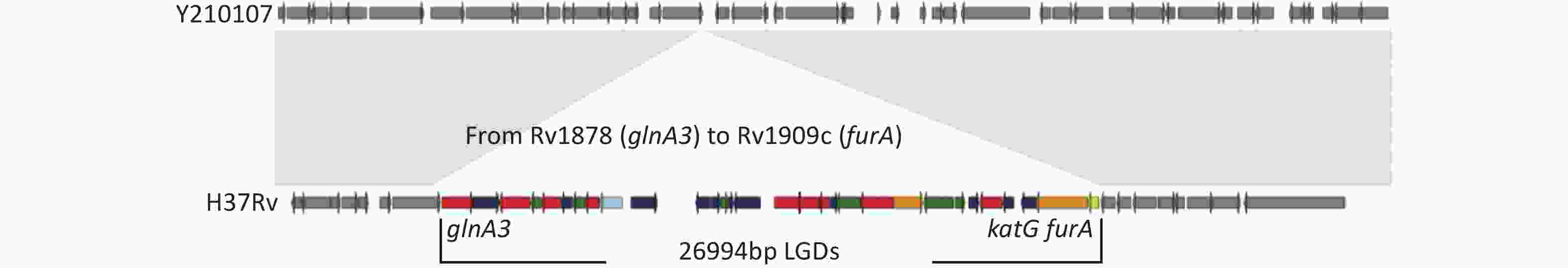
Figure 1. The arrangement of 26,994 bp LGDs missing in strain Y210107 compared with the H37Rv genomic region from Rv1978 (glnA3) to Rv1909c (furA).
In conclusion, we analyzed the frequency of RIF and INH mutations in 62 MDR-TB strains isolated from rural areas of Hangzhou using next generation sequencing. As a result, a high prevalence of rpoB (S450L) and katG (S315T) was observed. All 62 MDR-TB strains contained RIF mutations in RRDR, indicating that the rapid diagnostic technique GeneXpert was effective in the identification of RIF-resistant strains in the suburban districts of Hangzhou from July 2018 to June 2022. We also observed a novel katG mutation type in the form of a 26,994-bp large genomic deletions compared to H37RV. This is the first study to investigate the frequency of RIF and INH gene mutations in Hangzhou using whole-genome sequencing. Based on our findings, we aim to continue characterizing the resistance genes of other first- and second-line drugs to further understand the drug-resistant genotypes of MDR-TB in Hangzhou. In addition, our results provide a basis for patients with TB who have not recovered.
-
HUANG Yin Yan and XIE Li designed the study; HUANG Yin Yan supervised all the experiments; HUANG Yin Yan, WU Yi Fei, JIA Qing Jun, and BAI Xue Xin performed the experimental studies; CHENG Qing Lin, LI Qing Chun, and AI Liyun performed the data analysis; Huang Yin Yan drafted the manuscript; and XIE Li revised the manuscript for intellectual content.
-
Obtained.
-
None of the tests were performed on patients. All experiments were performed on strains. Informed consent was obtained from all patients for the use of clinical samples. This ethical review was approved by the Ethics Committee of the Hangzhou Center for Disease Control and Prevention.
-
Not commissioned; externally peer-reviewed.
-
Gene Categorya Product glnA3 7 Glutamine synthetase GlnA Rv1879 10 Hypothetical protein cyp140 7 Cytochrome P450 Cyp140 lppE 3 Lipoprotein LppE Rv1882c 7 Short-chain type dehydrogenase/reductase Rv1883c 10 Hypothetical protein rpfC 3 Resuscitation-promoting factor RpfC Rv1885c 7 Chorismate mutase fbpB 1 Diacylglycerol acyltransferase/mycolyltransferase Ag85B Rv1887 10 Hypothetical protein Rv1890c 10 Hypothetical protein Rv1891 10 Hypothetical protein Rv1892 3 Membrane protein Rv1893 10 Hypothetical protein Rv1894c 10 Hypothetical protein Rv1895 7 Zinc-binding alcohol dehydrogenase Rv1896c 7 S-adenosyl-L-methionine-dependent methyltransferase Rv1897 7 D-tyrosyl-tRNA(Tyr) deacylase Rv1898 10 Hypothetical protein lppD 3 Lipoprotein LppD lipJ 7 Lignin peroxidase LipJ cinA 0 Competence damage-inducible protein CinA nanT 3 Sialic acid-transport integral membrane protein NanT Rv1903 3 Membrane protein Rv1904 10 Hypothetical protein aao 7 D-amino acid oxidase Rv1906c 10 Hypothetical protein Rv1907c 10 Hypothetical protein katG 0 Catalase-peroxidase furA 9 Ferric uptake regulation protein FurA Note. a0, virulence, detoxification; 1, lipid metabolism; 3, cell wall and cell processes; 7, intermediary metabolism and respiration; 9, regulatory proteins; 10, hypothetical protein. Table S2. Genes of 26,994 bp LGDs in strain Y210107 compared to H37Rv
HTML
 23095+Supplementary Materials.pdf
23095+Supplementary Materials.pdf
|

|







 Quick Links
Quick Links
 DownLoad:
DownLoad: