-
Lassa fever (LF) is an acute viral hemorrhagic disease caused by the Lassa virus (LASV), with an incubation period typically ranging from 5 to 21 days. The overall case fatality rate (CFR) of Lassa fever is approximately 1%. In endemic regions, around 80% of LASV infections are asymptomatic, though the CFR among hospitalized patients can reach as high as 15%[1-3]. The primary reservoir host for LASV is the multimammate rat species (Mastomys natalensis)[4,5]. Other rodents, such as the house mouse and grass rats, have also been identified as viral hosts[6-9]. These animals excrete the virus through their urine and feces, and humans can become infected through direct contact with these contaminated materials[10]. LASV can also spread from person to person, mainly through contact with bodily fluids like blood, urine, saliva, feces, and semen of infected individuals[11].
LASV is a small (80–200 nm) asymmetric enveloped single-stranded RNA virus belonging to the Arenaviridae family[10,12]. The viral genome is composed of two segments of negative-strand RNA, designated as S RNA and L RNA. The S RNA, which is approximately 3,400 bases long, encodes the nucleoprotein (NP) and glycoprotein (GP)[13]. The L RNA, about 7,200 bases in length, encodes the Z protein (a ring finger protein) and the L protein (an RNA-dependent RNA polymerase, RdRP)[14]. Genetic analysis has identified seven lineages (I-VII) of LASV that are currently circulating in West Africa[10].
LF is mainly prevalent in West Africa[15]. Since the first case of LF was reported in Nigeria in 1969, outbreaks have occurred in regions such as Taraba, Yobe, and Salawa states[16-18]. Especially since 2016, outbreaks of Lassa fever have occurred annually during the dry season in Nigeria, with increasingly severe consequences. In 2023, the worst outbreak in a decade was recorded: 9,155 suspected cases and 227 deaths were reported. Out of Nigeria’s 36 states, 28 were affected[19]. As of November 3, 2024, Guinea, Liberia, and Nigeria have reported 9,829 suspected cases and 186 deaths since the beginning of the year[20]. Meanwhile, over the past decade, the endemic area has expanded, with new virus lineages emerging in some previously unaffected countries such as Ivory Coast, Ghana, and Togo[21,22]. Ivory Coast, Benin, Burkina Faso, Ghana, Togo, and Mali are now known as endemic countries[23]. LASV is estimated to infect approximately 897,700 people annually, resulting in about 18,000 deaths in West Africa each year[24]. Alarmingly, with increasing global mobility, LF is being imported into countries that have not previously reported cases. Imported cases have been documented in nations such as the United States, Germany, the United Kingdom, Japan, Israel, and Canada[25,26]. Here, we report the first imported case of LF in China and analyze the molecular characteristics of the virus.
-
On August 3, 2024, a hospital in Mianyang City, Sichuan Province, reported a case of “suspected Lassa fever” and notified the disease control agency to conduct epidemiological investigations and sample collection. Cerebrospinal fluid (CSF), blood, saliva, and urine were collected from the patient between August 3 and September 17, 2024. Whole blood, urine, and saliva samples were collected from close contacts between August 6 and August 24, 2024.
-
Blood samples were centrifuged at 1,500 rpm for 5 min to separate the serum from blood cells. The centrifuged blood, along with CSF, urine, and saliva samples, was inactivated at 60 °C for 1 h. After inactivation, 200 μL of each sample was taken in a biosafety level 2 (BSL-2) biosafety cabinet for total viral RNA extraction using the CqEx-DNA/RNA Virus Nucleic Acid Extraction Kit (Tianlong, Xi’an, China), following the manufacturer’s instructions. Two LASV nucleotide detection kits, produced by Guangzhou DaAn Gene Co., Ltd., and BioGerm (Qingdao) Medical Technology Co., Ltd., were used for RT-qPCR testing, following the respective manufacturers’ protocols.
-
LASV RNA was reverse-transcribed into cDNA using SuperScript IV Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). Whole-genome capture amplification was then performed using Q5® Hot Start High-Fidelity 2X Master Mix (NEB, Massachusetts, MA, USA) and specific amplicon primers designed by our laboratory (Supplementary Table S1). The amplification products were purified using the AMPure XP Purification Kit (Beckman Coulter, Brea, CA, USA).
The DNA sequencing library was prepared using the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions, and sequencing was conducted on the MiniSeq platform (Illumina, San Diego, CA, USA). The sequencing data underwent quality control, cleaning, and sequence assembly using the CLC Genomics Workbench 23.0.4 (QIAGEN, Düsseldorf, NRW, Germany) to generate a consensus sequence. Through NCBI BLAST, the sequences with the highest similarity to LASV (> 86%) were selected. Based on published references[21,22,27,28], a total of 28 viral S and L segment sequences (Supplementary Table S2) were chosen from GenBank as reference sequences for subsequent analysis, considering different genotypes, epidemic regions, and collection times. All those sequences were nearly full length(> 92%). Multiple sequence alignment was performed using MAFFT software (version 7.450), and homology analysis was conducted with MEGA software (version 7.0). Evolutionary trees for the S and L segments were constructed using the neighbor-joining method, with a bootstrap value of 1,000.
-
The patient’s epidemiological history is as follows: In March 2024, the patient traveled to Guinea, Africa, to work as a chef. On July 17, while in Guinea, the patient experienced a high fever, loss of appetite, and other symptoms, prompting her to seek medical attention at a local hospital. She was diagnosed with “malaria and typhoid fever”, and her condition improved after treatment.
On July 23, the patient departed Guinea for China. Upon arrival in China on July 24, she began experiencing symptoms such as lower back pain, abdominal pain, nausea, and vomiting. She sought treatment at Mianyang Central Hospital and later returned home. On July 30, the patient exhibited symptoms such as slow reaction, hearing loss, slurred speech, incontinence, and difficulty swallowing, and she collapsed at home. She was transported by ambulance to Jiangyou People’s Hospital. On August 1, she was transferred to Mianyang 404 Hospital, where blood samples were collected for testing. Malaria was excluded as a diagnosis. On August 2, CSF samples were collected and sent to a third-party detection institution for high-throughput sequencing. Later that day, the patient was transferred to West China Hospital of Sichuan University. After admission, the patient received antiviral treatment with aciclovir and ribavirin, antibacterial treatment with piperacillin-tazobactam, and symptomatic supportive care. However, the patient still had a fever. Her blood samples were collected and sent to the Sichuan Center for Disease Control and Prevention (Sichuan CDC) for real-time fluorescent quantitative PCR (RT-qPCR) testing. On the morning of August 5, the patient was transferred to Chengdu Public Health Clinical Center and admitted to the ICU for isolation and treatment. Upon admission, the patient presented with lethargy; hearing loss; ecchymosis at the intravenous infusion site on the upper limbs; positive neck stiffness; mild edema in both lower limbs; weakened knee reflexes; and significantly elevated levels of lactate dehydrogenase, hydroxybutyrate dehydrogenase, blood amylase, and blood lipase[29]. During isolation, the patient was treated with ribavirin for antiviral therapy, dexamethasone for anti-inflammatory treatment, vitamin B complex for nourishing the nerves, and enoxaparin sodium for anticoagulation therapy after platelet transfusion. Additionally, the patient received symptomatic and supportive treatments, including anti-infective therapy, human serum albumin infusion, nutritional support, blood glucose control, and psychological counseling[29]. After two weeks of active treatment, the clinical symptoms improved markedly. Her hearing was partially recovered, and her platelet count and liver and kidney functions returned to normal. There was no sign of hemolysis or bleeding[29].
-
The first CSF sample was collected on August 2, 2024. High-throughput sequencing performed by a third-party institution indicated a “suspected case of LF”. The Sichuan CDC immediately conducted RT-qPCR tests on the CSF and blood samples collected on August 2 and 3, both of which tested positive for LASV (Table 1). On August 5, 2024, clinical samples were sent to the Chinese Center for Disease Control and Prevention (China CDC) for re-examination, and the results also confirmed LASV infection. Between August 8 and September 17, clinical samples, including blood, saliva, and urine, were collected from the patient for RT-qPCR testing by Sichuan CDC and Mianyang Center for Disease Control and Prevention (Mianyang CDC). The results indicated that urine samples collected on August 9, 12, 16, 21, and September 3 and 10 tested positive, whereas all other samples were negative (Table 1). From August 6 to August 24, multiple samples from close contacts were also collected and tested via RT-qPCR, with all results returning negative. Additionally, the Chengdu Center for Disease Control and Prevention (Chengdu CDC) conducted RT-qPCR tests on the sewage from the aircraft on which the patient had traveled, and these results were negative.
Sample number Sample source Sample type Collection date Test result Ct value LSR24-1 case CSF 2024/8/2 positive 33.25 LSR24-2 case blood 2024/8/3 positive 36.66 LSR24-6 case blood 2024/8/8 negative − LSR24-22 case blood 2024/8/9 negative − LSR24-23 case urine 2024/8/9 positive 32.45 LSR24-24 case saliva 2024/8/9 negative − LSR24-26 case blood 2024/8/12 negative − LSR24-27 case urine 2024/8/12 positive 31.62 LSR24-28 case saliva 2024/8/12 negative − LSR24-63 case urine 2024/8/16 positive 31.52 LSR24-64 case urine 2024/8/21 positive 33.70 LSR24-65 case urine 2024/8/30 positive 34.90 LSR24-66 case urine 2024/9/3 positive 36.30 LSR24-67 case urine 2024/9/10 positive 36.60 LSR24-68 case urine 2024/9/13 negative − LSR24-69 case urine 2024/9/17 negative − Table 1. Nucleotide detection results of case samples
-
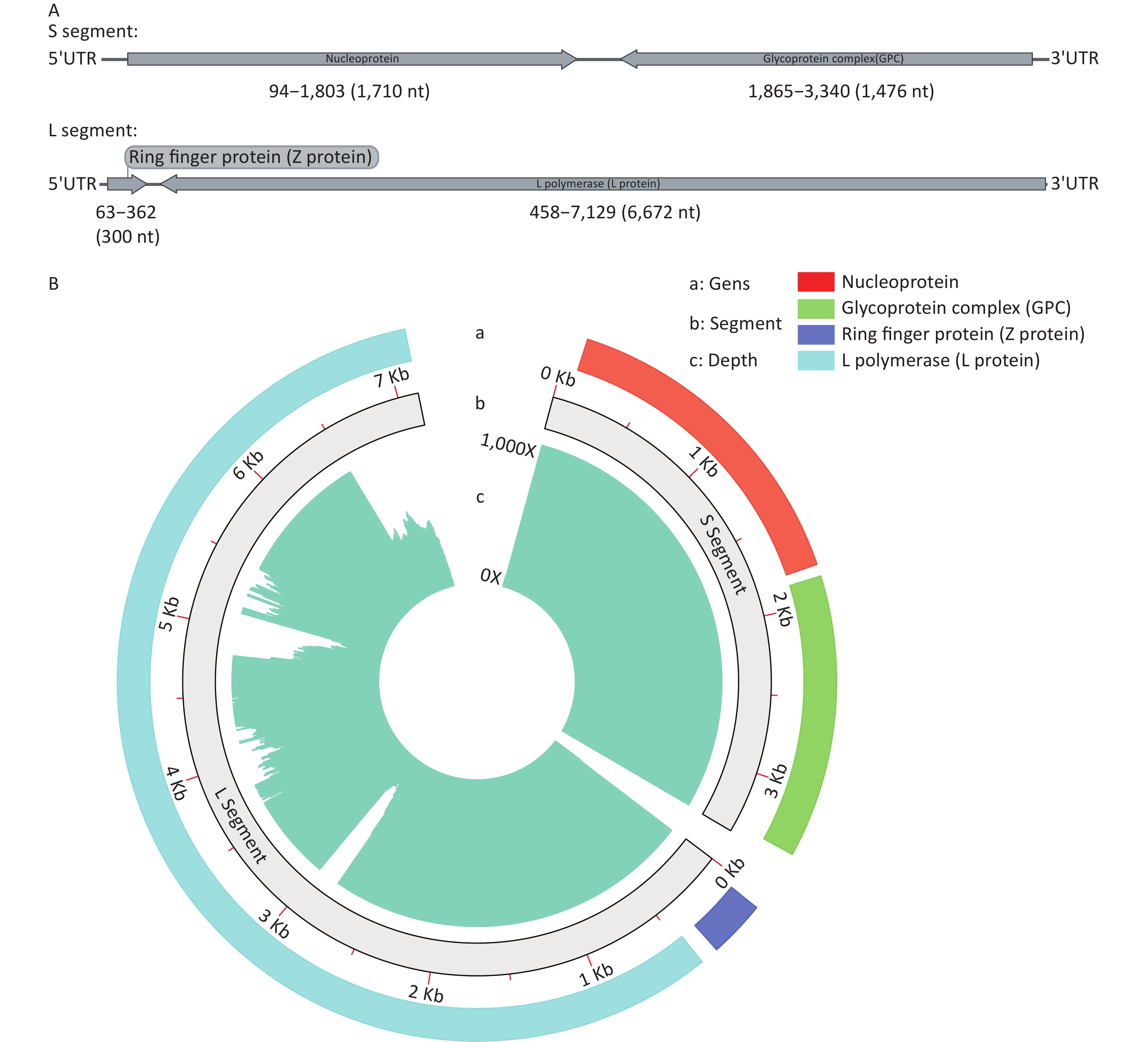
Whole-genome sequencing of the LASV was performed using targeted next-generation sequencing (tNGS). The data were subjected to reference-based assembly, resulting in two complete genome segments, designated S and L (Figure 1A). The coverage reached 99.36% (Figure 1B). The strain identified from the sequencing was named SCLSR24-01, with the S and L segments labeled as SCLSR24-01-S and SCLSR24-01-L, respectively. The S segment, with a length of 3,395 bp, contains two open reading frames (ORFs): one from positions 94 to 1,803 (1,710 nt), encoding the nucleoprotein, and the other from 1,865 to 3,340 (1,476 nt), encoding the glycoprotein complex (GPC). The L segment, with a length of 7,130 bp, also contains two ORFs: one from positions 63 to 362 (300 nt), encoding the Z protein, and the other from 458 to 7,129 (6,672 nt), encoding the L protein. Both genome segments have been uploaded to the Genbase database with accession numbers SCLSR24-01-S (C_AA084676.1) and SCLSR24-01-L (C_AA084677.1).
-
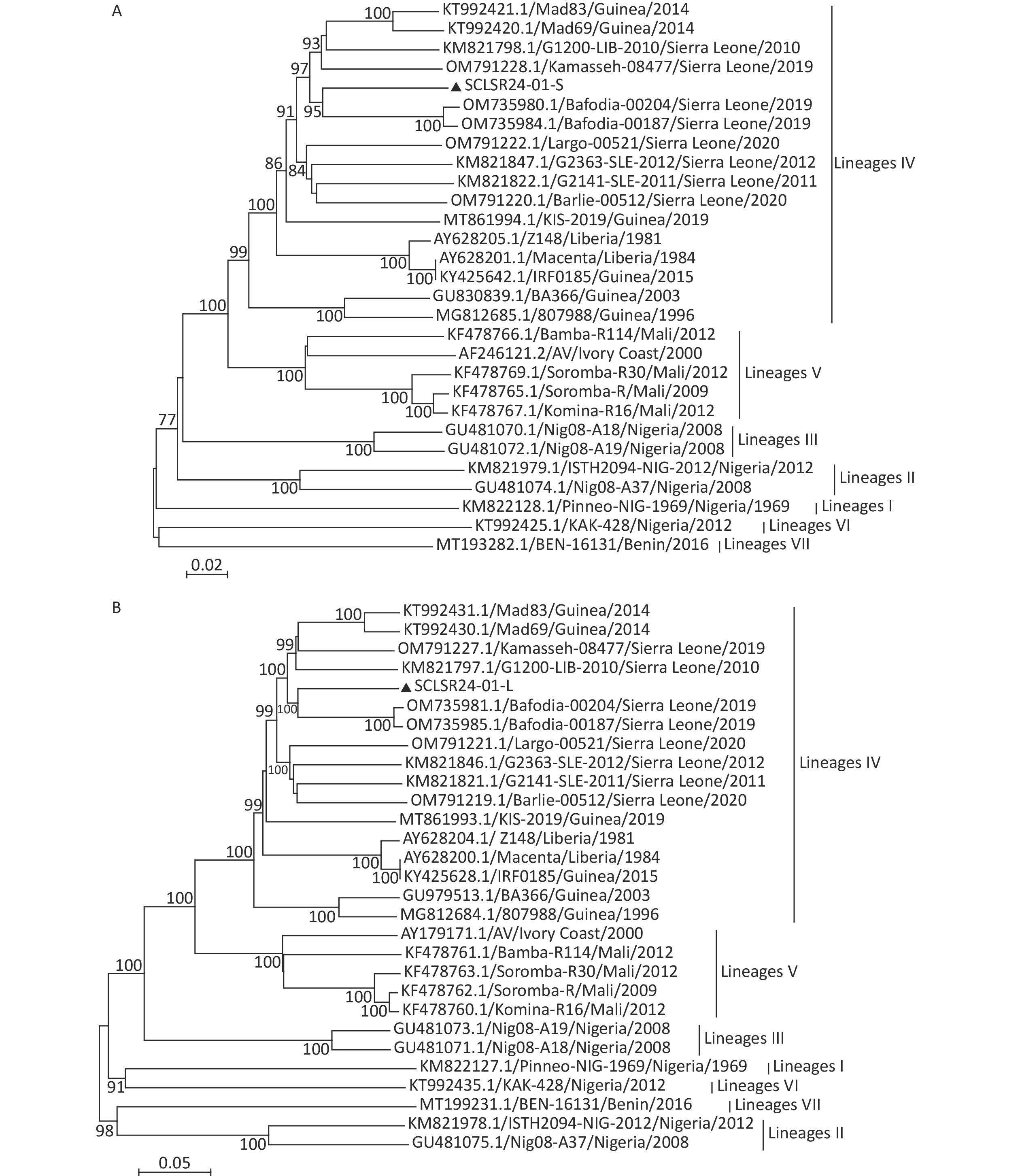
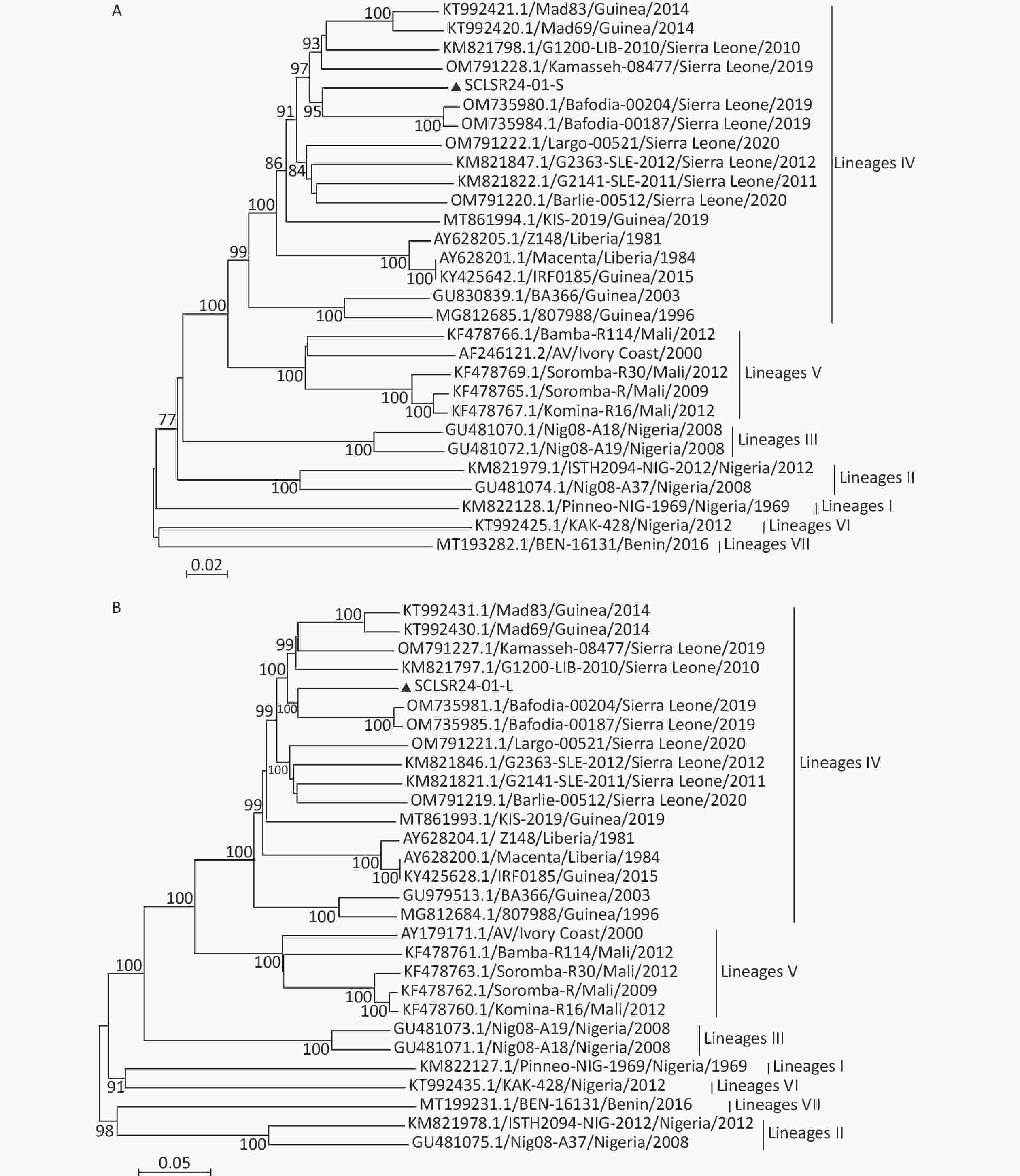
Phylogenetic trees were constructed using full-genome sequences of LASV from lineages I to VII, along with the viral genome assembled in this study. The results showed that both the S and L segments belong to lineage IV. The virus is genetically closest to the 2019 Sierra Leone strains Bafodia-00204 and Bafodia-00187, clustering within the same small clade. In contrast, it is more distantly related to Guinea strains KIS-19, Mad83, Mad69, RF0185, BA366, and 807988, which belong to different evolutionary sub-branches within the same lineage (Figure 2).
-
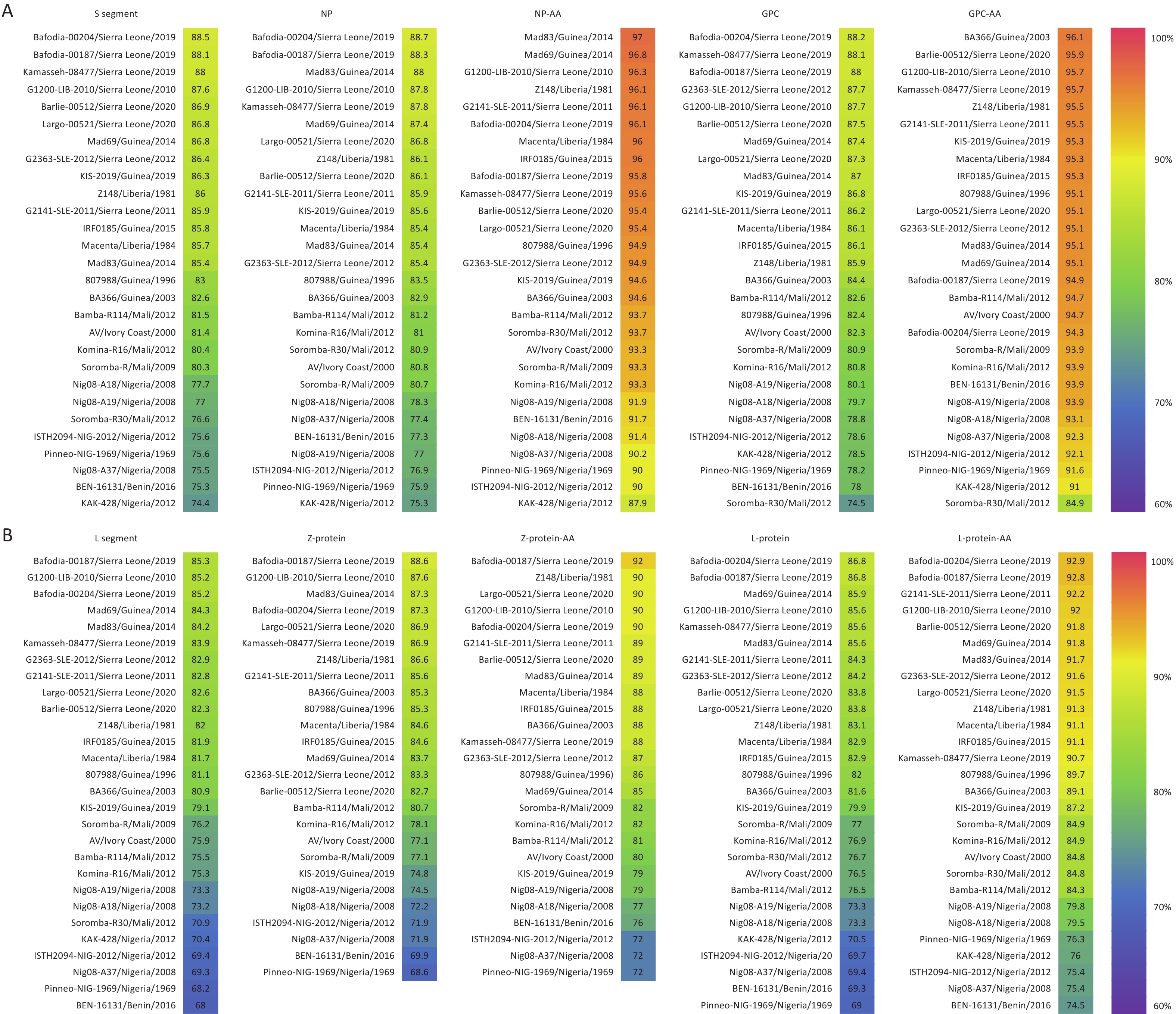
Homology analysis was conducted between SCLSR24-01-S, SCLSR24-01-L, and reference strains. The nucleotide homology of the S segment ranged from 74.4% to 88.5%, whereas that of the L segment ranged from 68% to 85.3%, with the S segment showing higher homology than that of the L segment. For the S segment, the nucleotide homology of NP ranged from 75.3% to 88.72%, and the amino acid sequence homology ranged from 87.9% to 97%. The nucleotide homology of the GPC ranged from 74.5% to 88.2%, with amino acid homology ranging from 84.9% to 96.1%. Among the reference strains, the Sierra Leone strain Bafodia-00204 exhibited the highest nucleotide homology with the S segment, NP, and GPC of SCLSR24-01. The Guinea strain Mad83 exhibited the highest amino acid sequence homology with NP, whereas the Guinea strain BA366 showed the highest amino acid sequence homology with GPC. For the L segment, the nucleotide homology of the Z protein ranged from 68.6% to 88.6%, and the amino acid sequence homology ranged from 72% to 92%. The nucleotide homology of the L protein ranged from 69% to 86.8%, and the amino acid homology ranged from 74.5% to 92.9%. The Sierra Leone strain Bafodia-00187 displayed the highest nucleotide and amino acid homology with the L segment and Z protein of SCLSR24-01. Additionally, the highest nucleotide and amino acid homology for the L protein was with the Sierra Leone strain Bafodia-00204. Overall, the homology of the two protein sequences encoded by the S segment was significantly higher than that of the proteins encoded by the L segment (Figure 3).
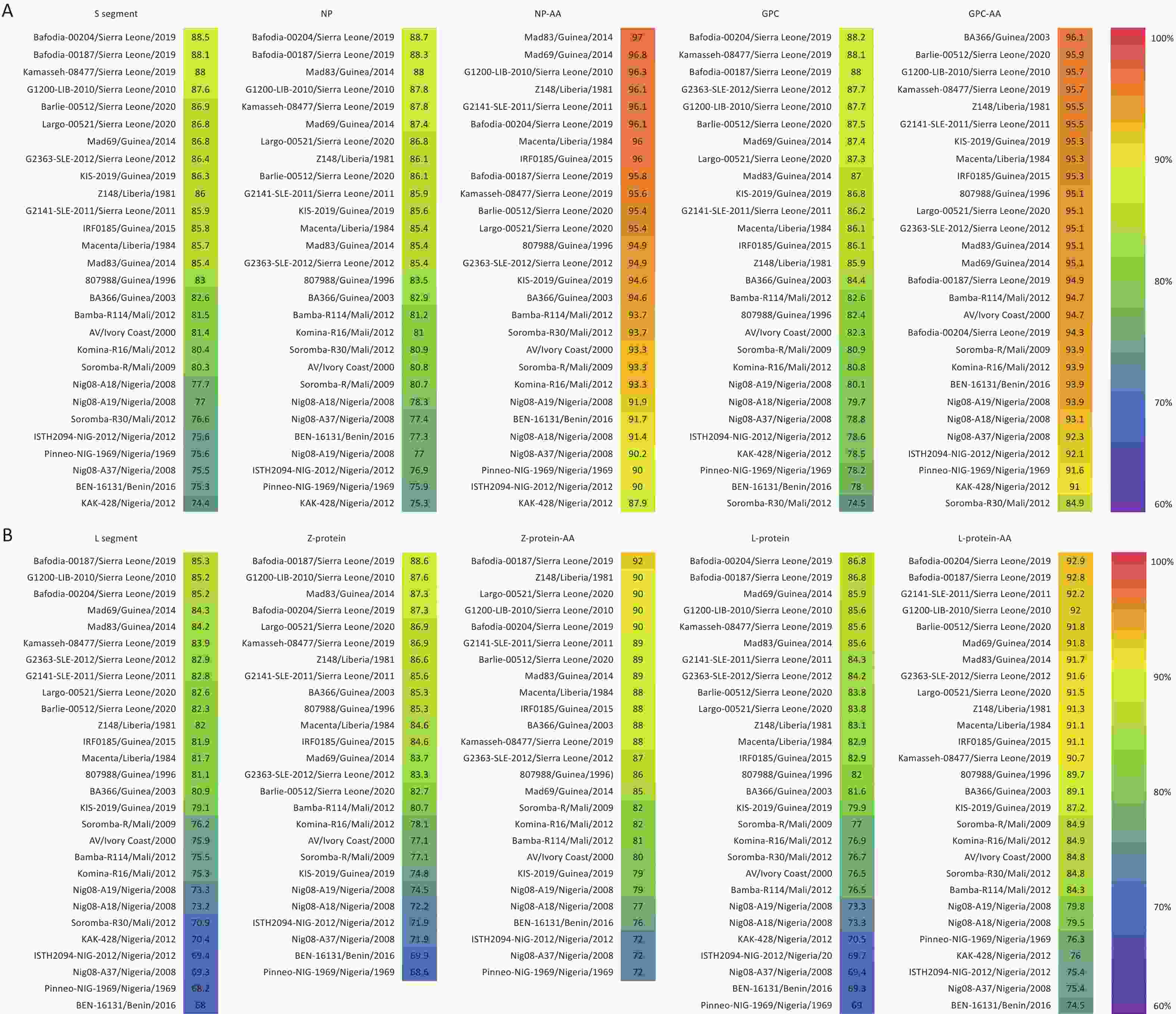
Figure 3. The result of homology analysis of the whole genome sequence of the lassa virus strain from the first imported case of lassa fever in Sichuan Province. The resulting figure for the S segment (A) and L segment (B). S segment: Nucleotide Sequence Homology of S Segment, NP: Nucleotide Sequence Homology of NP, NP-AA: Amino Acid Sequence Homology of NP, GPC: Nucleotide Sequence Homology of GPC, GPC-AA: Amino Acid Sequence Homology of GPC, L Segment: Nucleotide Sequence Homology of L Segment, Z-protein: Nucleotide Sequence Homology of Z-protein, Z-protein-AA: Amino Acid Sequence Homology of Z-protein, L-protein: Nucleotide Sequence Homology of L-protein, L-protein-AA: Amino Acid Sequence Homology of L-protein.
-
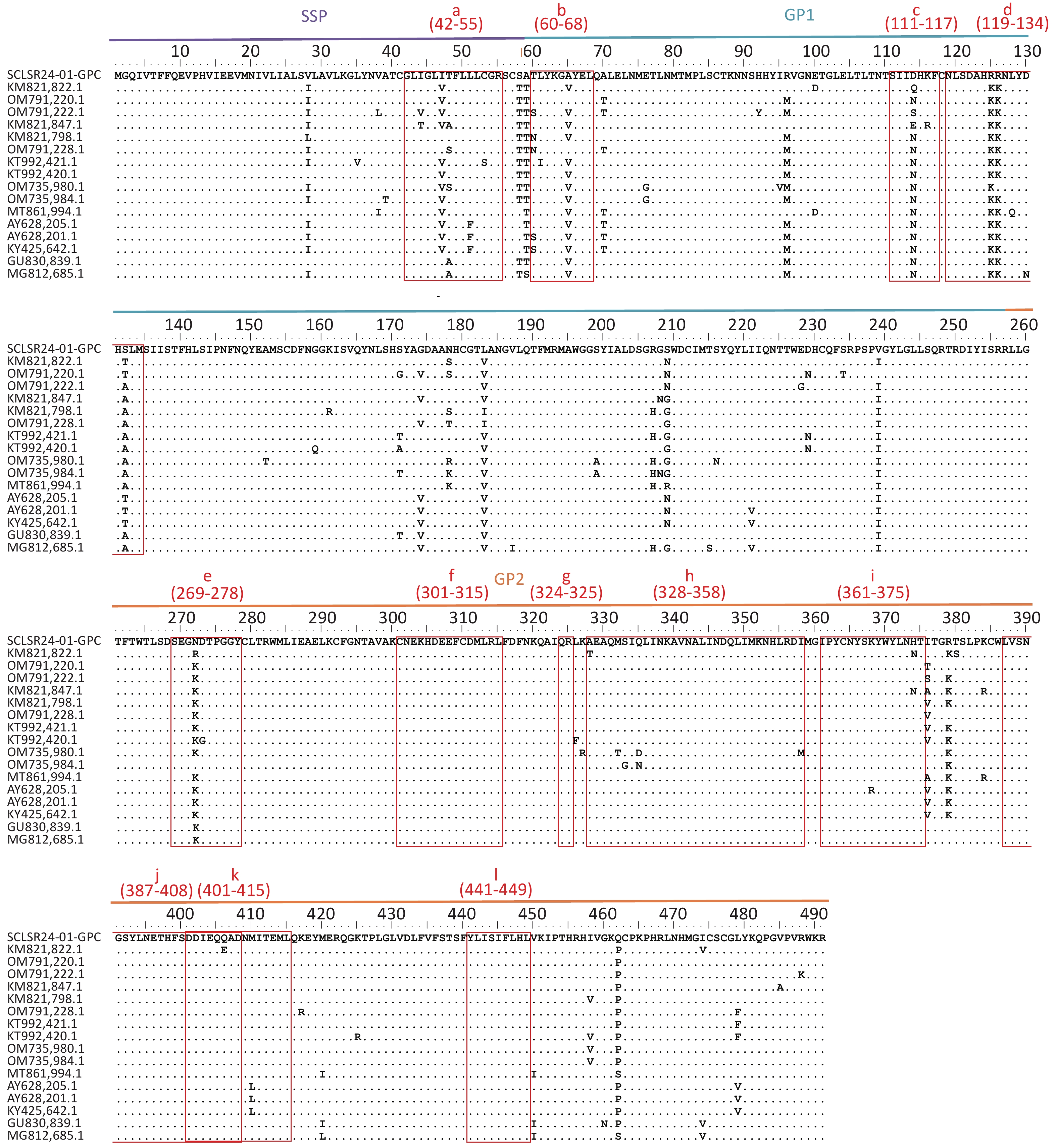
The GPC amino acid sequence of SCLSR24-01 was analyzed for antigenic sites by aligning it with reference sequences from lineage IV. The analysis focused on B-cell and T-cell epitopes within GPC (Figure 4). The B-cell epitopes included the following regions: site A (60-68AA, 387-408AA) (b&j), site B (269-278AA, 324-325AA) (e&g), GPC-A (60-68AA, 269-278AA) (b&e), GP1-A (111-117AA, 328-358AA) (h), GP2-B (119-134AA) (d), GP2-L1 (301-315AA) (f), GP2-L2 (361-375AA) (i), and GP2-L3 (401-415AA) (k). The T-cell epitopes included the stable signal peptide (SSP: 42-55AA) (a), GP1 (60-68AA) (b), and GP2 (441-449AA) (l)[30,31]. The results indicated that the GPC of SCLSR24-01 was relatively conserved in the B-cell epitopes GP2-L1 (f), GP2-B (h), GP2-L2 (i), site A (j), GP2-L3 (k), and the T-cell epitope GP2 (l). These findings were consistent with most reference strains from lineage IV. However, significant amino acid variations were observed in the T-cell epitope SSP (a), as well as in the B-cell epitopes GPC-A (b), GP1-A (c), GP2-B (119-134AA) (d), and site B/GPC-A (e). Notably, epitope c at position D114 and epitope d at positions R125 and S132 differed from all reference sequences, with a simultaneous mutation of R at position 125–126 in epitope d.
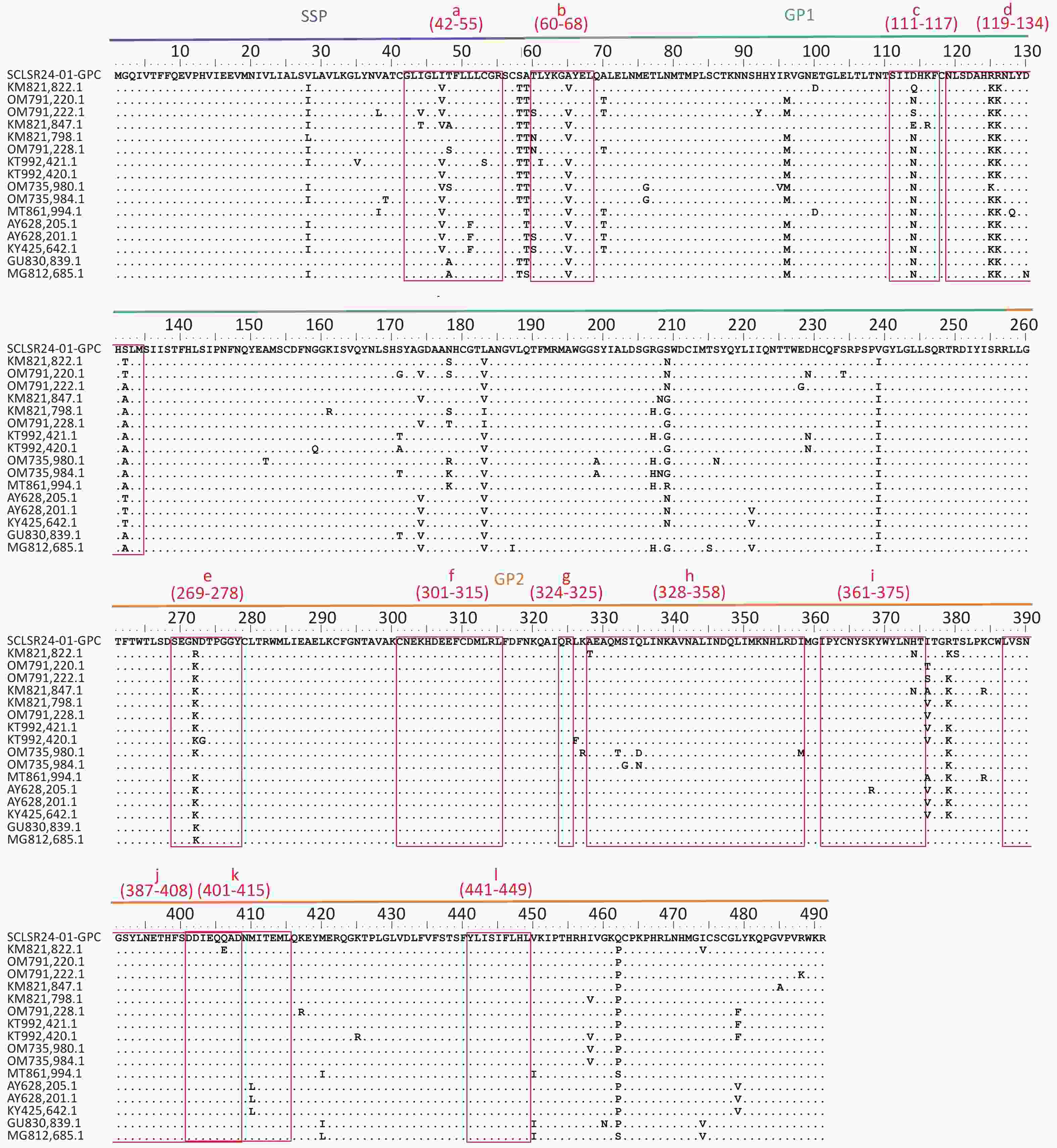
Figure 4. Aligned GPC amino acid sequence of lassa virus (LASV) confirmed and proposed lineage IV sequences. multiple sequence alignment of lineage IV LASV strains selected from the literature (Supplementary Table S2) created with clustal W in BioEidt 7.0.5.1 and Manually Annotated. LASV GP B-cell Epitopes include: 37.7H Epitope Site A (b & j) and Site B (e & g), GPC-A (b & e), GP1-A (c), GP1-B(d), GP2-B (h), GP2-L1(f), GP2-L2 (i), and GP2-L3 (k). LASV GP T-cell Epitopes include: SSP Epitope 42-50AA (a), GP1 Epitope 60-68AA (b), and GP2 Epitope 441-449AA (l).
-
LASV is an Old World arenavirus endemic to West Africa. The World Health Organization (WHO) lists it among the top five diseases requiring priority research due to its pandemic potential[32]. Initially, the primary transmission host of LASV was believed to be the multimammate rat[5], one of the most common large rodent species in equatorial regions. However, subsequent research has identified other rodent species, including those belonging to the genera Rattus, Mus, Lemniscomys, and Praomys, as hosts[6], many of these have a global distribution. With increasing global population, urbanization, and globalization, an increasing number of countries are reporting imported cases of LF[25,26]. The expansion of LASV transmission hosts raises the risk of the virus spreading to new areas with suitable hosts, potentially broadening the epidemic range of LF. This highlights the importance of monitoring imported cases more closely.
Clinical manifestations of LASV infection include fever, headache, fatigue, sore throat, chest pain, back pain, abdominal pain, vomiting, diarrhea, and bleeding from various body sites[12,33]. These symptoms are often difficult to differentiate from other diseases such as malaria, typhoid fever, dengue fever, and yellow fever, as well as other viral hemorrhagic fevers[34]. Definitive diagnosis requires laboratory testing. In this study, the patient initially presented with symptoms such as high fever, anorexia, back pain, abdominal pain, nausea, and vomiting, leading to an initial diagnosis of malaria and typhoid fever. However, subsequent RT-qPCR testing confirmed LASV infection. Research indicates that the virus can be isolated from bodily fluids such as blood, urine, saliva, feces, and semen in LF cases[11]. In this case, LASV was detected in blood, urine, and CSF, with the presence of the virus in CSF being less commonly reported. This underscores the widespread presence of LASV in human bodily fluids. Viremia in LASV infection typically lasts around 20 days, whereas virus shedding in urine can persist for 3 to 9 weeks[35,36]. In this case, LASV was detected in CSF 16 days after the onset of symptoms, whereas subsequent blood tests gradually became negative. However, by September 10, the urine test remained positive 55 days after symptom onset, aligning with reports of prolonged viral excretion in urine. This suggests that the virus may have a longer excretion period through urine, warranting closer attention to laboratory testing and patient care during this phase.
To analyze the virus, we selected the recently prevalent strain as a reference sequence (Supplementary Table S2) for designing multiple amplification primers used in whole-genome amplification and NGS sequencing. The sequence assembly, with over 99% coverage, allowed us to obtain the complete genome of LASV. Phylogenetic analysis of the first LASV strain imported into Sichuan Province revealed that it belongs to the lineage IV strain, with the highest homology to strains from Sierra Leone. This is consistent with epidemiological data showing that lineage IV strains are predominantly found in West African countries like Sierra Leone and Guinea[10], and the fact that this case was imported from Guinea. LASV contains two single-stranded RNA segments, each encoding two proteins, NP and GPC, and L protein and Z protein, repsectively[37]. These proteins serve various functions[38-41]. Homology analysis of the SCLSR24-01-S and SCLSR24-01-L sequences and their encoded proteins revealed nucleotide sequence differences in the S and L segments ranging from 11.5% to 25.6% and 14.7% to 32%, respectively, compared to seven other strains. These differences align with reported values of 25% and 32% for the S and L segments in the literature[42], indicating significant strain diversity. Among the four proteins, NP and GPC exhibited higher homology compared to the others. Notably, GPC variability ranged from 3.9% to 15.1%, broader than the 4.9% to 11% range observed in a 2020 study by Ibukun FI[30]. This suggests that while NP and GPC are more conserved, GPC diversity continues to increase.This may be attributed to GPC acting as a surface antigen involved in the immune response[43], subjecting it to greater selective pressure. This could have adverse implications for vaccine development.
GPC serves as the primary surface membrane protein and sole surface antigen of LASV, making it the main target for protective humoral immune responses and T-cell immunity[43-46]. After translation, GPC is cleaved by the host cell’s subtilisin protease SKI-1/S1P into three components: receptor-binding glycoprotein 1 (GP1), glycoprotein 2 (GP2, a class I membrane fusion protein), and a myristoylated stable signal peptide (SSP)[47]. Comparative analysis of the GPC amino acid sequence from the SCLSR24-01 strain revealed that the GP1 region exhibits more active mutations compared to the GP2 region, consistent with previous reports[48]. An analysis of various immunogenic epitopes showed that the T-cell epitope SSP(a) in the SSP region, the B-cell epitope GPC-A(b) in the GP1 region, along with the site B epitope and GPC-A(e) of 37.7H, display amino acid differences from most reference strains. These mutations contribute to greater diversity in these regions, which may affect antibody responses.
In particular, the surface conformational region of GP1, spanning amino acids 111–117 in site B, contains a unique mutation at position D114, which differs from all other lineage IV strains. GP1-A monoclonal antibodies (MAb group) neutralize the virus by targeting a conformational epitope on residues 111–117 of the GP1 subunit[44]. Additionally, the E2 and E4 epitopes on the GPC surface include positions I112 and N114, which are key for antibody binding[28]. The mutation from N to D at position 114 in the SCLSR24-01 strain could potentially impact the binding efficacy of the GP1-A MAb group neutralizing antibodies. The D114 mutation is a deamidation variant that introduces a negative charge, while also altering the protein’s three-dimensional conformation, directly leading to changes in antigen–antibody binding affinity[49]. This might explain why SCLSR24-01 binds to GP1-A MAb with an impact; however, further exploration is warranted. Furthermore, within the conformational epitope spanning residues 119–134 of site B, the amino acids R125 and S132 differ from all reference sequences. Mutations at positions 125–126 disrupt the previously conserved region, which may affect the interaction with GP1-B MAb group non-neutralizing antibodies[45].
LASV is classified as a WHO risk group 4 pathogen, requiring biosafety level 4 (BSL-4) containment for handling[50]. The high-risk nature of this pathogen necessitates stringent biosafety measures. These measures include adopting appropriate personal protective equipment and strictly conducting corresponding specimen testing in laboratories of the required level. If a reduction in protective levels is necessary, samples must be inactivated, and the results of the inactivation must be assessed. Rapid diagnosis and monitoring under such conditions are essential for effective pathogen tracing, epidemic response, and risk assessment. In this case, continuous nucleotide detection of close contacts was conducted throughout the isolation period, and no positive cases or environmental samples were identified. This indicates that the case did not lead to secondary cases or viral spread, also signifying that the handling of the case to block the transmission of the virus was successful, effectively preventing an epidemic.
This study provides valuable insights for the detection and research of imported pathogens with high hazard levels, such as LASV. Additionally, the fact that this imported case was not detected at the initial point of entry underscores the importance of employing advanced screening technologies in the post-COVID-19 era. With globalization increasing the movement of people, especially in non-endemic areas for Lassa fever, it is crucial to enhance the detection capabilities of entry and exit inspection agencies and the diagnostic abilities for this disease at medical institutions. Utilizing detection technologies such as RT-qPCR for rapid screening and diagnosis and promptly implementing control measures for cases are essential for the prevention and control of the virus.
HTML
Sample Collection
Sample Processing and Nucleotide Detection
Virus Genome Sequencing and Analysis
Epidemiological History of the Patient
Laboratory Diagnosis
Whole-Genome Assembly
Gene Evolution Analysis
Homology Analysis
Protein Variation Analysis
Competing Interests The authors declare that there are no conflict of interest.
&These authors contributed equally to this work.








 Quick Links
Quick Links
 DownLoad:
DownLoad: