-
The “Treat All” program, implemented in China since 2016, guarantees universal access to antiretroviral therapy (ART) for people living with HIV, regardless of the stage of infection[1]. Expanded ART coverage has significantly reduced HIV transmission and decreased HIV-related morbidity and mortality. However, the scale-up of ART may also increase the risk of drug resistance mutations (DRMs), which can be transmitted to newly infected individuals[2,3]. Pretreatment drug resistance (PDR) has emerged as a significant concern in China[4], as it can limit treatment options for people living with HIV. PDR may be transmitted alongside with the initial infection or may result from previous exposure to antiretroviral drugs, leading to early virological failure[5]. Therefore, evaluating PDR is critical for optimizing ART programs and improving treatment outcomes.
Dehong Prefecture in Yunnan, China, a region that was a pioneer in implementing ART and where the first HIV epidemic in China was identified[6] — has reported a high proportion of drug-resistant strains. A survey conducted from 2017 to 2019 found that the prevalence of PDR in Dehong had reached a moderate level of 7.1%[7]. Dehong, which shares a border with Myanmar, has long been a hotspot for the recombination and dissemination of subtypes. Most of the HIV strains currently circulating in China have originated from these subtypes[8,9]. Understanding the impact of frequent recombination on the spread of DRMs among treatment-naive populations in Dehong is therefore of great importance. With the implementation of China’s opening-up policy, cross-border travel and business have become more frequent[10]. The ongoing cross-border transmission of HIV between Chinese and Burmese individuals has posed significant challenges to the prevention and control of the HIV epidemic in Dehong[11]. Previous studies have shown that onward transmission among ART-naive patients was the primary driver of the increasing prevalence of drug resistance[12,13]. With advances in molecular epidemiology, drug resistance genetic cluster analysis has contributed to a clear understanding of the transmission characteristics of PDR, thereby providing novel insights into the development of preventive and control strategies[4,14]. To date, the role of DRM in the PDR genetic network has not been explored in Dehong. This study aimed to elucidate the prevalence of PDR in Dehong from 2020 to 2023 and to utilize a molecular network to determine the transmission dynamics of PDR within a network.
-
In total, 948 HIV pol gene sequences were successfully amplified from 1,178 plasma samples collected in Dehong from 2020 to 2023. The eligibility criteria were as follows: (1) confirmed HIV-positive status, (2) treatment-naive at the time of sampling, and (3) provision of signed informed consent. Epidemiological information such as nationality, sex, mode of transmission, and CD4+ cell count was obtained from the China Information System for Disease Prevention and Control. All plasma samples were linked to the epidemiological data using anonymous numerical codes.
-
The reference sequence used for alignment was HXB2. All sequences were assembled, edited using LaserGene 7.1, aligned using BioEdit 7.0, and refined manually. Processed sequences were submitted to the China HIV Gene Sequences Database for subtyping (https://nmdc.cn/hiv/tool/sequence)[15]. DRMs and resistance levels were determined using the Stanford HIVDB Program (https://hivdb. stanford. edu/hivdb/bysequences/). Mutations that could lead to low or higher resistance were classified as PDR. PDR was defined based on resistance to one or more of the following drugs: five non-nucleotide reverse transcriptase inhibitors (NNRTIs, doravirine (DOR), efavirenz (EFV), etravirine (ETR), nevirapine (NVP), and rilpivirine (RPV)), seven nucleotide reverse transcriptase inhibitors (NRTIs, abacavir (ABC), zidovudine (AZT), stavudine (D4T), didanosine (DDI), emtricitabine (FTC), lamivudine (3TC), and tenofovir (TDF)) and eight proteinase inhibitors (PIs, atazanavir/r (ATV/r), darunavir/r (DRV/r), fosamprenavir (FPV/r), indinavir/r (IDV/r), lopinavir/r (LPV/r), nelfinavir (NFV), saquinavir/r (SQV/r), and tipranavir (TPV/r)). Based on the genotypic susceptibility score, detected PDR was categorized as high-level resistance (score ≥60), medium resistance (score 30–59), or low-level resistance (score 15–29)[16].
-
The molecular transmission network was inferred based on pairwise genetic distances using HIV-TRACE (Transmission Cluster Engine)[17]. The Tamura-Nei 93 nucleotide substitution model (TN93) was used to calculate the pairwise distances between all pairs of HIV pol sequences. Ambiguous nucleotides in all sequences were < 5%. Network data were visualized using the HIV-TRACE website (https://veg.github.io/hivtrace-viz/#). Clusters are defined as connected components of a network comprising two or more nodes. To identify all possible associations between individuals, nodes were linked to each other if their pairwise genetic distance was up to 1.5% substitutions per site, based on the genetic distance threshold recommended by the Chinese Center for Disease Control and Prevention[14].
-
All statistical analyses were performed using R version 4.3.2. Categorical variables were compared using the chi-square (χ2) test. Factors associated with PDR were evaluated using logistic regression analysis. Univariate logistic regression analysis was performed, and variables with a P-value < 0.10 were subsequently chosen for multivariate logistic regression. The unadjusted odds ratio (OR), adjusted OR (aOR), and 95% confidence interval (CI) were reported. A two-tailed P-value < 0.05 was considered statistically significant.
-
The median age of the 948 individuals with pol sequences was 35 years (interquartile range, 27–47 years). Of these, 543 (57.3%) were from China and 405 (42.7%) were from Myanmar. Additionally, 597 (63.0%) were men, 412 (43.5%) were married, 344 (36.3%) were of Han ethnicity, and 807 (76.2%) had a middle school education or below. Heterosexual contact accounted for 82.4% (781/948) of transmissions. Among the 935 participants with CD4+ cell count data, 481 (51.4%) presented with counts 200–499 cells/μL.
-
Among the 948 pol sequences, 36 HIV subtypes were detected, including subtypes B and C, 33 circulating recombinant forms (CRFs), and unique recombinant forms (URFs). URFs strains were the predominant subtypes, accounting for 18.8% (178/948), followed by CRF01_AE (18.5%, 175/948), CRF07_BC (10.9%, 103/948), subtype B (10.1%, 96/948), subtype C (9.5%, 90/948), CRF08_BC (8.6%, 82/948), and other subtypes (23.6%, 224/948). The difference in the prevalence of PDR between the recombinant and non-recombinant strains was not statistically significant (χ2 = 2.878, P = 0.090).
-
The overall prevalence of PDR was 12.4% (118/948), with annual rates from 2020 to 2023 of 11.9% (38/320), 15.2% (25/164), 13.6% (27/199) and 10.6% (28/265), respectively. NNRTIs, NRTIs, and PIs resistance mutations were detected in 10.7% (n = 101), 1.3% (n = 12), and 1.6% (n = 15) of patients, respectively (Figure 1). High levels of NNRTI-related resistance to EFV (n = 46) and NVP (n = 58) were observed. The most common NNRTI mutations were K103N (n = 31) and E138A (n = 20). Among the NRTI-related drug resistance cases, high-level resistance was detected in 58.3% (7/12) of patients, with PDR to FTC and 3TC presenting the highest levels, which was related to the M184I/V/MV mutation. Even in the PR region, NFV (n = 12) was the most frequent PI-related drug and M46L/MI/I was the most common mutation associated with PI. Moreover, 92.4% (109/118) of the patients with pretreatment drug resistance mutations (PDRM) displayed single-class resistance. Only eight strains harbored dual-class resistance (PI + NNRTI, n = 1; NRTI + NNRTI, n = 7), and one HIV strain with a PDRM harbored resistance to three classes of drugs.
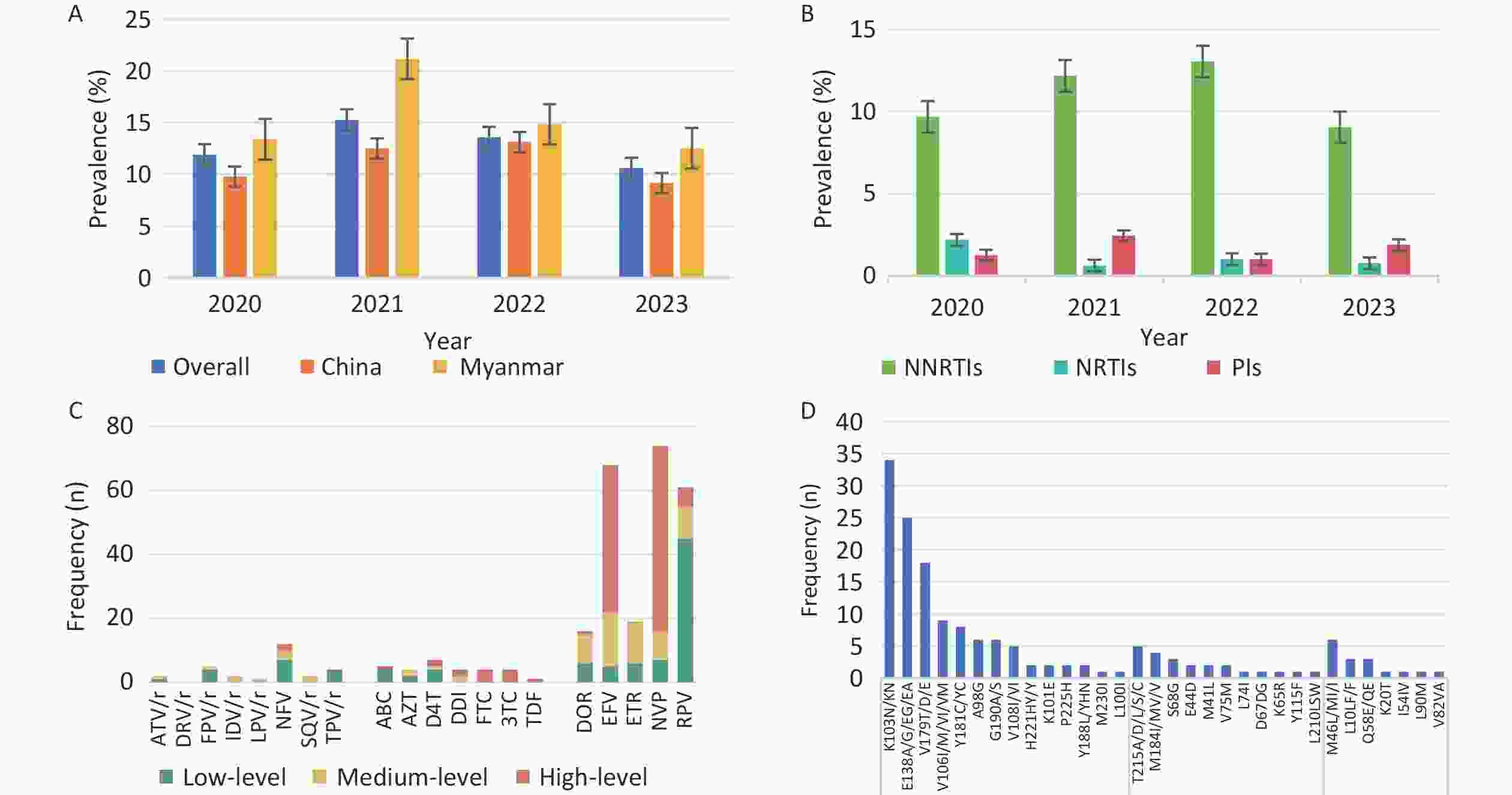
Figure 1. Prevalence and frequency of HIV PDR and DRM in Dehong, 2020–2023. (A) Prevalence trend of PDR from 2020 to 2023 by nationality. (B) Prevalence trend of PDR from 2020 to 2023 by three classes drugs. (C) Frequency and resistance level of PDR to 20 antiretroviral drugs. (D) Frequency of HIV PDRM according to gene sites. PDR, pretreatment drug resistance; PDRM, pretreatment drug resistance mutation; DRM, drug resistance mutation; NNRTIs, non-nucleotide reverse transcriptase inhibitors; DOR, doravirine; EFV, efavirenz; ETR, etravirine; NVP, nevirapine; RPV, rilpivirine; NRTIs, nucleotide reverse transcriptase inhibitors; ABC, abacavir; AZT, zidovudine; D4T, stavudine; DDI, didanosine; FTC, emtricitabine; 3TC, lamivudine; TDF, tenofovir; PIs, proteinase inhibitors; ATV/r, atazanavir/r; DRV/r, darunavir/r; FPV/r, fosamprenavir; IDV/r, indinavir/r; LPV/r, lopinavir/r; NFV, nelfinavir; SQV/r, saquinavir/r; TPV/r, tipranavir.
Univariate logistic regression analysis showed that sex, ethnicity, mode of transmission, and CD4+ cell count were independent factors associated with PDR. Multivariate logistic regression analysis did not reveal any significant factors (Table 1). The prevalence of PDR among Burmese individuals was higher than that among Chinese individuals, although there was no statistically significant difference in the prevalence of PDR between Chinese and Burmese individuals (χ2 = 2.278, P = 0.131).
Characteristics All PDR [n (%)] Univariate analysis Multivariate analysis OR (95% CI) P-value aOR (95% CI) P-value Nationality China 543 60 (11.0) Ref. Myanmar 405 58 (14.3) 1.346 (0.913–1.981) 0.132 Sex Male 597 65 (10.9) 0.687 (0.466–1.017) 0.059 0.767 (0.500–1.176) 0.223 Female 351 53 (15.1) Ref. Ref. Age at diagnosis (years) < 35 459 59 (12.9) Ref. ≥ 35 489 59 (12.1) 0.930 (0.632–1.369) 0.713 Marital status Married 412 44 (10.7) Ref. Unmarried 321 46 (14.3) 1.399 (0.899–2.180) 0.136 Divorced or widowed 215 28 (13.0) 1.252 (0.749–2.065) 0.383 Ethnicity Han 344 37 (10.8) Ref. Ref. Dai 216 24 (11.1) 1.037 (0.595–1.777) 0.896 1.001 (0.570–1.731) 0.997 Jingpo 269 46 (17.1) 1.712 (1.076–2.739) 0.024 1.537 (0.945–2.511) 0.084 Other 119 11 (9.2) 0.845 (0.399–1.664) 0.641 0.822 (0.385–1.631) 0.592 Education level Middle school or below 807 104 (12.9) Ref. High school or above 141 14 (9.9) 0.745 (0.397–1.302) 0.328 Mode of transmission Heterosexual 781 101 (12.9) Ref. Ref. IDUs 71 9 (12.7) 0.977 (0.442–1.933) 0.951 0.984 (0.425–2.068) 0.967 MSM 82 5 (6.1) 0.437 (0.151–1.005) 0.081 0.561 (0.189–1.341) 0.238 Other 14 3 (21.4) 1.836 (0.410–5.999) 0.357 1.915 (0.419–6.461) 0.335 Subtype B 96 14 (14.6) 1.428 (0.616–3.387) 0.408 C 90 16 (17.8) 1.808 (0.798–4.233) 0.160 URFs 178 19 (10.7) 0.999 (0.462–2.259) 0.999 CRF01_AE 175 22 (12.6) 1.203 (0.568–2.681) 0.638 CRF07_BC 103 11 (10.7) Ref. CRF08_BC 82 12 (14.6) 1.434 (0.595–3.489) 0.420 Other 224 24 (10.7) 1.004 (0.482–2.212) 0.992 CD4+ cell count (cells/μL) < 200 243 38 (15.6) Ref. Ref. 200– 481 50 (10.4) 0.626 (0.398–0.989) 0.043 0.655 (0.415–1.041) 0.070 ≥ 500 211 29 (13.7) 0.860 (0.506–1.446) 0.571 0.879 (0.514–1.490) 0.634 Unknown 13 1 (7.7) 0.450 (0.024–2.385) 0.449 0.470 (0.025–2.532) 0.476 Note. MSM, men who have sex with men; IDUs, intravenous drug users; Unknown, data are not available; OR, odds ratio; aOR, adjusted odds ratio; CI, confidence interval; Ref., reference; PDR, pretreatment drug resistance; ART, antiretroviral therapy; URFs, unique recombinant forms. Table 1. Factors associated with PDR among newly diagnosed and ART-naive individuals in Dehong
-
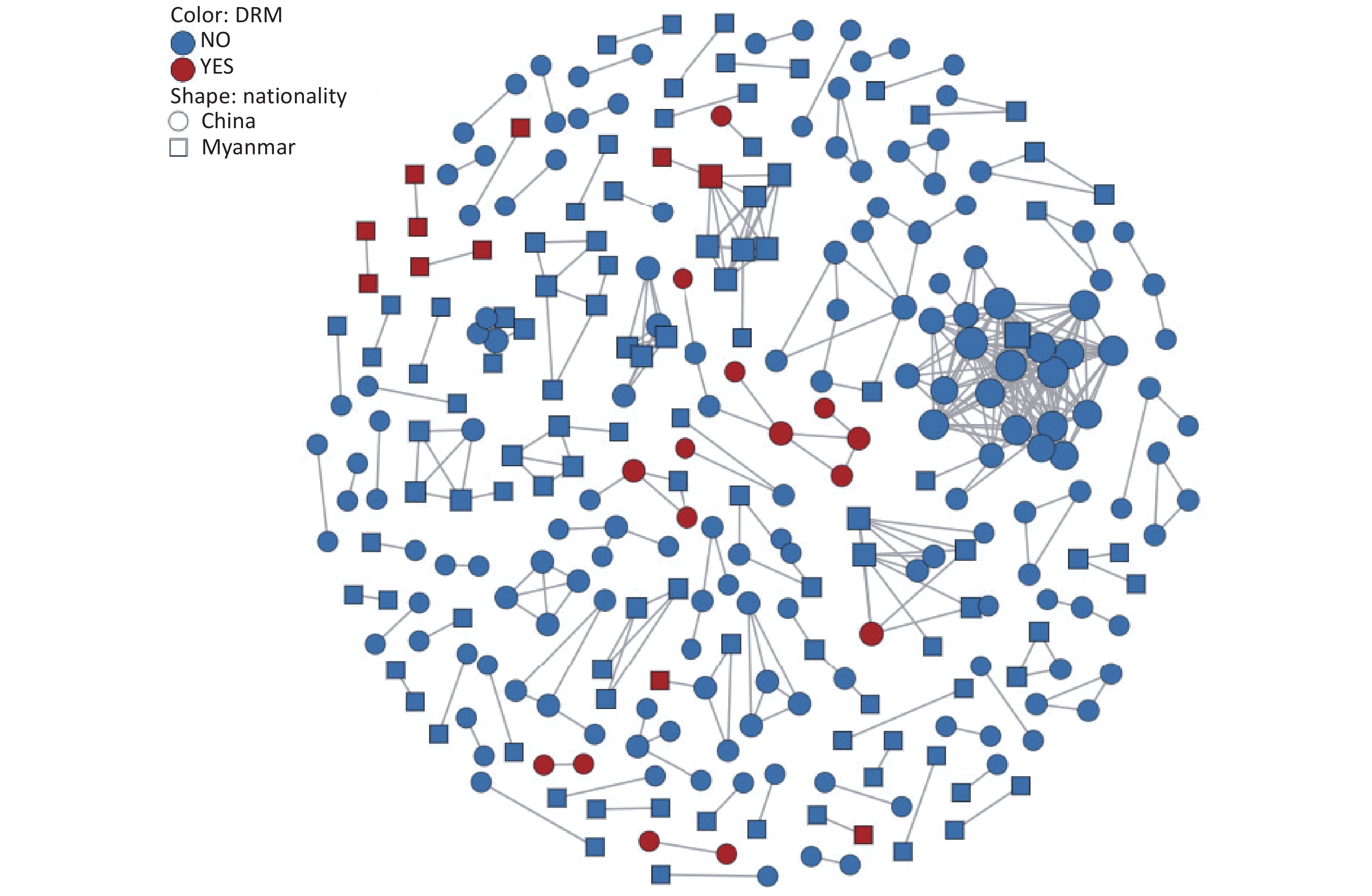
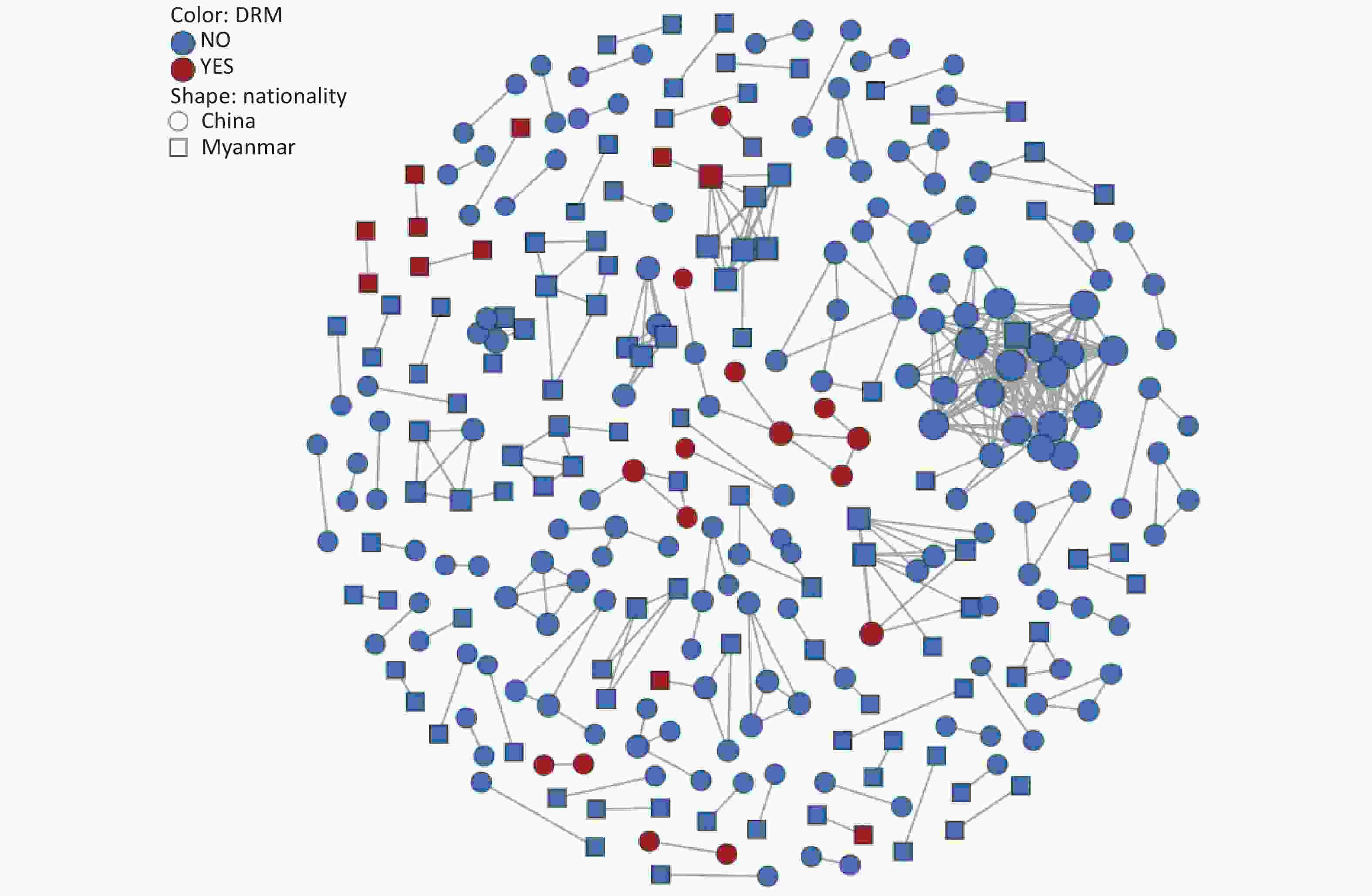
To explore transmission dynamics, a molecular network was constructed. A total of 91 transmission clusters were identified under the genetic distance threshold of 0.015 comprising 287 of the 948 participants (30.3%) (Figure 2). Individual PDR status was significantly associated with cluster membership (22.0% [26/118] vs. 31.4% [261/830]; χ2 = 4.335, P = 0.037). We identified 26 individuals with PDR from 14 clusters, including 15 Chinese and 11 Burmese individuals. Among the clustered individuals with PDR, 73.1% (19/26) were genetically linked to each other in the eight PDR clusters, suggesting PDR transmission among ART-naive patients. Most PDR strains linked to these clusters were resistant to NNRTIs (80.8%, 21/26). The most prevalent shared PDRM was K103N (57.9%, 11/19), followed by V179T (36.8%, 7/19), and E138A (21.1%, 4/19). The largest PDR-related cluster, CRF01_AE consisted of eight individuals. Among these, six individuals developed drug resistance with PDRM propagation of K103N and V179T. Additionally, five of the six participants had initial CD4+ cell counts of less than 200 cells/μL.
-
This study analyzed the prevalence and dissemination of PDR in Dehong from 2020 to 2023. It not only revealed a high prevalence of PDR, but also identified PDR clusters via molecular network analysis, thereby providing evidence-based insights for the accurate prevention and control of HIV.
The current study identified a variety of subtypes in Dehong, featuring a high proportion of CRFs and URFs, which are more complex than those observed in most regions of China[16,18]. Previous research has indicated that frequent recombination facilitates HIV evasion of the immune system and development of drug-resistance[19-21]. Contrary to expectations, this study did not demonstrate a statistically significant increase in the prevalence of PDR among the recombinant subtypes compared to the non-recombinant strains. This discrepancy may be due to the fact that previous study focused on treated patients[21]. Recombination permits the viral genome to integrate preexisting beneficial mutations, potentially boosting viral survival in the presence of antiretroviral drugs. The participants in our study were treatment-naive without drug selection pressure. This may explain the lack of a significant association between PDR and the recombinant subtype.
Another significant finding was that the overall prevalence of PDR in Dehong was 12.4%, a moderate level (5%–15%) that surpassed previous local data from 2017 to 2019[7] and the average PDR prevalence in Yunnan[4]. Annually, the resistance rate exceeded World Health Organization’s (WHO) 10% warning threshold. This may be attributed to the prolonged HIV epidemic, potentially causing an accumulation of resistant mutations, or extensive use of ART in the region for over two decades. Additionally, PDR was first identified in an IDU population in Dehong, where poor adherence to medications and a multitude of high-risk behaviors were identified as factors contributing to drug resistance[6]. Importantly, this study revealed that the prevalence of PDR among Burmese individuals was higher than among Chinese individuals, although the difference was not statistically significant. The prevalence of drug resistance has been reported to be 12.8% among therapy-naive Burmese travelers entering Dehong port[22]. Frequent China-Myanmar cross-border transmission may accelerate the spread of drug-resistant strains in different regions. It is imperative to monitor the transmission of drug-resistant strains among Burmese nationals in Dehong and to explore coordinated transnational intervention approaches with the Burmese government.
Notably, HIV strains resistant to NNRTIs (10.7%) were more prevalent than those resistant to NRTIs and PIs, which is consistent with the results in most regions of China[4]. This may be related to mutations associated with reduced susceptibility to NNRTIs that emerged rapidly in the early selection process with a low genetic barrier and their extensive application as first-line regimens in China[23,24]. Among these mutations, K103N and E138A remain the most frequent, resulting in a high level of resistance to EFV and NVP. Research conducted in Argentina and Luanda revealed that K103N is the most frequent DRM among the NNRTIs employed as first-line antiviral drugs[25,26]. Additionally, this study found that the prevalence of PI-related PDR (1.6%) was higher than that of NRTI (1.3%), consistent with a study in Guangxi[16]. Moreover, the PDR rates for PI in Dehong were higher than the WHO’s estimated regional PDR rates in Southeast Asia (PI PDR: 0.4%), Africa (PI PDR: 0.3%), and the Americas (PI PDR: 0.8%)[24]. Further research is required to identify the underlying causes of high PI-related PDR rates in Dehong. These results highlight the need for resistance testing before ART initiation.
The primary contribution of this study was the identification and characterization of PDR clusters. The investigation revealed that a lower proportion of drug-resistant strains entered the network than non-resistant strains. One possible explanation is that the transmission potential of most drug-resistant strains is weaker than that of the non-resistant wild-type strains[27]. The largest PDR-related cluster contained eight CRF01_AE strains, six of which exhibited drug resistance. The CRF01_AE cluster with PDR carried K103N and V179T, indicating the transmissibility of CRF01_AE strains harboring these mutations. To our knowledge, this is the first report of clustered PDR transmission among patients newly diagnosed with HIV infection in Dehong. Of particular concern was the observation that five individuals with PDR in this cluster had low CD4+ cell counts less than 200 cells/μL. Real-time monitoring and public health responses should be conducted in this cluster to prevent further expansion.
This study has some limitations. First, it focused solely on drug-resistance testing among newly diagnosed individuals with eligible sequences, which may have overestimated or underestimated the prevalence of PDR. Nonetheless, given the relatively high coverage of HIV testing in local region[28], the current level of drug resistance can still provide some insights into public health policies. Second, the links between individuals in clusters do not reflect the true transmission relationships. The actual causes of PDR require further elucidation through epidemiological investigations. Nonetheless, the common characteristics exhibited by individuals within the clusters could inform future epidemiological studies.
In conclusion, this study reveals the diverse and complex distribution of HIV subtypes in Dehong. PDR is moderately prevalent (12.4%) from 2020 to 2023, and there is evidence of the transmission of CRF01_AE strains carrying the K103N and V179T mutations. Routine surveillance of PDR and strengthening control measures to prevent its development are essential to guide first-line ART regimens in Dehong.
HIV Pretreatment Drug Resistance and Transmission Clusters among Newly Diagnosed Patients in the China-Myanmar Border Region, 2020–2023
doi: 10.3967/bes2025.080
- Received Date: 2024-09-15
- Accepted Date: 2025-04-07
-
Key words:
- HIV /
- Drug resistance /
- Subtype /
- Cluster analysis
Abstract:
The authors declare no conflict of interest related to this study.
This study was approved by the Institutional Review Board of the National Center for AIDS/STD Control and Prevention, Chinese Center for Disease Control and Prevention (No. X231018772).
&These authors contributed equally to this work.
| Citation: | Huan Liu, Yuecheng Yang, Xing Duan, Yichen Jin, Yanfen Cao, Yi Feng, Chang Cai, Hehe Zhao, Houlin Tang. HIV Pretreatment Drug Resistance and Transmission Clusters among Newly Diagnosed Patients in the China-Myanmar Border Region, 2020–2023[J]. Biomedical and Environmental Sciences, 2025, 38(7): 840-847. doi: 10.3967/bes2025.080 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: