HTML
-
Platelet inhibition shows wide inter-individual variation in patients with acute coronary syndrome (ACS) undergoing percutaneous coronary intervention (PCI), despite the administration of dual antiplatelet therapy (DAPT) with aspirin and clopidogrel. High residual platelet reactivity (HRPR) is associated with increased short-and long-term risk of cardiovascular ischemic events, such as myocardial infarction and in stent thrombosis[1-5]. The individual variability of antiplatelet response to clopidogrel may be related to environmental factors, such as patients' noncompliance and drug interactions[6]. However, genetic polymorphism plays crucial roles. Cytochrome P450 2C19 (CYP2C19) polymorphism is linked to clopidogrel responsiveness and ischemic events in ACS patients or PCI-treated subjects[7-9]. With the introduction of more potent purinergic receptor P2Y12 antagonists, such as prasugrel or ticagrelor, the individual antiplatelet variability has been reduced[10-11], but substantial variability still exists[12], suggesting that other genetic polymorphisms are the underlying cause of the response variability of this class of antiplatelet drugs. Circulating catecholamine plays an important role in platelet function through the activation of α2A-AR on platelet membrane surface[13-14]. The α2A-AR is encoded by ADRA2A on chromosome 10. In healthy volunteers, a genetic variant (6.3-kb variant) of ADRA2A is responsible for the increased platelet reactivity with catecholamine[15]. Peace AJ et al.[16]demonstrated that α2A-AR promotes P2Y12 receptor functionality and contributes to HRPR in stable coronary artery disease in patients taking DAPT with aspirin and clopidogrel[17]. However, Cuisset T et al.[18] reported that the 6.3-kb variant of ADRA2A did not show any major impact on residual platelet reactivity in ACS patients. Therefore, the interaction between the genetic variants of ADRA2A and platelet reactivity with DAPT is still unclear. This study aims to examine the potential association of ADRA2A genetic variants with residual platelet reactivity in Chinese patients on DAPT undergoing PCI.
-
Study Population Patients presenting to the Fuwai Hospital of Chinese Academy of Medical Sciences and Peking Union Medical College between March 1, 2011, and March 30, 2013, were consecutively enrolled in this prospective observational study. Inclusion criteria for enrollment were patients with coronary heart disease (CHD) who have undergone PCI. The exclusion criteria were: age < 18 years; hemodynamic instability; active bleeding and bleeding diatheses; use of glycoprotein IIb/IIIa inhibitor; oral anticoagulation therapy; usage of intensified antiplatelet agents other than the standard DAPT with aspirin and clopidogrel; contraindication to antiplatelet therapy; non-cardiac disease with a life expectancy of < 1 year; disagreement with platelet function test and ADRA2A genotyping; or inability to follow the protocol. The Institutional Review Board approved the study protocols, and the patients provided written informed consent for participation.
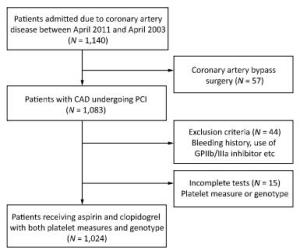
Study Design If patients was not taking long-term aspirin or clopidogrel, they received a 300-mg loading dose of clopidogrel and 300-mg loading dose of aspirin at least 12 h before undergoing PCI, followed by 75 mg/day clopidogrel for 1 year and 100 mg/day maintenance dose of aspirin for life. The decision for PCI was based on the coronary angiography results and all interventions were conducted according to the 2010 European Society of Cardiology/ European Association for Cardio-Thoracic Surgery (ESC/EACTS) guidelines on myocardial revascularization[19], and 2011 American College of Cardiology/American Heart Association Task Force on Clinical Practice/ Society for Cardiovascular Angiography and Interventions (ACCF/AHA/SCAI) guidelines for PCI[20]. The types of stents used were chosen by the operators. Anticoagulation with low molecular weight heparin (enoxaparin) or unfractionated heparin was initiated before angiography in all patients. The flow diagram for the trial is shown in Figure 1.
Genotyping Four single nucleotide polymorphisms (SNPs) ADRA2A were selected based on the analysis of HapMap and Chinese Han Beijing Databank using Haploview 4.2 software. HapMap selection criteria included minor allele frequency to be greater than 0.05. Blood samples were obtained from a peripheral vein from each patient and stored in 4 mL evacuated collection tubes containing ethylenediaminetetraacetic acid (EDTA). Genomic DNA was extracted from white blood cells according to the standard salting-out method and was stored in 200 μL of TE (10 mmol/L Tris/HCl and 1.0 mmol/L EDTA, pH 8.0). The selected ADRA2A SNPs were rs11195419 (C > A), rs13306146 (A > G), rs553668 (A > G), rs3750625 (C > A), and CYP2C19*2 rs4244285 (G > A) were genotyped using the ligase detection reaction (LDR) and a commercially available detection system (ABI 3730XL DNA Analyzer System; Applied Biosystems, Foster City, CA, USA). Repeat genotyping was performed on random duplicate samples (n = 45), and Sanger sequencing was used for quality control.
Thromboelastograph Platelet Mapping Assay Blood was collected in evacuated collection tubes, containing 3.2% trisodium citrate and the inner walls coated with lithium heparin, 12-24 h after undergoing PCI. A predetermined volume of blood was drawn, and the tube was inverted 3-5 times to ensure complete mixing with the anticoagulant. Modified thromboelastography (TEG®) uses four channels to detect the effects of antiplatelet therapy via the arachidonic acid (AA) and adenosine diphosphate (ADP) pathways. A detailed description of this method has been outlined previously[21]. The TEG Hemostasis Analyzer (Haemonetics Corp, Braintree, MA, USA) and automated analytical software were used to measure the physical properties.
The average percentage of platelet inhibition by clopidogrel was computed as the contribution of ADP-stimulated platelets to the maximal clot strength (ADP inhibition) by the TEG software: 100 -100 × [(MAAA or ADP-MAfibrin)/ (MAthrombin-MAfibrin)], where MAAA is the AA-induced clot strength (measurement of the aspirin effect), MAADP is the ADP-induced clot strength (measurement of the ADP effect), MAfibrin is the activator-induced clot strength (measurement of the fibrin contribution), and MAthrombin is the thrombin-induced clot strength (maximum clot strength).
Statistical Analysis Continuous variables were expressed as mean ± standard deviation (SD) and compared using Student's t-test or one-way analysis of variance (ANOVA) was appropriate. After demonstrating significant differences among variables by the ANOVA test, post hoc comparisons between the groups were performed with the Student-Newman-Keuls test for multiple comparisons. Categorical variables were expressed as frequencies and percentages and were compared with a chi-square test (χ2) or Fisher's exact test. All SNPs evaluated in our study were tested for deviation from the Hardy-Weinberg equilibrium using the chi-square test. Any deviation between observed and expected frequencies was tested for significance using the χ2 test. Linear regression analysis was performed to assess the association between SNPs and ADP-induced platelet aggregation. Bonferroni correction was used to correct the multiple comparisons problem. In the multivariate model, we adjusted for potential confounders including CYP2C19*2, age, sex, BMI, platelets count, diabetes mellitus, hypertension, hypercholesterolemia, current smoking, ACS, angiotensin converting enzyme inhibitors/ angiotensin Ⅱ receptor blockers (ACEI/ARB), calcium channel blockers (CCB), statins, β-blocker, and PPIs use. Subgroup analyses were performed according to sex. Additive model was assumed to test the association between SNPs and ADP inhibition. Statistical analyses was mainly performed with SPSS version 22.0 (SPSS Inc., Chicago, IL, USA), and a two-tailed P value less than 0.05 was considered statistically significant.
-
From March 2011 to March 2013, 1, 024 patients who underwent PCI and received post intervention aspirin and clopidogrel, were enrolled in the study. All these patients also completed platelet function measures and genetic sampling tests. The minor allele frequencies of the ADRA2A SNPs (rs11195419, rs3750625, rs553668, and rs13306146) and CYP2C19*2 (rs4244285) were 18.1%, 17.2%, 43.7%, 25.4%, and 32.1%, respectively. The homozygous groups of the SNPs were 2.7%, 2.7%, 19.5%, 8.0%, and 10.4%, respectively. Genotypic distributions of these SNPs conformed with the Hardy-Weinberg equilibrium. The allele and genotype frequencies are reported in Table 1. The average age of the patients was 58.5 ± 10.5 years, and > 75% were males. Less than half of the patients presented with ACS (31.8%). PCIs were mostly performed using drug-eluting stents (99.2%). History of diabetes was significantly different across rs11195419 (P = 0.023) and rs3750625 (P = 0.023) genotypes. Platelets count (P = 0.019), current smoking history (P = 0.048), and previous myocardial infarction history (P = 0.049) were also significantly different across the rs553668 genotypes. The baseline characteristics of SNPs (rs11195419 and rs3750625) are presented in Table 2 and the SNPs (rs553668 and rs13306146) are reported in the Appendix (available in www. besjournal.com).
Characteristics RS11195419 N (%) RS3750625 N (%) RS553668 N (%) RS13306146 N (%) Allele frequency, n = 2, 048 Major allele 1, 677 (81.9) 1, 695 (82.8) 1, 160 (56.6) 1, 528 (74.6) Minor allele 371 (18.1) 353 (17.2) 888 (43.4) 520 (25.4) Genotype frequency, n = 1, 024 Wild type 682 (66.6) 700 (68.4) 334 (32.6) 587 (57.3) Heterozygote 313 (30.6) 295 (28.8) 492 (48.0) 354 (34.6) Homozygote 29 (2.8) 29 (2.8) 198 (19.3) 83 (8.1) Table 1. Allele and Genotype Frequency of ADRA2A Genetic Variants
Variables RS11195419 RS3750625 WT
(N = 682)HE
(N = 313)HO
(N = 29)P WT
(N = 700)HE
(N = 295)HO
(N = 29)P Age, years 58.2 ± 10.6 59.2 ± 11.6 59.0 ± 7.6 0.692 58.2 ± 10.6 59.2 ± 10.6 59.8 ± 8.2 0.273 Male, n (%) 525 (77.0) 227 (72.5) 21 (72.4) 0.293 537 (76.7) 216 (73.2) 20 (69.0) 0.358 BMI, kg/m2 26.0 ± 3.2 26.2 ± 3.2 25.8 ± 3.7 0.739 26.0 ± 3.2 26.2 ± 3.3 26.0 ± 3.7 0.531 Platelet, × 109/L 206.4 ± 53.3 204.3 ± 56.7 183.8 ± 55.4 0.089 207.0 ± 53.5 202.2 ± 56.0 189.2 ± 61.1 0.126 ACS, n (%) 228 (33.4) 92 (29.4) 6 (20.7) 0.190 232 (32.1) 88 (29.8) 6 (20.7) 0.252 DM, n (%) 186 (27.3) 110 (35.1) 12 (41.4) 0.017 192 (27.4) 104 (35.3) 12 (41.4) 0.020 Hypertension, n (%) 425 (62.3) 192 (61.3) 16 (55.2) 0.725 436 (62.3) 180 (61.0) 17 (58.6) 0.873 HCL, n (%) 573 (84.0) 270 (86.3) 22 (75.9) 0.285 589 (84.1) 254 (86.1) 22 (75.9) 0.318 Current smoking, n (%) 261 (38.3) 114 (36.4) 8 (27.6) 0.463 267 (38.1) 109 (36.9) 7 (24.1) 0.306 CHD family history, n (%) 7 (1.0) 4 (1.3) 1 (3.4) 0.484 7 (1.0) 4 (1.4) 1 (3.4) 0.458 Previous MI, n (%) 124 (18.2) 45 (14.4) 5 (17.2) 0.332 127 (18.1) 41 (13.9) 6 (20.7) 0.230 Previous PCI, n (%) 103 (15.1) 39 (12.5) 5 (17.2) 0.491 103 (14.7) 39 (13.2) 5 (17.2) 0.749 Previous CABG, n (%) 3 (0.4) 2 (0.6) 0 (0) 0.852 3 (0.4) 2 (0.7) 0 (0) 0.814 LAD, n (%) 379 (55.6) 192 (61.3) 20 (69.0) 0.107 386 (55.1) 185 (62.7) 20 (69.0) 0.040 LCX, n (%) 197 (28.9) 90 (28.8) 7 (24.1) 0.858 204 (29.1) 82 (27.8) 8 (27.6) 0.904 RCA, n (%) 314 (46.0) 141 (45.0) 11 (37.9) 0.678 322 (46.0) 133 (45.1) 11 (37.9) 0.684 LM, n (%) 31 (4.5) 19 (6.1) 0 (0) 0.272 32 (4.6) 18 (6.1) 0 (0) 0.275 Statin, n (%) 663 (97.2) 302 (96.5) 29 (100) 0.522 681 (97.3) 284 (96.3) 29 (100.0) 0.438 β-blocker, n (%) 604 (88.6) 278 (88.8) 27 (93.1) 0.750 621 (88.7) 261 (88.5) 27 (93.1) 0.750 ACEI/ARB, n (%) 408 (59.8) 183 (58.5) 13 (44.8) 0.268 418 (59.7) 173 (58.6) 13 (44.8) 0.277 CCB, n (%) 256 (37.5) 129 (41.2) 8 (27.6) 0.260 264 (37.7) 121 (41.0) 8 (27.6) 0.297 PPIs, n (%) 115 (16.9) 43 (13.7) 4 (13.8) 0.435 120 (17.1) 38 (12.9) 4 (13.8) 0.232 DES, n (%) 676 (99.1) 311 (99.4) 29 (100) 0.821 694 (99.1) 293 (99.3) 29 (100.0) 0.852 BMS, n (%) 5 (0.7) 1 (0.3) 0 (0) 0.668 5 (0.7) 1 (0.3) 0 (0) 0.713 Ballooning only, n (%) 1 (0.1) 1 (0.3) 0 (0) 0.824 1 (0.1) 1 (0.3) 0 (0) 0.791 Note. WT: Wild Type, HE: Heterozygote, HO: Homozygote, ADRA2A: the α2A-adrenergic receptor gene, BMI: body mass index, ACS: acute coronary syndrome, DM: diabetes mellitus, CHD: coronary heart disease, HCL: Hypercholesterolemia, MI: myocardial infarction, PCI: percutaneous coronary intervention, CABG: coronary artery bypass grafting, LAD:left anterior descendens, LCX: left circumflex, RCA: right coronary artery, ACEI: angiotensin conversion enzyme inhibitor, ARB: angiotensin receptor blocker, CCB: calcium channel blocker, PPI: proton pump inhibitor, DES: Drug-eluting stent, BMS: Bare mental stent. Table 2. Baseline Characteristics by ADRA2A Genotypes (rs11195419 and rs3750625)
-
The median level of ADP inhibition was 52.7% ± 29.0%. ADP inhibition was significantly different among genotypes with rs11195419 (adjusted P = 0.022) and rs3750625 (adjusted P = 0.016) in additive model and the homozygous groups of these SNPs had the lowest ADP inhibition (39.3% ± 19.5% and 37.5% ± 18.6%, respectively). However, there we did not observe differences in ADP inhibition among genotypes with rs13306146 and rs553668. We did not identify any association with AA inhibition and ADRA2A SNPs either (Table 3). We also found the ADP inhibition was significantly different according to the CYP2C*2 genotype (P = 0.005). (The allele, genotypes and ADP inhibition of CYP2C19*2 are presented in Table 4). We conducted a multivariate analysis including CYP2C19*2 genotype and other potential confounders to identify whether the two significantly associated SNPs of ADRA2A by univariate analysis were independently related to ADP inhibition. The multivariate analysis indicated that rs11195419 (95% CI: -21.3 to -1.0; P = 0.033) and rs3750625 (95% CI: -22.3 to -2.0; P = 0.020) were independent predictors of ADP inhibition (Table 5). A significant association of ADP inhibition was also observed with CYP2C19*2, male, age and hypertension.
SNPs ADP Inhibition, % Additive Model P Value Additive Model Adjusted P Value AA Inhibition, % Additive Model P Value Additive Model Adjusted P Value Wild type Heterozygote Homozygote Wild type Heterozygote Homozygote RS11195419 54.2 ± 29.7 50.7 ± 27.7 39.3 ± 19.5 0.011 0.022 88.5 ± 20.0 89.3 ± 18.9 85.3 ± 20.7 0.351 1.404 RS3750625 54.0 ± 29.8 51.1 ± 27.4 37.5 ± 18.6 0.004 0.016 88.8 ± 19.3 88.8 ± 19.8 85.4 ± 20.7 0.356 0.712 RS13306146 51.8 ± 28.1 54.3 ± 30.3 52.1 ± 29.4 0.857 0.857 87.6 ± 21.0 90.0 ± 17.7 90.3 ± 18.1 0.428 0.571 RS553668 52.3 ± 30.1 54.4 ± 29.3 50.5 ± 27.7 0.812 1.083 87.6 ± 19.9 89.3 ± 20.0 88.9 ± 19.0 0.810 0.810 Note. SNP: single nucleotide polymorphism; ADP: adenosine diphosphate, AA: arachidonic acid. Table 3. Association of ADP Inhibition with ADRA2A Genotypes
Allele Frequency
(N = 2, 048)N (%) Genotype Frequency (N = 1, 024) N (%) ADP Inhibition , % P Value Major allele 1, 391 (67.9) Wild type 474 (46.3) Wild type 55.2 ± 28.7 Minor allele 657 (32.1) Heterozygote 443 (43.3) Heterozygote 51.7 ± 28.9 0.005 Homozygote 107 (10.4) Homozygote 45.5 ± 29.2 Note. ADP: adenosine diphosphate. Table 4. Allele, Geneotype Frequency and Association of ADP Inhibition with CYP2C19*2
Variables RS11195419 RS3750625 Univariate Analysis Multivariate Analysis Univariate Analysis Multivariate Analysis Coef (95% CI) P Coef (95% CI) P Coef (95% CI) P Coef (95% CI) P SNPs -2.5 (-24.5 to -3.1) 0.011 -2.1 (-21.3 to -1.0) 0.033 -2.5 (-24.5 to -3.1) 0.011 -2.3 (-22.3 to -2.0) 0.020 CYP2C19*2 -2.7 (-13.8 to -2.2) 0.007 -3.1 (14.3 to -3.3) 0.002 -2.7 (-13.8 to -2.2) 0.007 -3.1 (-14.3 to -3.3) 0.002 Male 6.8 (10.0 to -18.1) < 0.001 4.2 (5.1 to 13.9) < 0.001 6.8 (10.0 to -18.1) < 0.001 4.2 (5.0 to 13.9) < 0.001 Age (year) -4.4 (-0.5 to -0.2) < 0.001 -2.8 (-0.4 to -0.1) 0.005 -4.4 (-0.5 to -0.2) < 0.001 -2.8 (-0.4 to -0.1) 0.005 BMI (kg/m2) 2.3 (0.1 to 1.2) 0.018 2.0 (0.1 to 1.1) 0.047 2.3 (0.1 to 1.2) 0.018 2.0 (0.1 to 1.1) 0.043 Hypertension -2.8 (-8.8 to -1.5) 0.006 -2.0 (-8.0 to -0.1) 0.047 -2.8 (-8.8 to -1.5) 0.006 -2.0 (-7.9 to -0.2) 0.049 Hypercholesterolemia 2.10 (0.3 to 10.1) 0.036 2.0 (0.1 to 9.5) 0.044 2.10 (0.3 to 10.1) 0.036 2.0 (0.1 to 9.4) 0.045 Acute coronary syndrome 4.5 (5.03 to 12.9) < 0.001 3.5 (3.1 to 10.7) < 0.001 4.5 (5.03 to 12.9) < 0.001 3.5 (3.1 to 10.7) < 0.001 Note. SNP: single nucleotide polymorphism; BMI: body mass index. Table 5. Association of ADRA2A SNPs with ADP Inhibition by Multivariate Linear Regression Analysis
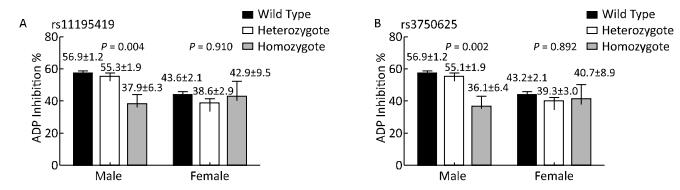
Additional analysis stratified by sex showed that in male patients, ADP inhibition was significantly different among the genotypes carrying rs11195419 (P = 0.003) and rs3750625 (P = 0.002) in additive model, and the homozygous groups had the lowest ADP inhibition. However, this difference was not found in female patients (Figure 2). Multivariate analysis also showed that rs11195419 and rs3750625 were predictors of ADP inhibition (95% CI: -28.6 to -4.2; P = 0.008 and 95% CI: -30.4 to -5.5; P = 0.005, respectively) in male patients, which was not observed in female patients (Table 6).
Characteristics Univariate Analysis Multivariate Analysis Coef (95% CI) P Coef (95% CI) P Male RS11195419 -2.9 (-31.3 to -6.3) 0.003 -2.6 (-28.6 to -4.2) 0.008 RS3750625 -3.2 (-33.3 to -7.8) 0.002 -2.8 (-30.4 to -5.5) 0.005 Female RS11195419 0.9 (-17.9 to 19.7) 0.928 0.1 (-17.5 to 20.2) 0.890 RS3750625 -0.2 (-19.2 to 16.3) 0.872 -0.1 (-19.2 to 16.6) 0.889 Table 6. Association of ADRA2A SNPs and ADP Inhibition by Univariate and Multivariate Linear Regression Analysis Stratificated by Sex
Genotype Distribution and Patient Characteristics
Association with ADP Inhibition and ADRA2A Genotypes
-
This study has reported several important findings. 1) ADRA2A SNPs (rs11195419, rs3750625, rs553668, and rs13306146) commonly exists in Chinese populations. 2) The SNPs of rs11195419 and rs3750625 were significantly associated with ADP-induced platelet aggregation in Chinese patients treated with aspirin and clopidogrel who have undergone PCI. 3) The SNPs of rs11195419 and rs3750625 were independent predictors of ADP-induced platelet aggregation in Chinese patients, especially males.
-
To the best of our knowledge, this is the first study to report ADRA2A polymorphisms in Chinese CHD patients, and is found quite prevalent in the study cohort. Approximately one-third of our CHD patients were either heterozygous or homozygous for SNPs at rs11195419, rs13306146, and rs3750625. The SNP rs553668 was previously detected in approximately one-third of healthy volunteers, patients with ACS and stable CHD[15-16, 18]. In this study involving Chinese population with CHD, the polymorphism of rs553668 was much more common. The frequency of the mutated allele A at rs553668 was 43.7% and more than half of the patients were either heterozygous or homozygous genotype.
-
Epinephrine is an important mediator of platelet aggregation by stimulating α2A-AR on the membrane surface. It is demonstrated that healthy volunteers exhibit higher platelet reactivity in response to epinephrine compared with another agonist such as ADP[22]. There is an increased platelet aggregation amplified by increased levels of circulating catecholamines in the first 3 h after arising and keeping the upright posture, consistent with an increased risk of the cardiovascular event at the same time[23-24]. The α2A-AR is a G protein-coupled receptor on platelet membrane surfaces. The activation of α2A-AR results in a reduction of the cyclic adenosine monophosphate (cAMP) level and release of cytosolic phospholipase-A2 (PLA2) and AA, which are essential to the aggregation of the platelet[25]. Meanwhile, unlike other cell types, the occupation of the G protein -coupled α2A-AR receptors on platelets does not result in phospholipase C activation, but rather in the modulation of the Ca2+ response by relieving cAMP-mediated suppression of InsP3-dependent Ca2+-induced Ca2+ release (CICR)[26]. A previous study[27] reported that in the presence of P2Y12 receptor blockade, activation of α2A-AR contributed to platelet aggregation response. It is also known that similar to the P2Y12 receptor (also a G protein-coupled receptor), α2A-AR activation causes deceleration of the deaggregation component and shifts the balance toward increased aggregation in platelets reactivity, and thus may increase thrombotic risk in certain disease states in healthy volunteers. In healthy volunteers, α2A-AR directly promotes platelet aggregability and a more functional P2Y12 receptor in vitro[27]. α2A-AR also contributes to HRPR in stable coronary artery disease patients taking DAPT with aspirin and clopidogrel[17]. The α2A-AR activity triggers platelet aggregation and may contribute to cardiovascular events despite DAPT[28]. It was also observed that patients with increased platelet aggregation in response to epinephrine had increased platelet aggregation in response to ADP. However, this significant interaction between epinephrine and ADP-induced platelet aggregation was not observed with other platelet activators, such as collagen, AA, and thrombin receptor-activating peptide. Therefore, this significant interaction of the two pathways may explain the effect of α2A-AR on HPPR with DAPT. ADRA2A is a protein-coding gene, 3, 876 bases pair long, which is located at chromosome 10 at 10q24-q26. This gene encodes α2A-AR and contains no introns in either of its coding or untranslated sequences. The markedly heterogeneous platelet aggregation in CHD patients on DAPT induced by epinephrine indicates that different α2A-AR functions might be dictated by ADRA2A genetic polymorphisms. To our knowledge, the current study was the first to find an association of ADRA2A polymorphisms with ADP-induced platelet aggregation in Chinese patients administered aspirin and clopidogrel and undergoing PCI. In univariate analysis, we found that the minor allele A at rs11195419 and rs3750625 was significantly associated with lower ADP inhibition. Peace et al.[16] reported a null effect of rs553668 on ADP-induced platelet aggregation, which was confirmed in our study. We also demonstrated the CYP2C19*2 genotypes and included it in multivariate analysis to find the influence of both CYP2C19 and ADRA2A SNPs on platelet reactivity. Multivariate linear regression model also confirmed the findings. The ADRA2A SNPs (rs11195419 and rs3750625) and CYP2C19*2 were both independent predictors of ADP inhibition. All SNPs in the current study were located at three prime untranslated regions (3′-UTR), suggesting the functional importance of 3′-UTR of ADRA2A in ADP-induced platelet aggregation. However, the underlying mechanism of the effect of these SNPs on α2A-AR function and platelet aggregation is still unclear and needs further investigation.
-
The α2A-ARs are the most prevalent adrenergic receptors in the brain, including cerebral cortex and locus coeruleus. Previous research found that ADRA2A SNPs were associated with inattention. In particular, ADRA2A C-1291G showed sex-specific association with attention deficit/hyperactivity syndrome symptoms in boys and girls[29] and with the perception of emotion in facial stimuli in males and females[30]. Although these studies focused on ADRA2A gene in the nervous system, they also showed that ADRA2A SNPs might have gender-specific effects. In our study, we found a gender-specific association of ADRA2A genotypes with ADP-induced platelets aggregation; males rather than females had low ADP inhibition with the minor allele at rs11195419 and rs3750625. According to the findings of multivariate analysis, sex was the strongest predictor of ADP inhibition (P < 0.0001). We conducted a subgroup analysis to determine the co-effect of sex and SNPs in ADP inhibition. In the subgroup analysis, we found that in male patients, ADP inhibition was significantly associated with rs11195419 and rs3750625 genotypes both in the univariate and multivariate analysis. However, we did not find the same association in female patients. The underlying mechanism of this phenomenon is still unknown. The α2A-ARs prevail on the platelet membrane, through which catecholamines potentiate the effects of other agonists (such as ADP). Previous studies have suggested that interaction of epinephrine with α2A-ARs occurs in the early phase of platelet activation, which is followed by platelet aggregation and is modulated by multiple pathways[31]. The higher basal level of catecholamines in males relative to females might cause platelet hyper-reactivity, which might be modulated by ADRA2A genotypes at rs11195419 and rs3750625. Therefore, despite the use of aspirin and clopidogrel, the later phase of ADP-induced platelet aggregation was still amplified in males compared with females, resulting in lower ADP inhibition on antiplatelet regimens. It may also explain the platelet hyper-reactivity during treatments in male patients carrying specific α2A-ARs SNPs. However, a larger sample size is needed and the exact underlying mechanism warrants further investigation.
Study Limitations There are several limitations of the current study. First, this is an observational study and platelet function was only evaluated by the TEG platelet mapping assay. Second, the mechanism of ADP inhibition and epinephrine-induced platelet aggregation remains unclear in the current study and it was difficult to explain the underlying mechanism of how the ADRA2A genotype might influence ADP-induced platelet aggregation, especially in males. Third, our investigation lacks a replication cohort. Lastly, the current study lacks the clinical follow-up to investigate the effect of ADRA2A SNPs on cardiovascular outcomes. Multi-centric, large, prospective clinical studies involving different ethnic populations to assess the clinical outcomes of high platelet reactivity are warranted in future.
Prevalence of ADRA2A Polymorphism
Association between ADRA2A Polymorphism and ADP-induced Platelet Aggregation
Association between ADRA2A Polymorphism and ADP-induced Platelet Aggregation Stratified by Sex
-
ADRA2A genetic variations were associated with ADP-induced platelet aggregation during administration of DAPT in Chinese patients undergoing PCI and the effect was more pronounced in males.
-
No conflict of interest to declare.
-
Variables RS553668 RS13306146 WT
(N = 334)HE
(N = 492)HO
(N = 198)P WT
(N = 587)HE
(N = 354)HO
(N = 83)P Age, years 59.1 ± 10.0 58.1 ± 10.4 58.6 ± 11.7 0.343 58.5 ± 10.2 58.6 ± 10.8 58.3 ± 12.3 0.974 Male, n (%) 242 (72.5) 385 (78.3) 146 (73.7) 0.134 438 (74.6) 269 (76.0) 66 (79.5) 0.601 BMI, kg/m2 26.1 ± 3.2 26.1 ± 3.2 25.9 ± 3.3 0.703 26.2 ± 3.2 25.9 ± 3.3 26.0 ± 3.6 0.325 Platelet, ×109/L 203.0 ± 52.2 202.5 ± 53.8 214.8 ± 59.2 0.019 202.8 ± 52.9 205.9 ± 54.9 217.3 ± 62.7 0.071 ACS, n (%) 98 (29.3) 159 (32.3) 69 (34.8) 0.399 175 (29.8) 116 (32.8) 35 (42.2) 0.070 DM, n (%) 116 (34.7) 141 (28.7) 51 (25.8) 0.059 189 (32.2) 99 (28.0) 20 (24.1) 0.181 Hypertension, n (%) 218 (65.3) 288 (58.5) 127 (64.1) 0.112 362 (61.7) 222 (62.7) 49 (59.0) 0.820 HCL, n (%) 285 (85.3) 414 (84.1) 166 (83.8) 0.866 494 (84.2) 299 (84.5) 72 (86.7) 0.830 Current smoking, n (%) 118 (35.3) 202 (41.1) 63 (31.8) 0.048 218 (37.1) 135 (38.1) 30 (36.1) 0.925 CHD family history, n (%) 4 (1.2) 4 (0.8) 4 (2.0) 0.411 9 (1.5) 2 (0.6) 1 (1.2) 0.409 Previous MI, n (%) 50 (15.0) 98 (19.9) 26 (13.1) 0.049 102 (17.4) 61 (17.2) 11 (13.3) 0.638 Previous PCI, n (%) 44 (13.2) 76 (15.4) 27 (13.6) 0.625 85 (14.5) 50 (14.1) 12 (14.5) 0.988 Previous CABG, n (%) 1 (0.3) 4 (0.8) 0 (0) 0.319 3 (0.5) 2 (0.6) 0 (0) 0.796 LAD, n (%) 199 (59.6) 273 (55.5) 119 (60.1) 0.379 349 (59.5) 190 (53.7) 52 (62.7) 0.140 LCX, n (%) 95 (28.4) 140 (28.5) 59 (29.8) 0.932 170 (29.0) 103 (29.1) 21 (25.3) 0.773 RCA, n (%) 151 (45.2) 227 (46.1) 88 (44.4) 0.913 263 (44.8) 168 (47.5) 35 (42.2) 0.597 LM, n (%) 17 (5.1) 22 (4.5) 11 (5.6) 0.818 29 (4.9) 18 (5.1) 3 (3.6) 0.851 Statin, n (%) 328 (98.2) 473 (96.1) 193 (97.5) 0.210 567 (96.6) 345 (97.5) 82 (98.8) 0.466 β-blocker, n (%) 293 (87.7) 438 (89.0) 178 (89.9) 0.722 522 (88.9) 312 (88.1) 75 (90.4) 0.832 ACEI/ARB, n (%) 205 (61.4) 285 (57.9) 114 (57.6) 0.554 341 (58.1) 214 (60.5) 49 (59.0) 0.776 PPIs, n (%) 66 (13.4) 58 (17.4) 38 (19.2) 0.109 94 (16.0) 55 (15.5) 13 (15.7) 0.980 DES, n (%) 331 (99.1) 488 (99.2) 197 (99.5) 0.878 584 (99.5) 350 (98.9) 82 (98.8) 0.522 BMS, n (%) 1 (0.3) 4 (0.8) 1 (0.5) 0.629 1 (0.2) 4 (1.1) 1 (1.2) 0.130 Ballooning only, n (%) 2 (0.6) 0 (0) 0 (0) 0.126 2 (0.3) 0 (0) 0 (0) 0.474 Note.*: P < 0.005; WT: Wild Type, HE: Heterozygote, HO: Homozygote, ADRA2A: the α2A-adrenergic receptor gene, BMI: body mass index, ACS: acute coronary syndrome, DM: diabetes mellitus, CHD: coronary heart disease, HCL: Hypercholesterolemia, MI: myocardial infarction, PCI: percutaneous coronary intervention, CABG: coronary artery bypass grafting, LAD:left anterior descendens, LCX: left circumflex, RCA: right coronary artery, ACEI: angiotensin conversion enzyme inhibitor, ARB: angiotensin receptor blocker, CCB: calcium channel blocker, PPI: proton pump inhibitor, DES: Drug-eluting stent, BMS: Bare mental stent. Table Appendix. Baseline characteristics by ADRA2A genotypes (rs553668 and rs13306146)








 Quick Links
Quick Links
 DownLoad:
DownLoad: