-
Sodium sulfite (Na2SO3) is a common industrial salt used in papermaking, food preservatives, and as an oxygen scavenger. Sodium sulfite is rarely used as an air pollutant[1,2]. Our previous work used sodium sulfite as a deoxidant in a hypoxic model and revealed that it could induce a steady hypoxic condition without chemical damage to living cells of Caenorhabditis elegans[3]. Compared to traditional deoxidants[4,5], sodium sulfite avoids cellular toxicity. Therefore, it would be valuable to apply it to microorganisms in anaerobic cultures or in hypoxic studies. Escherichia coli can extract energy from substrates without O2 via fermentation. Most studies have shown that the arcA/B, fnr, oxyR, and soxR/S systems are closely related to bacterial anoxia in E. coli[6,7]. These four hypoxia-related pathways act together when bacteria lack oxygen. However, the related genes may play different roles during hypoxia induced by sodium sulfite. The current study focused on revealing the different roles of these genes in hypoxia regulation using E. coli strains with relevant gene mutants.
Sodium sulfite and sodium sulfate (400 mmol/L) were prepared in Luria-Bertani (LB) liquid medium (1% bacto tryptone, 0.5% yeast extract, and 0.5% NaCl w/v) and then filtered using a 0.22 µm filter membrane. Before bacterial inoculation, LB medium with sodium sulfite and sodium sulfate was diluted to 1, 2.5, 5, 10, 20, 40, and 80 mmol/L, and the original LB medium was used as a control. After adding sodium sulfite and sodium sulfate, the pH values were adjusted to 7.0 using HCl. All mutants were developed from the E. coli strain MG1655, which is a wild-type strain of K-12. E. coli strains with hypoxic pathway-related gene mutations are listed in Table 1.
Name Genotype Source References arcA BW29363 arcA:kan E. coli Genetic Stock Center [8] arcB JW5536-1 arcB:kan E. coli Genetic Stock Center [9] fnr JW1328-1 fnr:kan E. coli Genetic Stock Center [10] soxR JW4024-1 soxR:kan E. coli Genetic Stock Center [11] soxS JW4023 soxS:kan E. coli Genetic Stock Center [9] oxyR JW3933-3 oxyR:kan E. coli Genetic Stock Center [9] sodA JW3879-1 sodA:kan E. coli Genetic Stock Center [9] sodB JW1648-1 sodB:kan E. coli Genetic Stock Center [9] sodC JW1638-1 sodC:kan E. coli Genetic Stock Center [9] Table 1. List of E. coli strains
All the bacterial strains were recovered on the day before the experiments and transferred to fresh LB medium (1:1,000) for 12 h at 37 °C on a shaker at 200 rpm. The bacterial strains were then inoculated into liquid LB medium (1:200) with different dilutions of sodium sulfite or sodium sulfate. The Bioscreen C Automated Microbiology Growth Curve Analysis System (Oy Growth Curves Ab Ltd., Switzerland) was used to provide a temperature and shaking condition for the bacterial growth with a real-time OD600 detection of the bacterial suspension once every hour. E. coli cells from the control, sodium sulfite-, and sodium sulfate-processed groups (approximately 2 mL bacilli) were centrifuged and washed three times with 0.5 mL ddH2O. The collected cells were fixed in 0.1 mol/L sodium cacodylate buffer (pH 7.4) containing 2.5% glutaraldehyde at 4 °C for 2 h and embedded in epoxy resin. Ultrathin sections were double stained with uranyl acetate and lead citrate. Bacterial subcellular structure images were captured using the AMT transmission electron microscopy TEM system (AMT Imaging, Woburn, MA, USA) and analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). All data are presented as mean ± SD. The results were statistically evaluated for significance using Student’s t test. Statistical significance was set at P < 0.05.
In the present study, with an increase in the concentration of sodium sulfite, the growth curve of E. coli MG1655 decreased, indicating inhibition of cell proliferation (Figure 1A). However, sodium sulfate did not affect bacterial growth (Figure 1B). The growth curve with sodium sulfite (≥ 40 mmol/L) showed a stable period of growth inhibition for at least 8 h (Figure 1A). Therefore, sodium sulfite could be used as a hypoxia revulsant for E. coli. The induced hypoxic period could be maintained for at least 8 h, and the concentration of sodium sulfite should exceed 40 mmol/L.
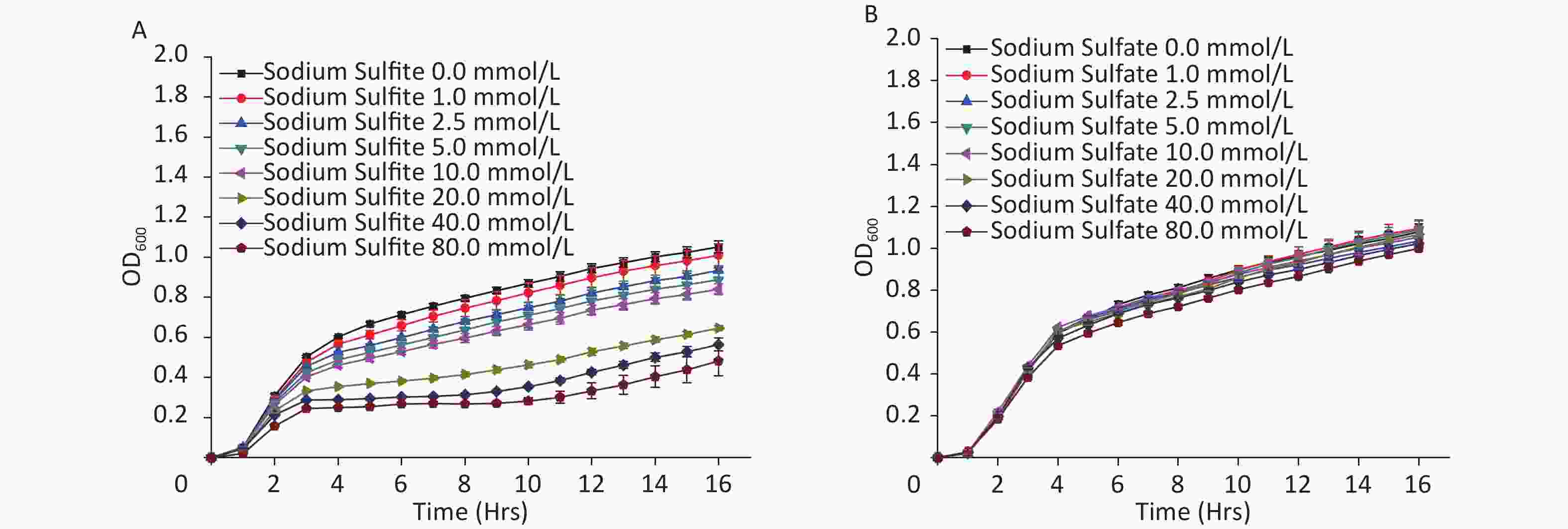
Figure 1. Growth curves of E. coli strain MG1655. Culture medium with (A) sodium sulfite and (B) sodium sulfate.
A complete membrane and high electron density nuclear area were observed using TEM in the control group (Figure 2A). The sodium sulfite treatment group did not show subcellular damage such as membrane lysis, cell shrinkage, or other cellular morphological changes of the inhibited bacterial cells, even with an increase in the concentration of sodium sulfite, indicating that sodium sulfite could inhibit the growth and proliferation of bacterial cells (Figure 2A). In the sodium sulfate group, the cell size was significantly smaller (Figure 2B and C). These results indicate that sodium sulfite can inhibit bacterial growth by inhibiting cell division rather than disrupting the structure of bacterial cells.
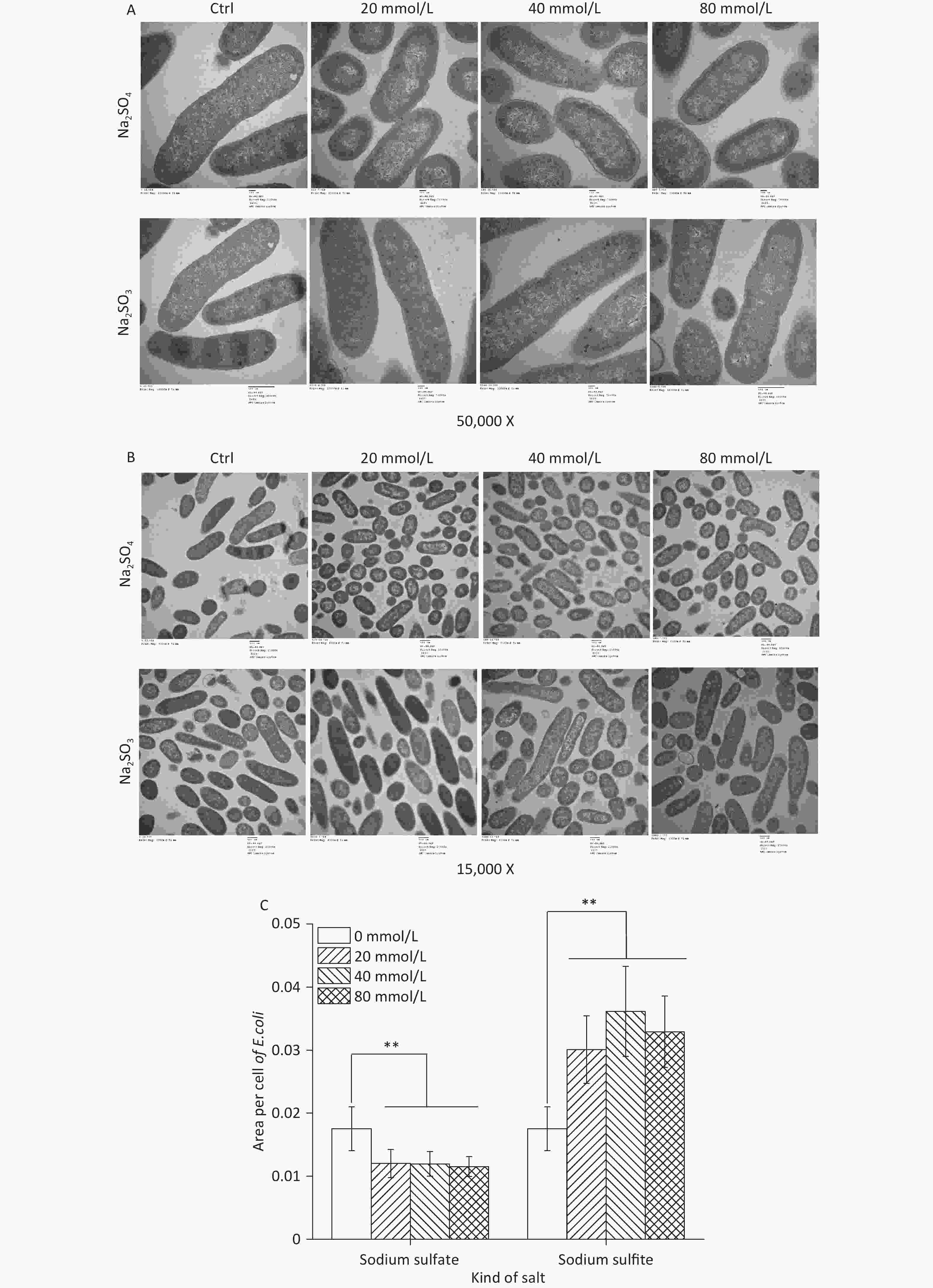
Figure 2. Ultrastructure and cell size of E. coli. (A and B) E. coli MG1655 cells were cultured in LB medium with sodium sulfite (20, 40, and 80 mmol/L) and sodium sulfate (20, 40, and 80 mmol/L). (C) Histogram showing size of E. coli MG1655 cells cultured in sodium sulfite and sodium sulfate. Data are shown as mean ± SD; **P < 0.01.
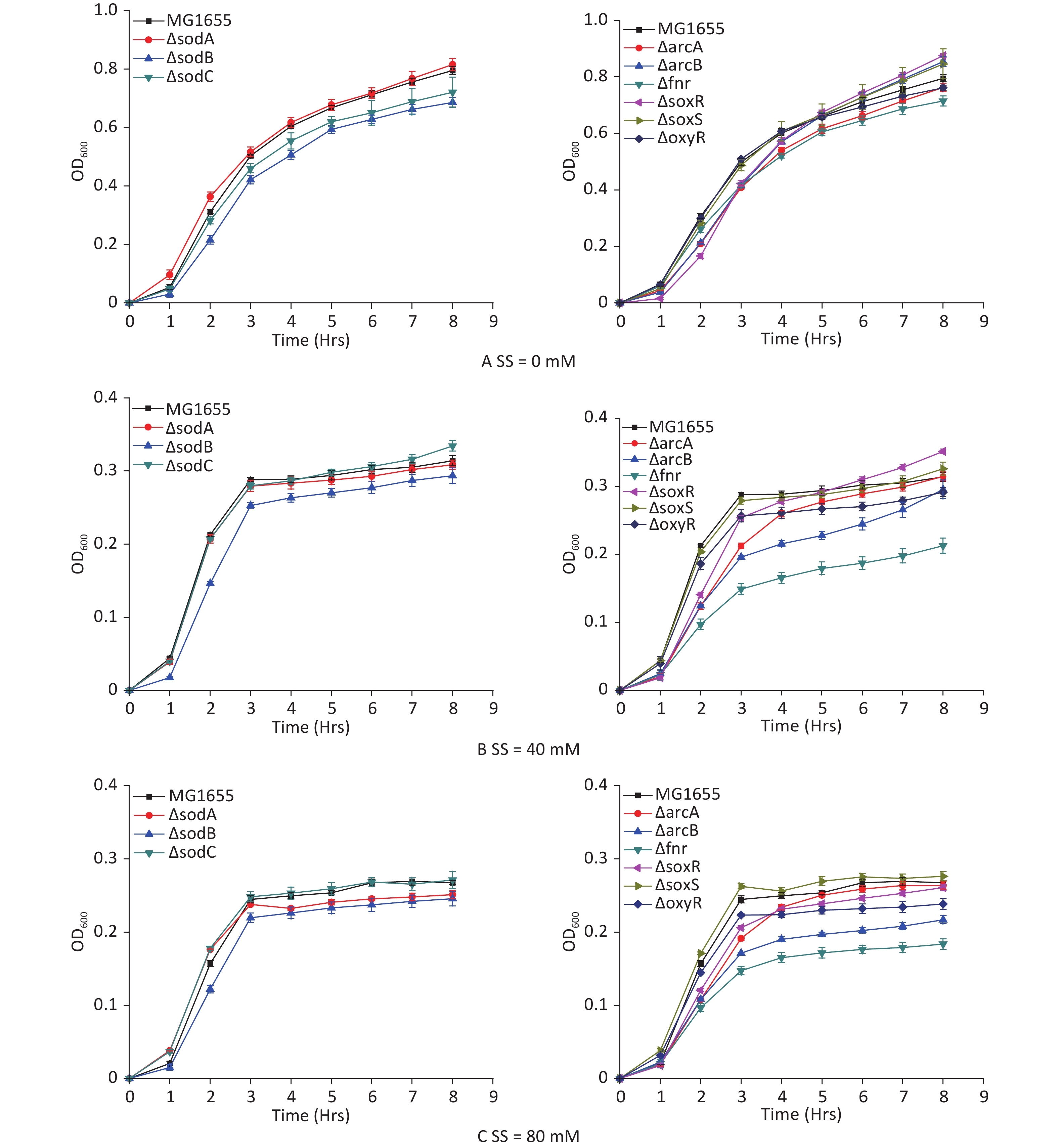
E. coli strains with hypoxia-related gene mutants were studied to analyze the hypoxic effects of sodium sulfite and the roles of various genes during the process. Sodium sulfite at 40 mmol/L (Figure 3B) and 80 mmol/L (Figure 3C) induced hypoxia for 8 h. The 8 h growth curves were detected for each mutant strain (Figure 3). Strains with arcB and fnr mutations were most strongly inhibited by sodium sulfite-induced hypoxia, demonstrating their important roles in the process.
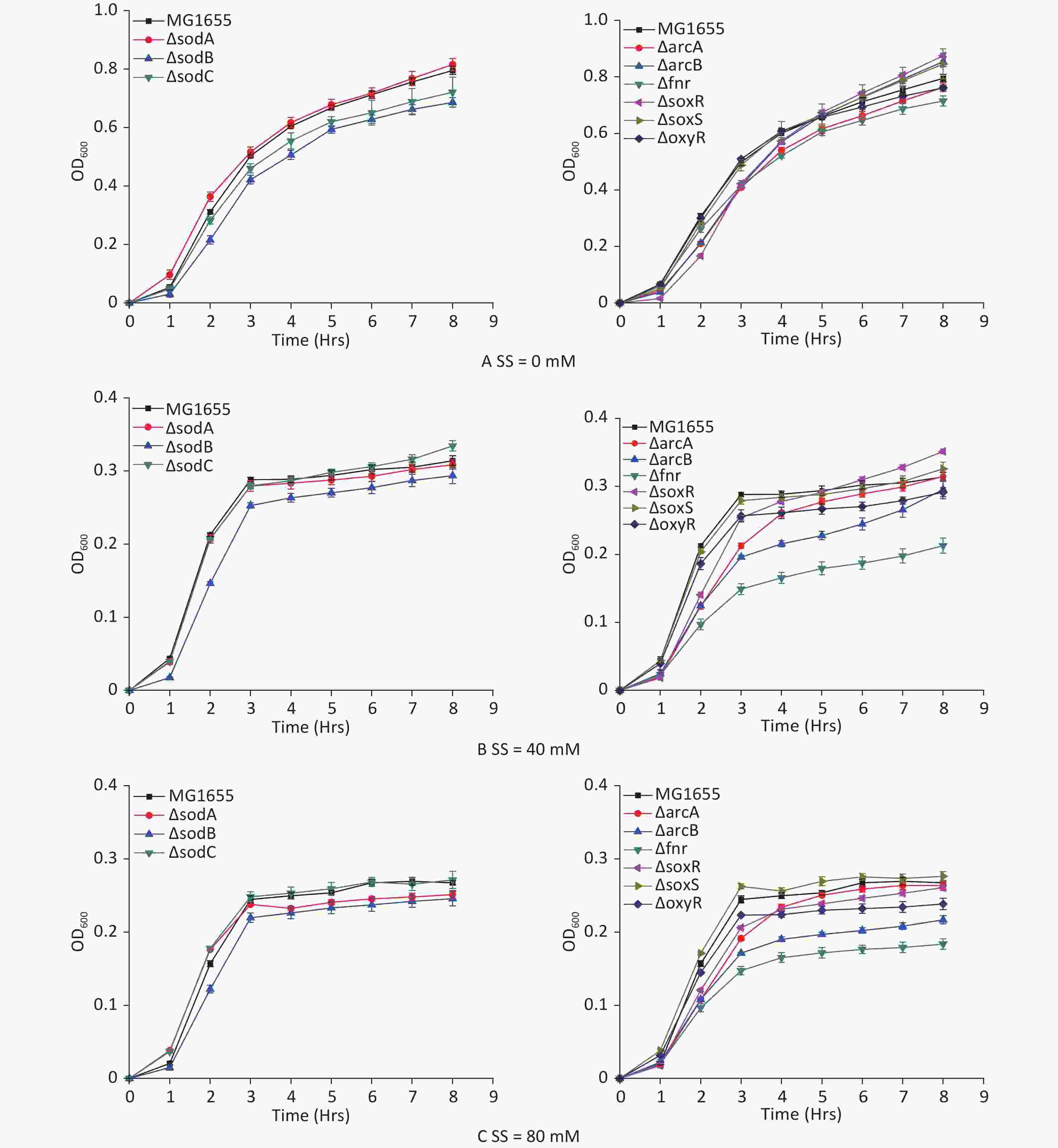
Figure 3. Growth curves of E. coli mutant strains. Mutant strains (arcA, arcB, fnr, oxyR, soxR, soxS, sodA, sodB, and sodC) were cultured in sodium sulfite at steady hypoxic concentrations (40 and 80 mmol/L).
The current study demonstrated that sodium sulfite inhibited bacterial growth at concentrations over 40 mmol/L for at least 8 h. Therefore, sodium sulfite could be used as a hypoxia revulsant in E. coli. Sodium sulfite, a reducing salt, consumes the oxygen in the medium to induce hypoxia. Although sodium sulfite can deprive dissolved oxygen during a short period (< 2 h)[4] in a closed system, E. coli growth inhibition is related to salt concentration. This may be due to the gas exchanged during shaking of the E. coli culture plate. These results demonstrate a novel and ideal method to induce bacterial hypoxia without toxicity or device dependence. The ultrastructural observation also demonstrated that the cellular structure under sodium sulfite-inhibiting conditions showed no significant changes, indicating that sodium sulfite did not kill the existing microorganisms but only inhibited their growth and proliferation. Furthermore, the cell size in the sodium sulfite group was significantly larger than that in the control and sodium sulfate groups, indicating the existence of a compensatory mechanism.
The ability of microorganisms to adapt to their environment is crucial. The transition between oxic and anoxic environments in E. coli has been studied extensively[12] and transition metabolism is mainly regulated by fnr and arcA/B[13,14]. Fnr was maintained at almost equal levels under oxic and anoxic conditions, whereas arcA, as a translation regulator needs to synthetize when oxygen levels are decreased. Moreover, fnr can sense oxygen directly[15], whereas the arcA/B system[16,17] can induce the expression of fermentation genes and inhibit aerobic pathways when E. coli encounters low-oxygen growth conditions[18,19].
In summary, this study demonstrated that sodium sulfite significantly decreased the bacterial growth rate and could be used as a hypoxia revulsant for E. coli. However, the bacterial cells were not damaged, and sodium sulfite only inhibited cell growth and proliferation. Furthermore, fnr and arcB played the most important roles in sodium sulfite-induced hypoxia. Under hypoxic conditions, the inhibition of bacterial growth may be attributed to the inhibition of cell division.
Sodium Sulfite as a Novel Hypoxia Revulsant Involved in Hypoxic Regulation in Escherichia coli
doi: 10.3967/bes2024.025
- Received Date: 2023-08-30
- Accepted Date: 2023-12-04
Abstract: As a reducing salt, sodium sulfite could deprive oxygen in solution, which could mimic hypoxic stress in Caenorhabditis elegans. In this study, the wild-type Escherichia coli strain MG1655 was used to examine the inhibition of sodium sulfite-induced hypoxia by observing the bacterial growth curves. We also analyzed the growth curves of mutant strains (for arcA/B, soxR/S, fnr, and oxyR) related to E. coli hypoxic pathways to reveal roles of the related genes during hypoxia. The ultrastructure of hypoxia-inhibited bacteria were also observed using transmission electron microscopy. Sodium sulfite could maintain hypoxic condition of bacterial culture for 8 h with concentrations over 40 mmol/L. Complete ultrastructure of the bacteria indicated sodium sulfite did inhibit bacterial growth and division. Among the hypoxia genes, fnr and arcB played key roles in sodium sulfite-induced hypoxia. This study showed that sodium sulfite could be used as a novel hypoxia revulsant for bacterial cultures.
&These authors contributed equally to this work.| Citation: | YE Qiao, HUO Jia Nan, LUO Yuan, MEI Zhu Song, FANG Long Mei, GUO Bing Qian, WANG Guang Yun. Sodium Sulfite as a Novel Hypoxia Revulsant Involved in Hypoxic Regulation in Escherichia coli[J]. Biomedical and Environmental Sciences, 2024, 37(2): 228-232. doi: 10.3967/bes2024.025 |








 Quick Links
Quick Links
 DownLoad:
DownLoad: