-
Colorectal cancer (CRC) is the leading cause of cancer-related morbidity and mortality worldwide, with 1.9 million new cases estimated in 2020[1]. Tumor metastasis and recurrence contribute to the high mortality rates in CRC[2]. Over 20% of patients with CRC are diagnosed at an advanced stage, with a 5-year survival rate of 14%[3]. The pathogenic mechanisms underlying the progression appear to be complex and heterogeneous. Neoplastic transformation and progression of CRC occur gradually[4]; instability of the genomic activation of oncogenes and mutational inactivation of tumor suppressor genes are associated with cancer development[5]. Hence, studying the molecular biology of CRC is essential for verifying diagnostic and therapeutic biomarkers and improving prognosis.
Copy number alterations are common somatic changes in cancer and are characterized by the gain or loss of copies of DNA fragments[6]. Genomic amplification of 8q24 is a frequent event in CRC[7,8]. Recent studies have indicated that the chromosome 8 open reading frame 76 (C8orf76) is located on chromosome 8q24.13, which is part of the gene desert region on the long arm of chromosome 8. C8orf76 is a nucleoprotein-coding gene that is preferentially amplified in primary gastric cancer compared to that in adjacent non-tumor mucosal tissues[9]. By activating the mitogen-activated protein kinase (MAPK) pathway through the long non-coding RNA (lncRNA) dual-specificity phosphatase 5 pseudogene 1 (DUSP5P1), C8orf76 plays an important role in the occurrence of gastric cancer and is an independent prognostic factor for patients with gastric cancer. Additionally, C8orf76 plays oncogenic roles in liver and breast cancers[9]. The expression level of the C8orf76 gene is frequently upregulated in liver cancer, wherein C8orf76 regulates ferroptosis by transcriptionally upregulating solute carrier family 7 member 11 (SLC7A11) and is significantly correlated with hepatocellular carcinoma (HCC) development. High expression of C8orf76 was observed in breast cancer tissues, which was significantly correlated with clinical stage. C8orf76 is an independent predictive factor for poor prognosis in patients with breast cancer[10,11].
However, the role of C8orf76 in CRC progression remains unclear. In this study, we performed a comprehensive analysis to verify the genes correlated with C8orf76 in CRC and found that C8orf76 positively correlated with cyclin-dependent kinase 4 (CDK4). Additionally, we constructed nomograms based on clinicopathological characteristics to predict the overall survival (OS) of patients with CRC and explored their prognostic significance.
-
The CRC cohort included 83 patients who underwent surgery at The First Hospital of Hebei Medical University, Shijiazhuang, China. The clinicopathological features of the patients are shown in Supplementary Table S1. The study was conducted in accordance with the principles of the Declaration of Helsinki, and was approved by the Ethics Committee of First Hospital of Hebei Medical University (20220227). Written informed consent was obtained from all participants.
Variable Cohort No. of patient 83 Age, mean ± SD 64.65±11.58 Gender Male 52 (62.65%) Female 31 (37.35%) Tumor location Colon 38 (45.78%) Rectum 45 (54.22%) Differentiation Moderate or high 65 (78.31%) Low 18 (21.69%) TNM Stage I+ II 49 (59.04%) III+IV 34 (40.96%) OS Alive
44 (53.01%)Dead 39 (46.99%) Table S1. Clinicopathologic features of CRC patients in our Cohort.
-
CRC cell lines (DLD-1, LOVO, HCT116, SW480, and SW1463) were generously provided by Prof. Jun Yu (Chinese University of Hong Kong). These cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; GIBCO, Grand Island, USA), supplemented with 10% fetal bovine serum (FBS; GIBCO, Grand Island, USA) and 1% penicillin-streptomycin (Invitrogen, MA, USA), in a 37 °C humidified incubator with 95% air and 5% CO2. The cells were grown for at most 25 passages in all experiments. The cell lines reached approximately 70% confluence.
-
We used the C8orf76-siRNA (GenePharma) and CDK4-siRNA (GenePharma). The sequences of C8orf76-siRNAs are as follows: C8orf76-siRNA: GUUCCAUACAGAGAUACAATT (sense: 5′–3′) and UUGUAUCUCUGUAUGGAACTT (anti-sense: 5′–3′). The sequences of CDK4-siRNAs are as follows: CDK4-siRNA: GCCAGUUUCUAAGAGGCCUTT (sense: 5′–3′) and AGGCCUCUUAGAAACUGGCTT (anti-sense: 5′–3′). A non-targeting RNA sequence served as a negative control. Cells were transfected with Lipofectamine 2000 (Invitrogen) (siRNA concentration used: 0.1 μmol/L per well in 6-well plates). The RNA oligo sequences used are listed in Supplementary Table S2.
RNA Oligo Sequence (5'-3') siC8orf76-sense GUUCCAUACAGAGAUACAATT siC8orf76-antisense
siCDK4-sense
siCDK4- antisenseUUGUAUCUCUGUAUGGAACTT
GCCAGUUUCUAAGAGGCCUTT
AGGCCUCUUAGAAACUGGCTTTable S2. RNA Oligo used in this study
-
The full-length open reading frame sequence of C8orf76 was subcloned into pcDNA3.1. Subsequently, pcDNA3.1-C8orf76 or empty vector pcDNA3.1 was transfected into DLD1 or LOVO cells using Lipofectamine 2000 (Invitrogen, Carlsbad, USA) to overexpress C8orf76 (Concentration of C8orf76 or empty vector used in the overexpression experiment: 2,000 ng per well in 6-well plates). After at least 2 weeks of selection with G418 antibiotics (GIBCO, Grand Island, USA), we obtained cell lines stably expressing C8orf76. The experiment was repeated three times and stable CRC cell lines were used for a maximum of 10 generations.
-
Total RNA was extracted by using the TRIzol™ Reagent (Thermo Fisher Scientific, MA, USA) (approximately 4.67×105 cells were used). Total RNA was reverse-transcribed to synthesize cDNA using a cDNA Reverse Transcription Kit (TransGen Biotech, Beijing, China) (Amount of RNA used for cDNA synthesis: 2000 ng per well). Real-time quantitative polymerase chain reaction (RT-qPCR) was performed using SYBR Green PCR Master Mix (Takara, Beijing, China) as per the manufacturer’s protocol. The final reaction was performed on a 7500 real-time PCR system (Thermo Fisher Scientific, MA, USA), and comprised the following stages: Hot-start DNA polymerase activation to 95 °C for 10 min; 40 cycles of 95 °C for 15 sec and 60 °C for 1 min. The primer sequences used are listed in Supplementary Table 3. Gene expression was normalized to β-actin and calculated using 2−ΔΔCt method.
Gene Primer Sequence C8orf76 Forward TTATACGAACCAGGCTTCTGC Reverse GCCAACACACTTCACCTCTG CDK4 Forward ATGGCTACCTCTCGATATGAGC Reverse CATTGGGGACTCTCACACTCT β-actin Forward CATCCACGAAACTACCTTCAACTCC Reverse GAGCCGCCGATCCACACG Table S3. Primers used in this study
-
Protein lysates from cells were obtained using a cocktail of RIPA buffer containing protease inhibitors (2.5×106 cells used). Total protein (60 ug) was loaded to a sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel (6% stacking gel and 10% separating gel). After gel electrophoresis, the proteins were transferred onto polyvinylidene difluoride membranes (PVDF) at 90 V for 2 h and then blocked with 5% bovine serum albumin (BSA) for 30 min at room temperature. Next, the PVDF membranes were incubated with primary antibodies specific to C8orf76 (1:1000, Bioss, Beijing, China), CDK4 (1:1000, Proteintech, Wuhan, China), and β-actin (1:1,000, Cell Signaling Technology, Boston, USA) at 4 °C overnight. On the following day, the PVDF membranes were incubated with secondary antibody. Protein signals were detected and scanned using the Odyssey CLx Imaging System (LI-COR Biosciences). The primary antibodies used are listed in Supplementary Table S4.
Antibodies Source Identifier Anti- C8orf76 Bioss Cat#bs-15301R Anti-CDK4 Proteintech Cat#11026-1-AP Anti-β-actin Cell Signaling Technology Cat#4970 Table S4. Antibodies used in this study
-
Briefly, 4-mm thick sections were cut from paraffin-embedded tissue blocks. The paraffin-embedded thin sections were deparaffinized with xylene and rehydrated with a gradient alcohol series, and then antigen retrieval was performed with 0.01 mol/L sodium citrate buffer (pH 6.0). Staining was performed using an immunohistochemical staining kit (ZSBG-BIO, Beijing, China). The sections were then incubated with Anti-human C8orf76 antibody (Bioss, 1:200 dilution) and Anti-human CDK4 antibody (Proteintech, 1:200 dilution) overnight at 4 °C. On the following day, secondary fluorescent antibodies were applied for 1 h at 37 °C. For immunohistochemical staining, color was developed using a diaminobenzidine (DAB) substrate solution. For IF staining, the paraffin-embedded thin sections were counterstained with 4',6-diamidino-2-phenylindole (DAPI). The positive percentage was scored as follows: 0, no positive staining; 1, between 1%–25% of cells; 2, between 26%–50% of cells; 3, between 51%–75% of cells; and 4, in > 75% of cells. Staining intensity was scored as follows: 0, negative; 1, weak; 2, moderate; and 3, high intensity. The final staining score was calculated as staining intensity score × percentage of positive cells. Two independent observers blindly evaluated the results. The antibodies used for IHC and IF staining are listed in Supplementary Table S4.
-
We downloaded CRC data from The Cancer Genome Atlas (TCGA) data portal (http://tcga-data.nci.nih.gov/tcga/findArchives.htm). Screening for genes that interact with C8orf76. Cytoscape 3.7 was used for generating the protein-protein interaction (PPI) network. The detailed descriptions are as follows:
Input the filtered genes into the STRING database(https://cn.string-db.org/) and retrieve the interaction network related to C8orf76. Click the "Exports" tab to download the file, and then import the file into Cytoscape 3.7. Adjust the network layout in Cytoscape, perform data analysis, and export the image.
-
All results were expressed as mean ± standard deviation (SD). Wilcoxon matched-pairs test or Student’s t-test was used to compare differences between the two groups. Pearson’s chi-square test was used to analyze the correlation between C8orf76 expression and clinicopathological features. Kaplan–Meier analysis was used to determine the relationship between C8orf76 expression and patient survival. Univariate and multivariate Cox proportional hazard regression analyses were used to estimate the prognostic significance of C8orf76 expression. All statistical tests were performed using GraphPad Prism 8.0, and P < 0.05 indicates statistical significance.
-
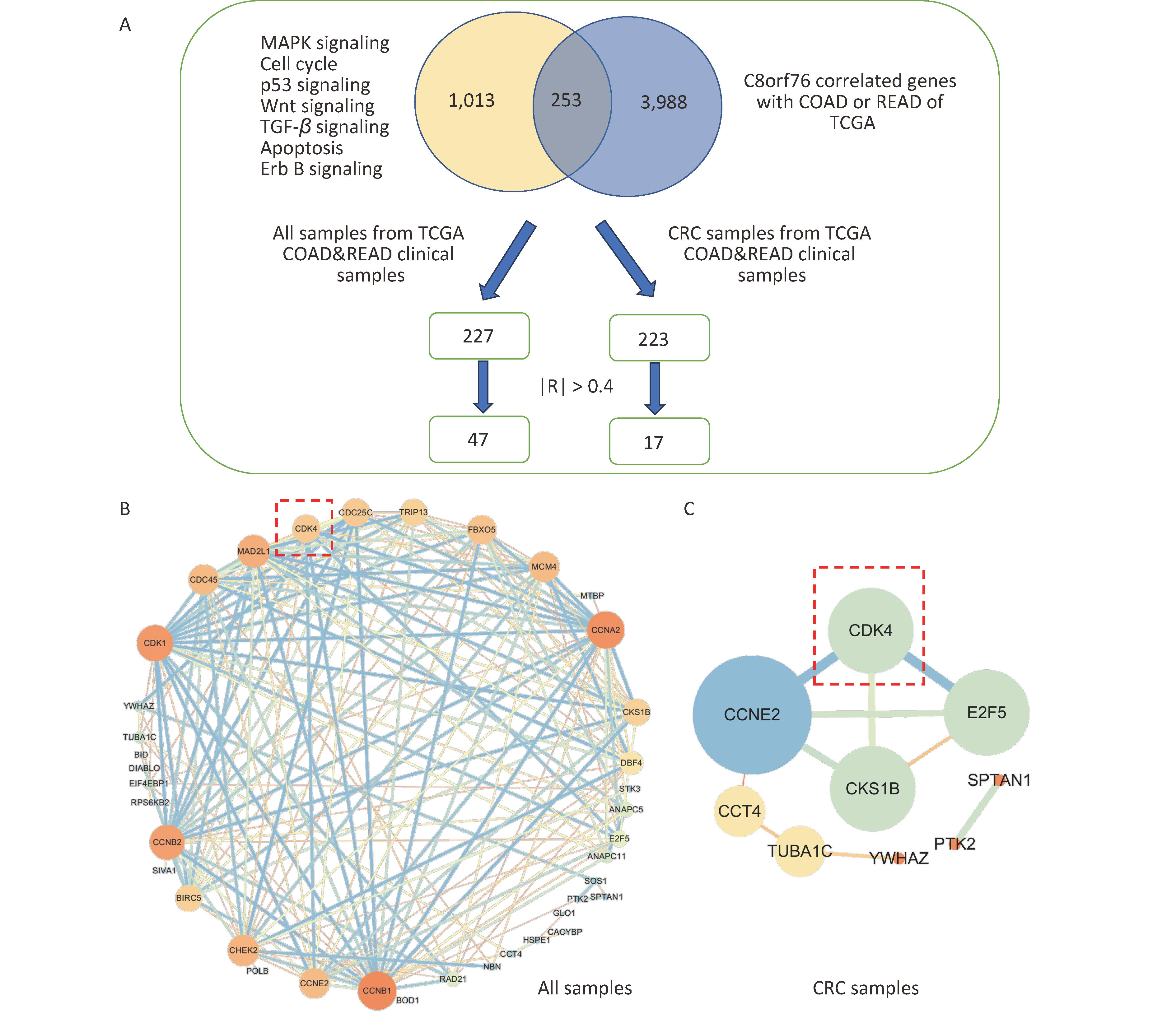
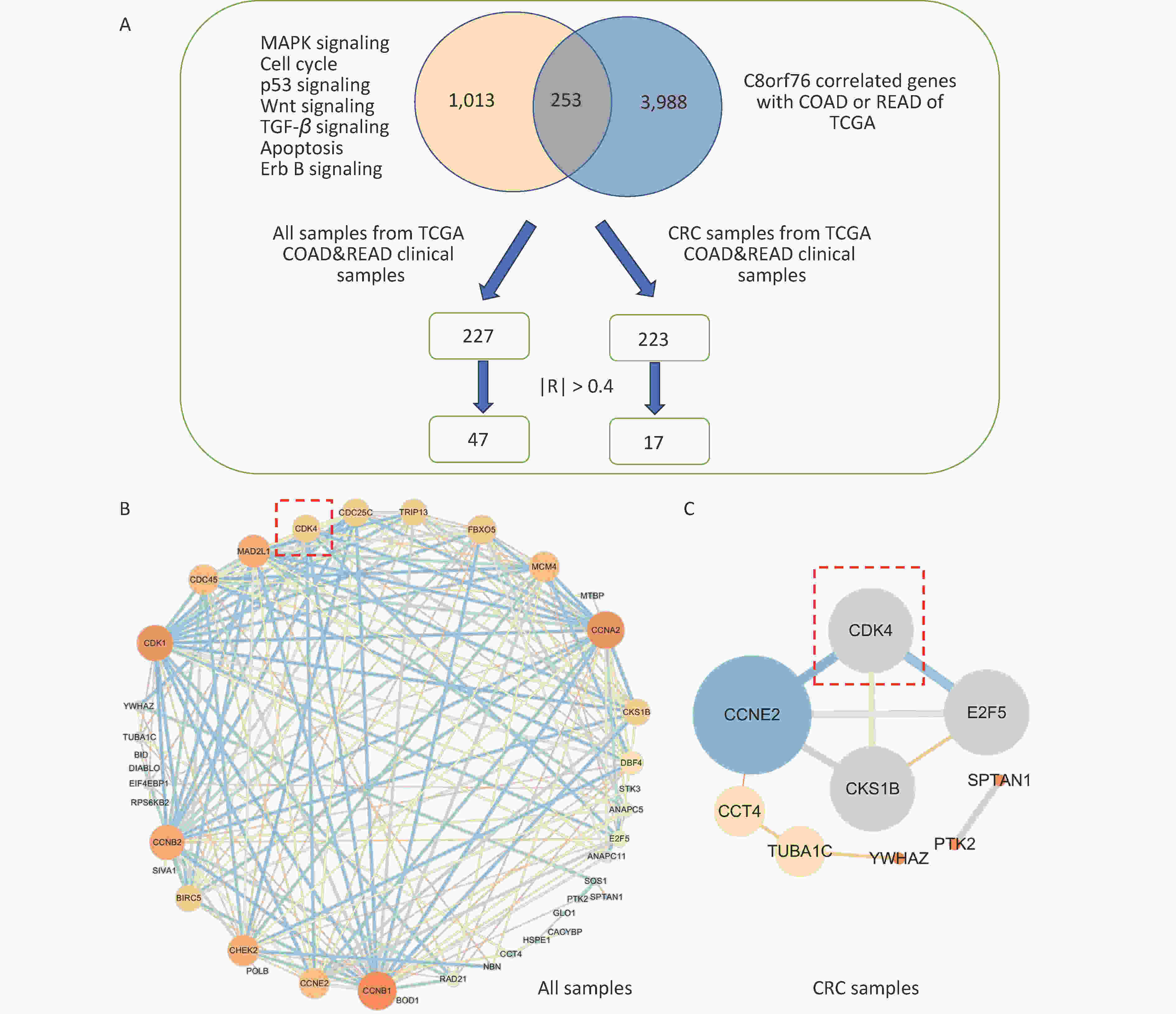
Firstly, we screened genes from C8orf76-enriched pathways (MAPK signaling, cell cycle, p53 signaling, Wnt signaling, TGF-β signaling, apoptosis, and Erb B signaling pathways) and C8orf76-related genes with Colon Adenocarcinoma (COAD) and Rectal Adenocarcinoma (READ) in the TCGA database. Pearson’s correlation coefficient analysis identified 253 genes (|R| > 0.3). Next, we compared these 253 genes with clinical CRC cases from the TCGA database. We selected the genes with |R| > 0.4 after Pearson analysis.
We found that 47 genes correlated with non-CRC patients (|R| > 0.4), whereas 16 genes correlated with CRC patients (|R| > 0.4; Figure 1A). Using Cytoscape, we identified interrelationships between the 47 associated genes in normal and CRC cases and 16 associated genes in CRC cases (Figures 1B and C). The results showed that there was an interaction between CDK4, Cyclin E2 (CCNE2), CDC28 Protein Kinase Regulatory Subunit 1 B (CKS1B), and E2F Transcription Factor 5 (E2F5) in CRC cases, with C8orf76 interacting the most with CDK4.
-
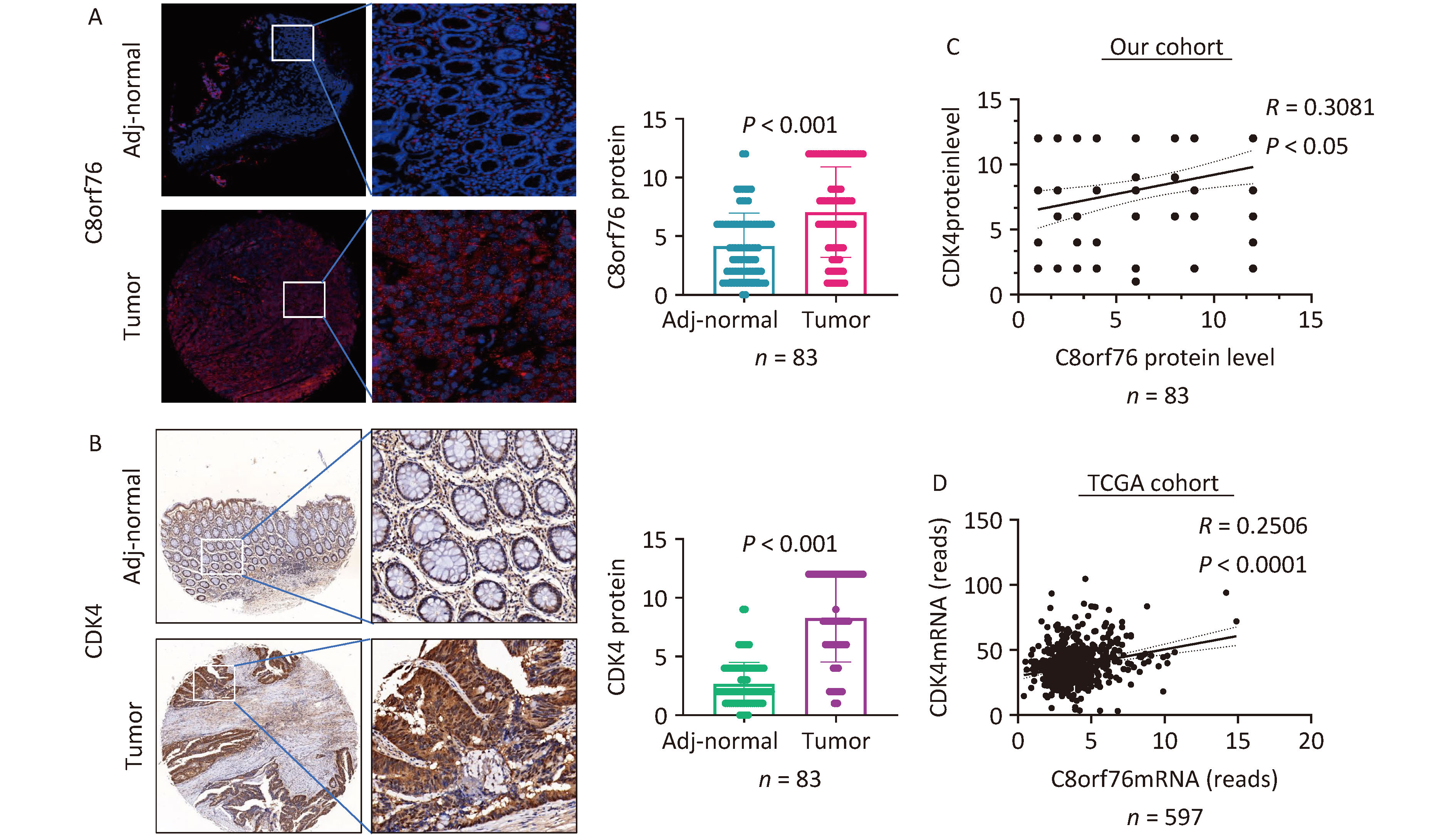
We found that the levels of C8orf76 and CDK4 proteins were significantly upregulated in CRC tissue samples compared to those in paired adjacent non-tumor tissues from our patient cohort (n = 83, P < 0.001) (Figures 2A and B). Furthermore, C8orf76 protein levels were positively correlated with CDK4 protein levels in the CRC samples (n = 83, R = 0.3081, P < 0.05) in our cohort (Figure 2C). Consistent with this result, C8orf76 mRNA expression was positively correlated with CDK4 mRNA expression in the CRC samples (n = 597, R = 0.2506, P < 0.001) in TCGA cohort (Figure 2D).
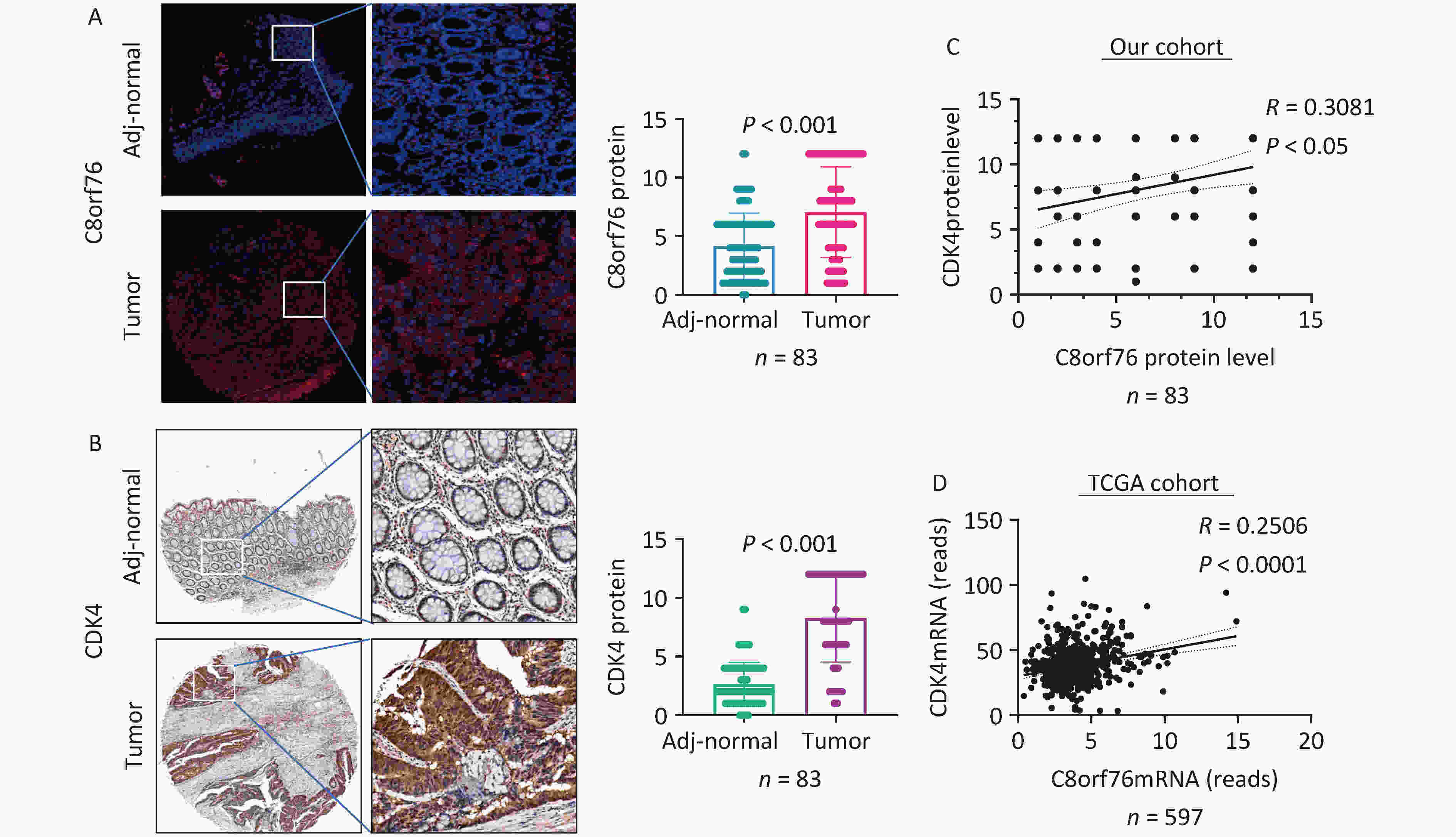
Figure 2. C8orf76 is positively correlated with CDK4 in our patient cohort as well as the TCGA cohort. (A) C8orf76 protein level is increased in CRC compared to paired adjacent normal tissues as determined from microtissue array (in our study cohort from Shijiazhuang, China). (B) CDK4 protein levels were increased in CRC compared to paired adjacent normal tissues as determined by microarray array (in our cohort from Shijiazhuang, China). (C) C8orf76 protein levels positively correlated with CDK4 protein levels. (D) C8orf76 mRNA levels positively correlated with CDK4 mRNA levels.
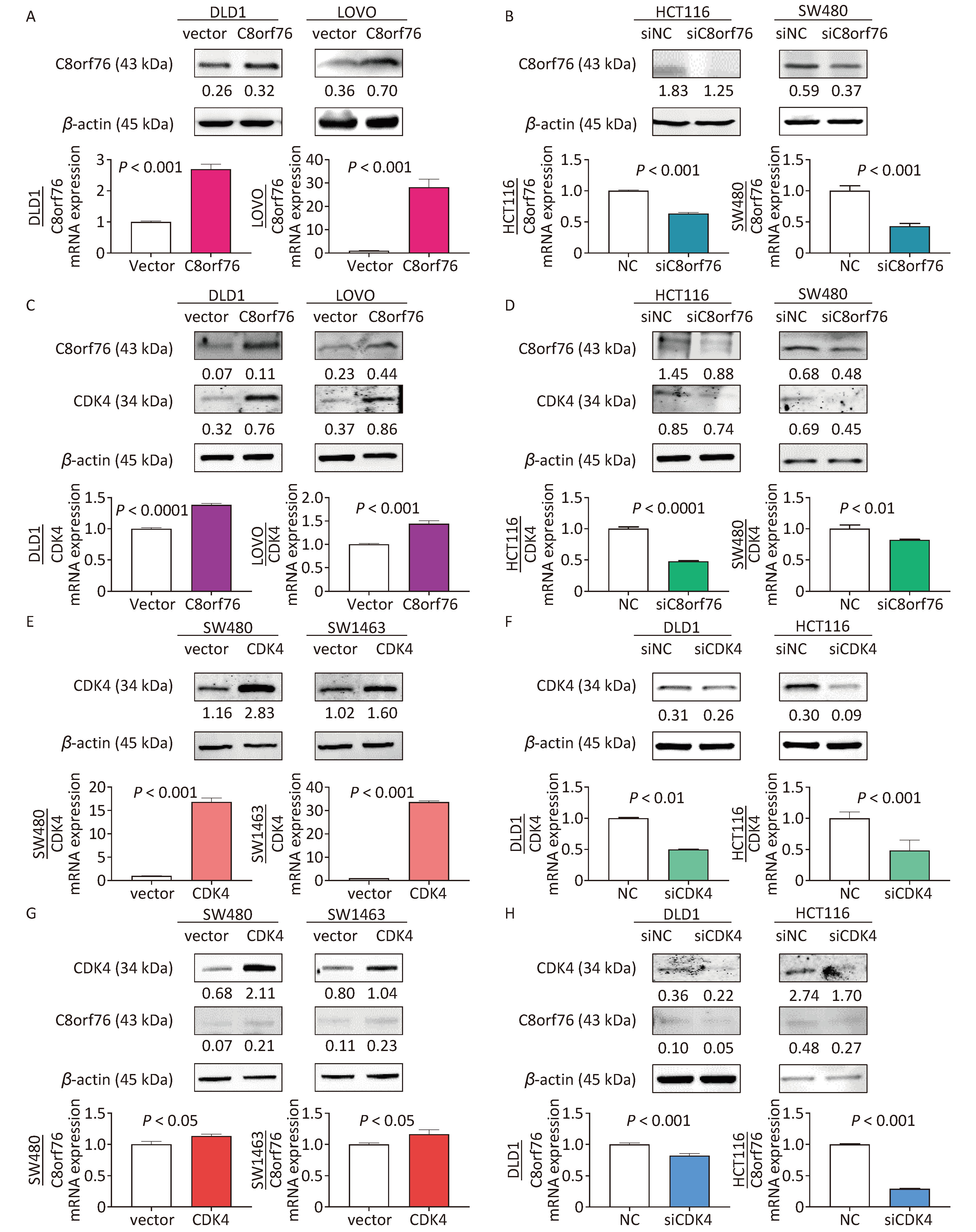
Next, we examined the expression of C8orf76 in CRC cell lines. Western blot analysis showed that samples from the C8orf76 overexpression group in the human CRC cell lines DLD1 and LOVO showed increased protein level of C8orf76 (Figure 3A). RT-qPCR analysis revealed that the ectopic expression of C8orf76 consistently increased its mRNA expression (Figure 3A). Conversely, western blotting showed that in HCT116 and SW480 cells, C8orf76 knockdown decreased its protein level (Figure 3B). Moreover, RT-qPCR revealed that the mRNA expression of C8orf76 was lower in the C8orf76 knockdown group than in the control group (Figure 3B).
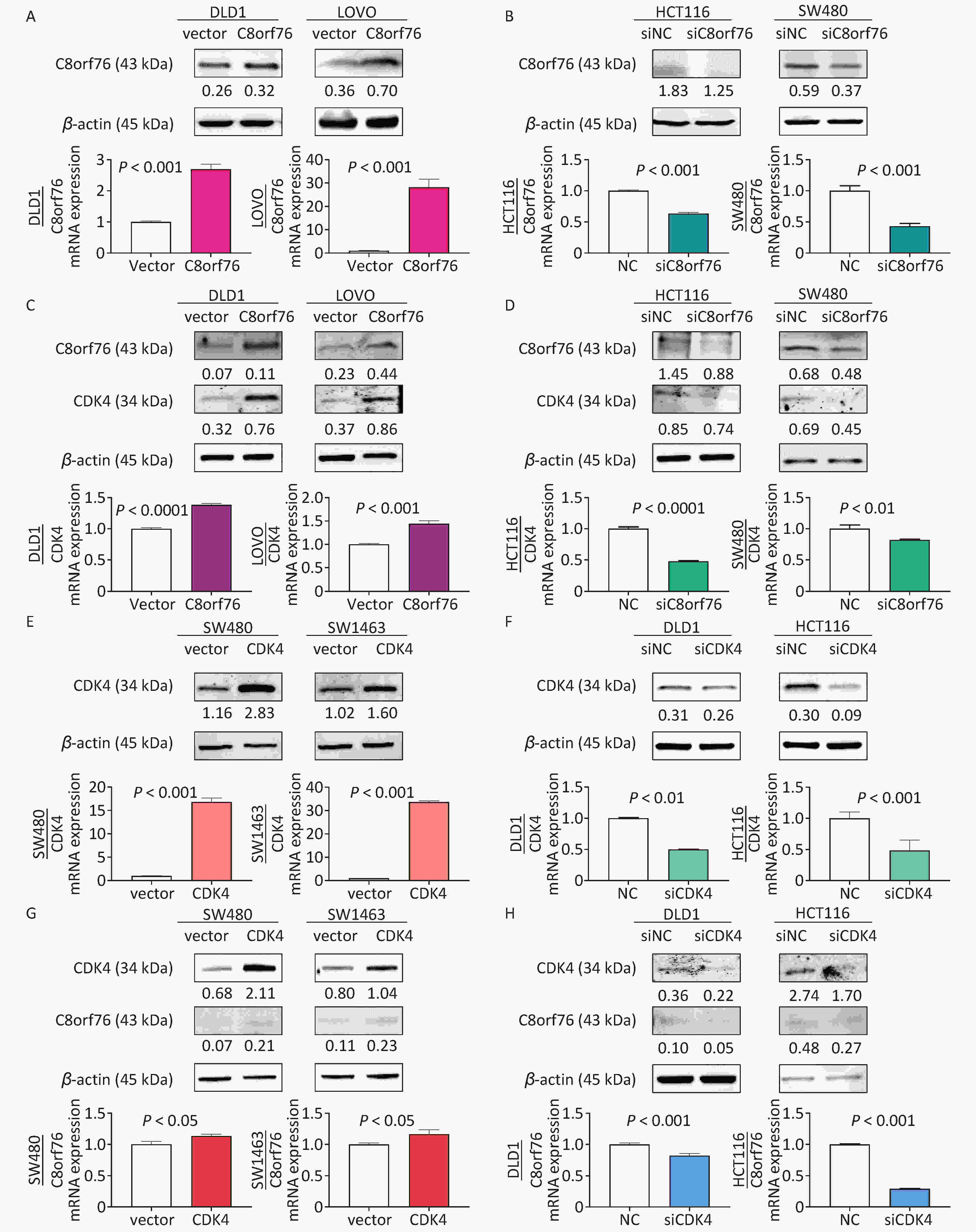
Figure 3. C8orf76 is positively correlated with CDK4 in CRC cell lines. (A) Overexpression of C8orf76 in DLD1 and LOVO cells was confirmed by western blotting and RT-qPCR. (B) Knockdown of C8orf76 in HCT116 and SW480 cells was confirmed by western blotting and RT-qPCR. (C and D) Overexpression of C8orf76 increased the protein and mRNA levels of CDK4 in DLD1 and LOVO cells, as seen from RT-qPCR and western blotting, whereas knockdown of C8orf76 decreases the expression of CDK4. (E) Overexpression of CDK4 in SW480 and SW1463 cells was confirmed by western blotting and RT-qPCR. (F) Knockdown of CDK4 in DLD1 and HCT116 cells as confirmed by western blotting and RT-qPCR. (G and H) Overexpression of CDK4 increases the protein and mRNA levels of C8orf76 in SW480 and SW1463 cells, as shown by RT-qPCR and western blotting, whereas knockdown of CDK4 decreases the expression of C8orf76. Error bars represent the mean ± SD.
Next, using western blotting, we found that the protein level of CDK4 in DLD1 and LOVO cells increased in the C8orf76 overexpression group (Figure 3C). RT-qPCR revealed that ectopic expression of C8orf76 consistently increased the CDK4 mRNA expression (Figure 3C). Conversely, western blotting revealed that C8orf76 knockdown decreased the CDK4 protein level in HCT116 and SW480 cells (Figure 3D). Additionally, RT-qPCR analysis showed that CDK4 mRNA expression decreased in the C8orf76 knockdown group (Figure 3D). We further explored the expression of CDK4 in CRC cell lines. Western blot analysis showed that ectopic expression of CDK4 increased its protein level in the human CRC cell lines SW480 and SW1463 (Figure 3E). RT-qPCR analysis revealed that ectopic expression of CDK4 consistently increased its mRNA expression (Figure 3E). Conversely, western blotting results showed that CDK4 knockdown decreased its protein level in DLD1 and HCT116 cells. Consistently, RT-qPCR results showed that CDK4 mRNA expression was decreased in the CDK4 knockdown group than in the control group (Figure 3F). In contrast, we found that the CDK4 overexpression increased the protein level of C8orf76 in SW480 and SW1463 cells, and RT-qPCR analysis showed that C8orf76 mRNA expression increased in the CDK4 overexpression group (Figure 3G). Conversely, CDK4 knockdown decreased the C8orf76 protein level in DLD1 and HCT116 cells, and RT-qPCR analysis showed that C8orf76 mRNA expression decreased in the CDK4 knockdown group (Figure 3H). In addition, western blot results showed that treatment with the CDK4 inhibitor LY2835219 (2 μmol/L) abolished the ectopic expression effect of C8orf76 (Figure S1). These results suggested that C8orf76 expression positively correlated with CDK4 expression.
-
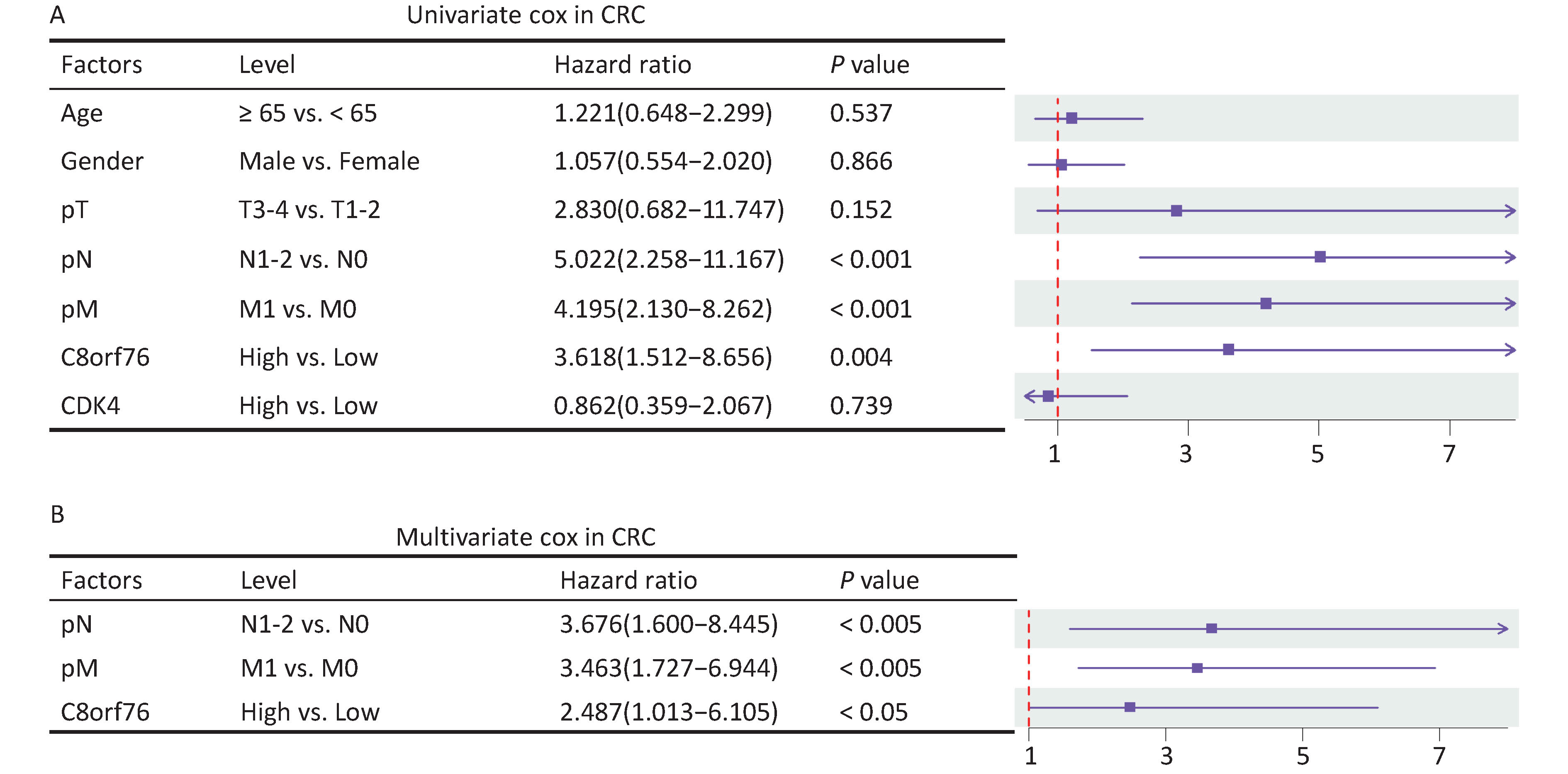
After adjusting for clinicopathological parameters such as age, gender, and T, N, and M stages, univariate and multivariate Cox regression analyses were performed on the cohort data. The results showed that the C8orf76 and CDK4 signatures were significantly correlated with OS, and these signatures were independent prognostic factors for OS in our cohort (Figures 4A and 4B). These results suggest that the C8orf76 and CDK4 signatures have important clinical significance as independent prognostic indicators.
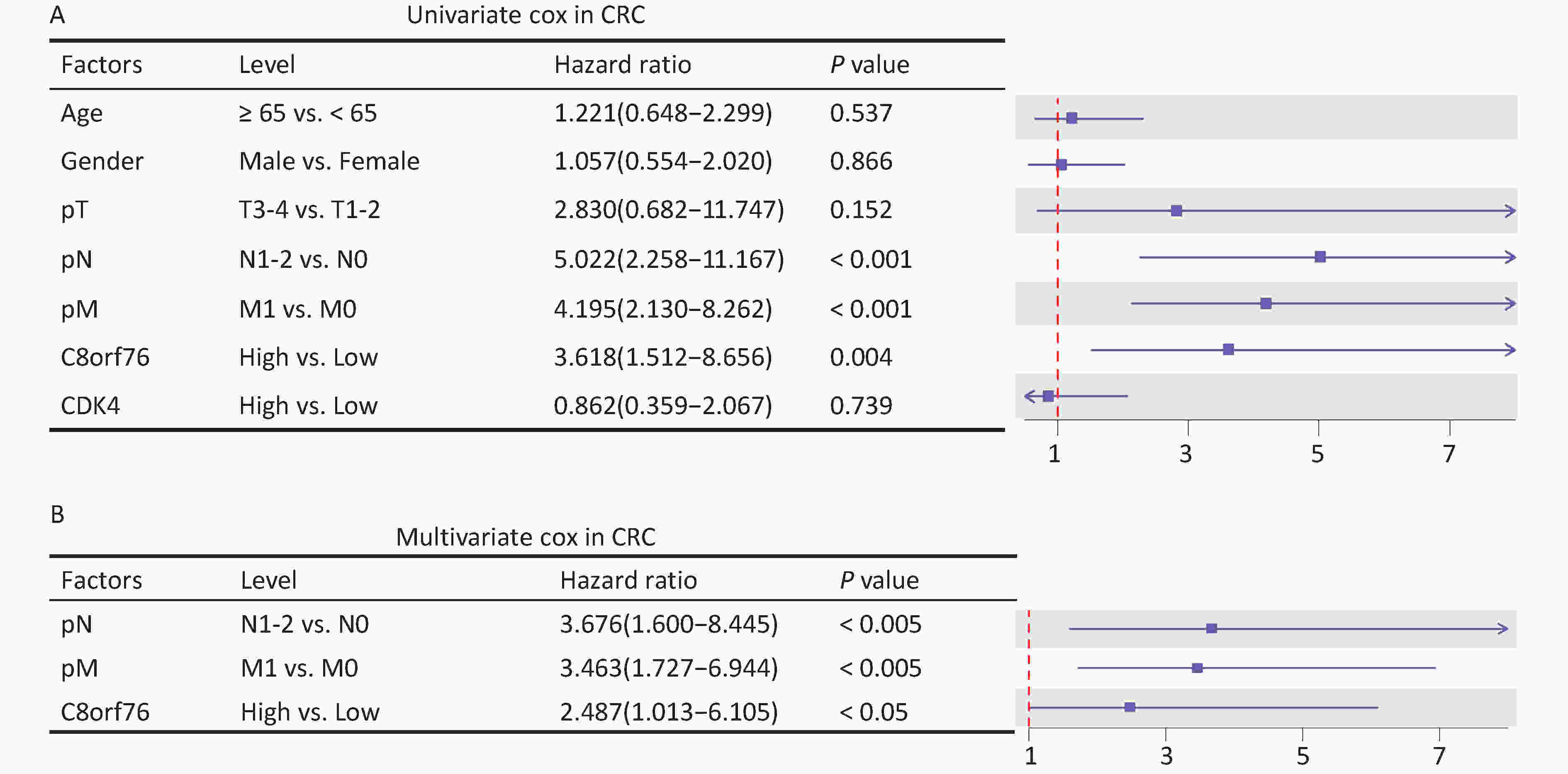
Figure 4. Association of the C8orf76 and CDK4 signatures with the overall survival (OS) of patients with CRC. (A) Univariate Cox regression analysis shows the clinicopathological parameters associated with the OS in patients with CRC in our cohort. (B) Multivariate Cox regression analysis shows the clinicopathological parameters associated with OS in patients with CRC in our cohort.
-
We constructed a nomogram to improve the accuracy of OS prediction in patients with CRC, including the N stage, M stage, C8orf76 scores, and CDK4 scores, to predict 1-year, 3-year, and 5-year OS probabilities in our cohort (Figure 5A). As shown in the calibration charts, the performance of the nomogram was appropriate for predicting 1-year, 3-year, and 5-year OS (Figures 5B, 5C, and 5D). The nomogram’s concordance index (C-index) in our cohort was 0.776.
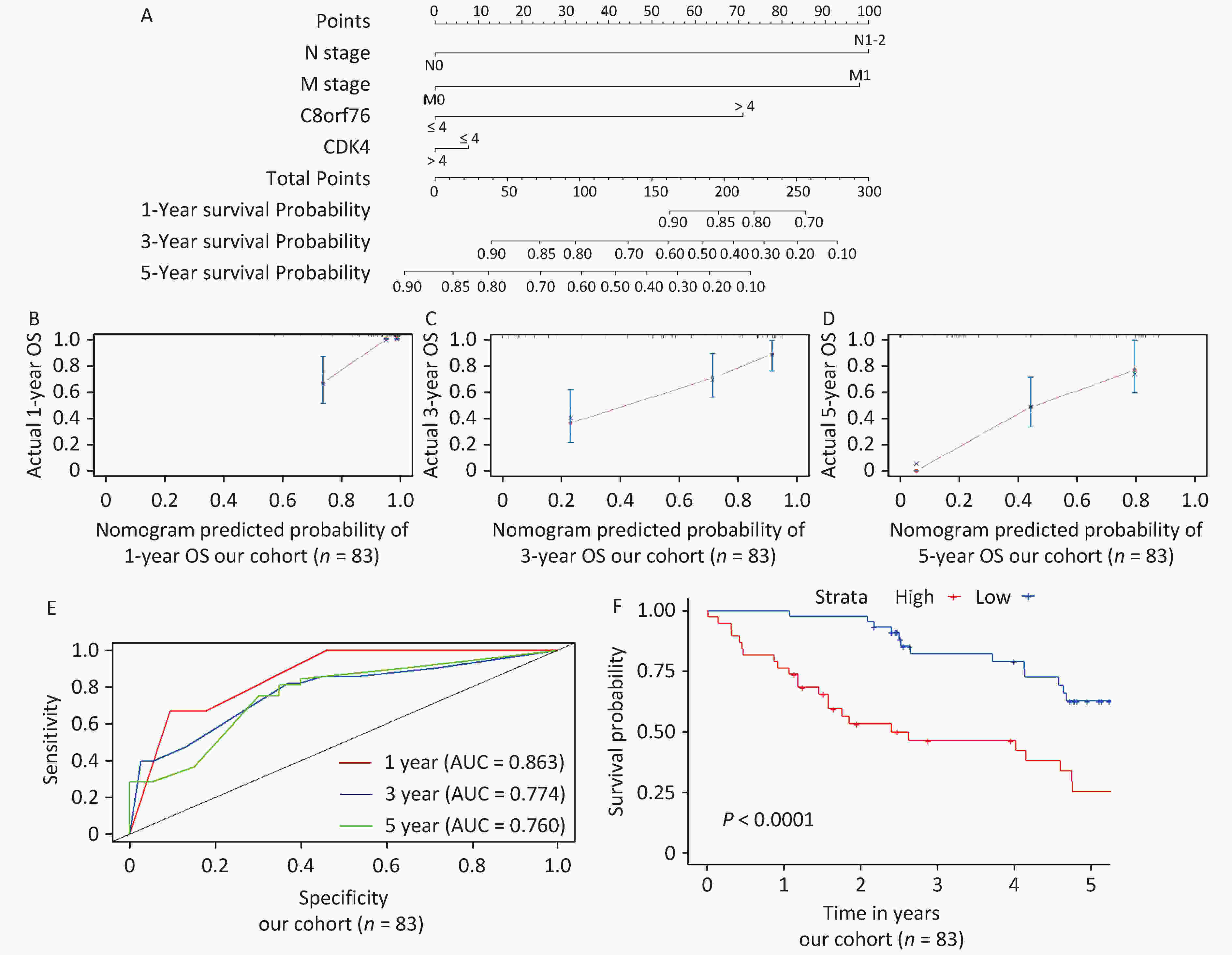
Figure 5. Establishment and validation of the nomogram for predicting overall survival (OS) of patients with CRC in our cohort. (A) Nomogram with N stage, M stage, C8orf76, and CDK4 scores for predicting 1-year, 3-year, and 5-year OS in patients with CRC. (B, C, and D) Calibration plot showing the comparison between the nomogram-predicted and actual 1-year, 3-year, and 5-year OS. (E) Time-dependent ROC curves showing the accuracy of OS prediction based on the nomogram. (F) Kaplan–Meier survival curves showing the OS of patients with CRC based on the nomogram.
Next, we used time-dependent receiver operating characteristic (ROC) curves to estimate the accuracy of the nomogram in predicting prognosis and found that the nomogram was significantly accurate in predicting 1-year, 3-year, and 5-year OS (area under the curve, AUC = 0.863, 0.774, and 0.760, respectively) (Figure 5E). Patients were divided into two groups according to quartiles, and the results showed that the prognosis of the high-risk group was significantly worse than that of the low-risk group (P < 0.0001) (Figure 5F).
-
The present study identified the potential value of C8orf76 and CDK4 in predicting CRC prognosis. Pearson’s correlation coefficient analysis was conducted to identify C8orf76-related genes in patients with CRC using the TCGA database. Among these, we selected 47 genes that correlated with non-CRC patients and 16 genes that correlated with CRC patients. PPI network analysis demonstrated that C8orf76 interacts with CDK4, CCNE2, CKS1B, and E2F5 in CRC, with CDK4 being the most critical interacting gene of C8orf76.
Chromosome 8q24.13, a common region of DNA copy number gain in CRC, and C8orf76, located on this chromosome, are associated with colorectal carcinogenesis[12]. Previous research has shown that C8orf76 promotes tumor growth in gastric cancer and is an independent prognostic factor for early-stage gastric cancer. This study assessed the clinical impact of C8orf76 in 592 patients with gastric cancer. C8orf76 was upregulated in two independent cohorts of gastric cancer and positively correlated with C8orf76 amplification. Moreover, gastric cancer patients with C8orf76 amplification or overexpression showed significantly reduced survival, particularly in the early stages of gastric cancer, indicating that C8orf76 serves as an independent prognostic factor in patients with early-stage gastric cancer. Biological functions of C8orf76 were investigated in vitro, in vivo, and in patient-derived organoid gastric cancer models. Mechanistically, dysregulation caused by aberrant expression of C8orf76 affected multiple signaling pathways, including MAPK/ERK signaling cascade along with cell cycle regulation, p53, Wnt, TGF-beta, apoptosis-related, and ErbB signaling pathways. The genes involved were mainly concentrated within the MAPK/ERK signaling cascade, which plays a crucial role in various cellular processes associated with tumorigenesis. C8orf76 binds directly to the AGGCTG motif located within the promoter region of lncRNA DUSP5P1, thereby activating DUSP5P1 transcription. DUSP5P1 induces MAPK/ERK signaling and promotes gastric tumorigenesis. Knockdown of DUSP5P1 abrogated the effects of C8orf76 on MAPK/ERK cascade activation and its tumor-promoting function[9].
Recently, Wang et al. demonstrated that C8orf76 was overexpressed in breast cancer tissues. Kaplan–Meier survival analysis showed that C8orf76 overexpression was associated with poor OS. Among the highly expressed phenotypes of C8orf76, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses showed significant enrichment in aromatase activity and the PPAR signaling pathway, whereas Gene Set Enrichment Analysis (GSEA) showed differential enrichment in receptor tyrosine kinase and PI3K/AKT signaling pathways, and some datasets related to the extracellular matrix and adhesion. Moreover, the reduced expression of C8orf76 led to reduced proliferation, increased apoptosis, and downregulation of pAKT and Bcl-2 in breast cancer cells. These findings suggested that high C8orf76 expression could serve as a potential biomarker for the diagnosis and prognosis of patients with breast cancer[11]. C8orf76 is also an oncogene in HCC, and it has been suggested that targeting the C8orf76/SLC7A11 pathway could improve anticancer therapy by inducing ferroptosis. By analyzing the publicly available TCGA database and their HCC patient cohorts, they observed higher mRNA expression of C8orf76 in HCC tissues than in adjacent non-tumor tissues. In addition, patients with increased C8orf76 levels exhibited worse OS. Furthermore, they found a positive association between C8orf76 overexpression and copy number amplification in HCC samples from the TCGA database. Downregulation of C8orf76 induced G1/S cell cycle arrest and inhibited cell proliferation. Moreover, they discovered that C8orf76 deficiency enhanced erastin- or sorafenib-induced ferroptosis by increasing the levels of lipid reactive oxygen species (ROS) levels. Furthermore, although C8orf76 overexpression have no effect on tumorigenesis under normal conditions, it promoted resistance to lipid disturbance and ferroptosis triggered by erastin or sorafenib. This ultimately facilitated HCC cell growth and tumor progression. Mechanistically, C8orf76 binds to the promoter region of SLC7A11 and transcriptionally upregulates SLC7A11 expression. SLC7A11-dependent cystine import results in sufficient glutathione synthesis and inhibition of lipid peroxidation, thereby accelerating tumor growth[10].
CDK4, located on chromosome 12q13, is a member of the serine/threonine kinase family and a key mediator in the cellular transition to S phase. CDK4 also serves as a bridge between extracellular signaling pathways and the cell cycle, and plays a critical role in the initiation, progression, and maintenance of many cancers[13,14].
CDK4 is overexpressed in various tumor types[15,16]. Various human tumors have lesions that hyperactivate cyclin D–CDK4/6[15]. For example, overexpression of cyclin D2, D3, or CDK4 or loss of p16INK4a promotes tumor formation[16], whereas knockdown of D-cyclins, CDK4, or CDK6 reduces tumor sensitivity[16]. CCND1- or CDK4-null or knock-in mice expressing kinase-inactive cyclin D1–CDK4/6 are resistant to human epidermal growth factor receptor 2 (HER2)-driven mammary carcinomas[17-20]. Likewise, in a KRAS-driven non-small-cell lung cancer mouse model, acute suppression of CDK4 inhibited proliferation and promoted senescence in tumor cells[21]. These observations indicate that CDK4 is a promising therapeutic target in cancer.
These findings underscore the importance of the identification of CDK4 inhibitors that regulate the cell cycle. Reports of the first CDK4/6 inhibitor, palbociclib, was published in 2004, and 11 years later, it was subsequently approved by the US Food and Drug Administration (FDA) for the treatment of breast cancer[22,23]. Since then, two other agents (ribociclib and abemaciclib) have been approved for the treatment of breast tumors, and all three have been well tolerated by patients[23]. Trilaciclib was recently approved to improve chemotherapy-induced bone marrow suppression in patients with small cell lung cancer[24,25].
In addition to cancer cell lines, other cell types including lymphocytes[26-28], fibroblasts[29], and endothelial cells[30], have also been investigated. In particular, CDK4/6 inhibitors can promote antitumor responses by directly stimulating the effector function of CD8+ T cells while inhibiting the proliferation of regulatory T (Treg) cells[26-28]. Several studies have highlighted the synergy between CDK4/6 inhibition and immunotherapy[26,27,31-33]. In a randomized trial, Trilaciclib-mediated immune stimulation improved the OS of patients with advanced triple-negative breast cancer[34]. In addition to CDK4/6 inhibitors, other immunotherapeutic strategies have shown promise in colorectal cancer. For instance, Liu et al. demonstrated that anti-OX40 antibody combined with HBc virus-like particles (VLPs) significantly delayed tumor growth in a mouse colon cancer model, highlighting the potential of immune modulation in CRC treatment[35].
Numerous studies have indicated that CDK4 may play an important role in the pathogenesis of several cancers; however, its interaction with C8orf76 during CRC progression remains unclear. Hence, we examined the correlation between C8orf76 and CDK4 and found that C8orf76 was positively correlated with CDK4 in the TCGA dataset as well as in our cohort. Accordingly, we analyzed the expression of C8orf76 in CRC cell lines. The results showed that ectopic expression of C8orf76 increased the mRNA and protein levels of CDK4, whereas C8orf76 knockdown decreased the mRNA and protein levels of CDK4. CDK4 knockdown decreased the mRNA and protein levels of C8orf76. These results suggest a positive correlation between C8orf76 and CDK4 expression.
The initiation and progression of CRC involve a highly heterogeneous tumor composition and complex oncogenic mechanisms involving a cascade of genetic and epigenetic alterations. Yang et al. demonstrated that miR-224-5p promotes CRC cell proliferation by targeting unc-51-like kinase 2 (ULK2) in a p53-dependent manner, demonstrating the complexity of CRC molecular mechanisms[36]. Therefore, it is crucial to develop individualized treatment strategies and evaluate patient prognosis. Hence, we validated the prognostic value of the C8orf76 and CDK4 signatures in CRC. Kaplan–Meier survival plots, ROC curve analyses, and univariate and multivariate Cox regression analyses showed that our signature could significantly predict OS in patients with CRC. We also found that the high-risk groups were correlated with more deaths, higher tumor stage, and lymph node and distant metastases in CRC. These clinicopathological features are considered determinants of OS.
Furthermore, we established a nomogram based on C8orf76 and CDK4 signatures and clinicopathological features to predict the prognosis of patients with CRC in our cohort. We included the expression of prognostic genes (C8orf76 and CDK4), pathological N, and pathological M, which could be used to predict the survival rate, in the nomograms from our cohorts. We validated these results in our cohort to verify their accuracy.
Although the signatures and nomograms constructed in this study using datasets from our patient database were robust, our findings need to be independently validated in additional CRC cohorts. In addition, our preliminary findings indicated that C8orf76 positively correlated with CDK4 expression. However, this study requires further in vivo validation; moreover, the upstream and downstream regulatory mechanisms of C8orf76 and CDK4 require further validation. Another limitation of this study was that the number of patients with CRC was relatively small.
-
In this study, we demonstrated a positive correlation between C8orf76 and CDK4 expression by verifying their expression in both the patient cohort and CRC cell lines. The expression of C8orf76 and CDK4 is closely associated with the malignant clinicopathological features of CRC. Clinical prediction charts were established by incorporating clinical characteristics, which revealed that the signatures of C8orf76 and CDK4 could contribute to the individualized prediction of CRC prognosis. This provides a basis for personalized and accurate treatment of patients with CRC. In summary, C8orf76 and CDK4 may serve as potential biomarkers for prognostic evaluation in CRC.
-
本研究有伦理审查添加Ethics ...; 本研究免于伦理审查请说明This study is exempt from ethical review.
Chromosome 8 open reading frame 76 (C8orf76) co-expressed with cyclin-dependent kinase 4 (CDK4) as a prognostic indicator of colorectal cancer
doi: 10.3967/bes2024.177
- Received Date: 2024-09-27
- Accepted Date: 2024-11-24
-
Key words:
- C8orf76 /
- CDK4 /
- Colorectal cancer /
- Nomogram /
- Prognosis signature
Abstract:
None of the authors report any financial or non-financial conflicts of interest.
&These authors contributed equally to this work.
| Citation: | Shang Guo, Chengcheng Liu, Zifeng Zhao, Zhongxin Li, Xia Jiang, Zengren Zhao. Chromosome 8 open reading frame 76 (C8orf76) co-expressed with cyclin-dependent kinase 4 (CDK4) as a prognostic indicator of colorectal cancer[J]. Biomedical and Environmental Sciences. doi: 10.3967/bes2024.177 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: