-
P. aeruginosa is a Gram-negative bacterium that often causes lung infections, trauma, and meningitis[1-2] in immunocompromised people. The pathogenicity of P. aeruginosa is often associated with toxicity and drug resistance. It has pathogenic structures such as lipopolysaccharides[3], flagella[4], and secretory systems represented by T6SS and T3SS[5-7]. These factors play important roles in invasion and colonization during host infection. Meanwhile, P. aeruginosa evades antibiotic attacks through complex drug resistance mechanisms. Owing to the resistance of P. aeruginosa and its prevalence in clinical settings, the World Health Organization has listed drug-resistant P. aeruginosa as a dangerous pathogen urgently in need of developing effective antibiotics[8-9].
Bacterial meningitis is an inflammatory disease of the central nervous system caused by bacteria[10]. The disease frequently affects people worldwide, with a mortality rate of up to 54%, and survivors often suffer from neurological sequelae, such as hearing loss and cognitive impairment[5,11-12]. The main clinical symptoms include headache, fever, and altered mental status[5]. The successful breach of the blood-brain barrier (BBB) by bacteria is critical for central nervous system infection and the development of meningitis[13-14]. The major cellular components of the BBB include microvascular endothelial cells, pericytes, and astrocytes[15-16]. In contrast, the tight junctions of endothelial cells and the highly restricted transcellular transport are key properties that enable the BBB to function[17-18]. Therefore, the successful damage to endothelial cells and subsequent penetration of the BBB are key processes in bacterial meningitis.
Among existing studies, research on the pathogenic mechanisms of bacterial meningitis has primarily focused on Streptococcus pneumoniae and Neisseria meningitidis. As the third most common pathogen causing bacterial meningitis, the virulence factors contributing to P. aeruginosa-induced meningitis should be further explored. In this study, we isolated P. aeruginosa A584 from clinical meningitis samples and deciphered its virulence factors and multi-drug resistance mechanism through bacterium-host interaction assays, whole genome sequencing, and bioinformatics analysis. It was found that A584 downregulated the expression of tight junction proteins Claudin-5 and Occludin in BBB-forming endothelial cells, altered the integrity of the BBB, and promoted the secretion of the inflammatory factors IL-1β and TNF-α in the brain. This study provides new insights into the mechanisms by which P. aeruginosa invades the BBB to cause bacterial meningitis and may guide the development of novel drug targets for this dangerous disease.
-
The P. aeruginosa strain A584 was isolated from the clinical cerebrospinal fluid of a patient with meningitis. Clinical strains P215, P220, P226, P232, C305, C324, C329, C348, and C393 were isolated from the CSF of patients without meningitis. Strains P215, P220, P226, and P232 were obtained from Tianjin Huanhu Hospital, and strains C305, C324, C329, A584, C348, and C393 were obtained from Tianjin Third Central Hospital. All of these strains are P. aeruginosa strains. These strains were stored in the Culture Collection of Nankai University. The standard strain PAO1 was obtained from the ATCC. The Staphylococcus aureus strain Sa1, which has a strong hemolytic capacity, was isolated from a clinical fecal sample. The strains were cultured on BHI blood agar, and fresh strains were incubated in BHI blood medium at 37 °C and 160 rpm overnight until they reached the logarithmic phase for subsequent experiments. The mouse endothelial bEnd.3 cells were cultured in DMEM medium (Dingguo, China) containing 10% fetal bovine serum and 1% penicillin-streptomycin in a humidity incubator 37 °C with 5% CO2.
-
Healthy female 5-week-old ICR mice (Huafukang, China) were randomly assigned to groups, with six mice per group. Five concentration gradients were established. Different concentrations of bacterial suspensions (104–108 CFU/ml) were injected via the caudal vein at a volume of 100 μL each. PBS was used as the control. Simultaneously, the bacterial solution was serially diluted to appropriate concentrations in a doubling ratio, and the number of viable bacteria was counted by plating. The mice were observed continuously for 7 days, and deaths were recorded every 12 h. The results are presented as the LD50, calculated using Karber's method. The experimental protocol was approved by the Animal Care and Use Committee of Nankai University.
The brain and lung tissues were retrieved from the mice. Some samples were fixed in 4% paraformaldehyde for 24 h and embedded in paraffin for sectioning. The sections were stained with H&E (Solarbio, China), and images were obtained using a light microscope. Another portion of the collected brain and lung tissue was homogenized in sterile water and diluted by 104 times. The diluted homogenates were plated onto solid YPD blood agars and cultured at 37 °C. After 24 h, colonies were counted.
-
The bEnd.3 cells were inoculated into the upper layer of the Transwell plate and cultured in 1 mL DMEM in the lower layer for 10 days. On day 10, the medium was changed to 1 mL of serum-free DMEM, and 10 μL of fluorescein Isothiocyanate fluorescence (FITC)-labeled dextran (1 mg/mL, BioLegend, China) was added to the upper layer. After being cultured at 37 °C for one hour without light, the lower medium was collected for fluorescence intensity assessment by enzyme-labeler to ensure the successful construction of a BBB model. A P. aeruginosa suspension (2 × 105 cells) was added to the upper layer of the Transwell, and the same volume of PBS was used as the control. FITC-labeled dextran (1:200, BioLegend, China) was added as described above and the cells were incubated in darkness for one hour. The lower medium was collected for fluorescence intensity assessment using a fluorescence microplate reader (Enpsire, USA).
-
After four hours of incubation of both the bEnd.3 and P. aeruginosa cells, the wells were washed with PBS. The infected endothelial cells were stained with LysoTracker Red (1:20,000, Beyotime, China), incubated at 37°C in darkness for 4 h, and stained at 37 °C in darkness for 30 min with DAPI (1:100, Sigma, USA). After staining, the cells were washed twice with PBS and observed under a confocal microscope (Nikon, Japan).
-
The bEnd.3 cells were seeded in 96-well plates at a density of 2 × 103 cells/well. Bacterial cells were suspended in PBS and inoculated into bEnd.3 wells at a density of 2 × 104 cells/well. PBS was added as a control and incubated for 12 h at 37 °C. According to the manufacturer's protocol, 20 μL LDH release reagent was added into the positive control well, blowing and mixing was repeated, and incubated for 1 h. The cells were then centrifuged at 400 × g for 5 min. The supernatant was transferred to a 96-well plate. Each well was added with 60 μL LDH working solution and incubated at 25 °C for 30 min in darkness. The absorbance was measured at 490 nm using a fluorescent microplate reader (Enpsire, USA).
-
Two microliters of P. aeruginosa suspension at a concentration of 1×10 CFU/mL were inoculated onto the BHI blood agar medium. Staphylococcus aureus was used as the positive control to compare the sizes of the hydrolytic rings around the strains. Meanwhile, precisely 200 μL of the bacterial suspension (1 × 10⁸ CFU/mL) and an equal volume of 200 μL of the red blood cell suspension were accurately added to a 24 - well plate. A suspension of red blood cells (RBCs) was added in equal volumes of PBS and solutions containing PAO1, A584, or other P. aeruginosa cells. After three hours of incubation, the sample was centrifuged at 1,200 rpm for 3 min, and 200 μL of supernatant was collected to measure the absorbance at 490 nm with a microplate reader.
-
One mL of the DMEM suspension containing 1 × 106 bacterial cells was inoculated into a 6-well plate containing 2 × 105 bEnd.3 cells and incubated for 4 h. Total RNAs was extracted from the cells using TRIzol (Dingguo, China) and then reverse-transcribed into cDNA. The SYBR Green qPCR SuperMix (TransGen Biotech, China) was used for RT-quantitative PCR, and the expression levels of the genes to be measured were normalized by that of the β-actin expression using the method 2−ΔΔCt.
-
Whole-genome sequencing of the A584 cells was performed. The protein sequences of A584 were compared with the virulence factor database, and the virulence genes were determined with a similarity > 60% and an E value < le-5[19]. The Comprehensive Antibiotic Resistance Database (CARD)[20] was used to identify drug resistance genes.
-
The SPSS software (IBM) was used for statistical analysis. All experiments were repeated thrice. All data are expressed as mean ± standard error. Student’s t-test or one-way analysis of variance (ANOVA) were used to calculate statistical significance between groups. Statistical significance was set at P < 0.05. significant.
-
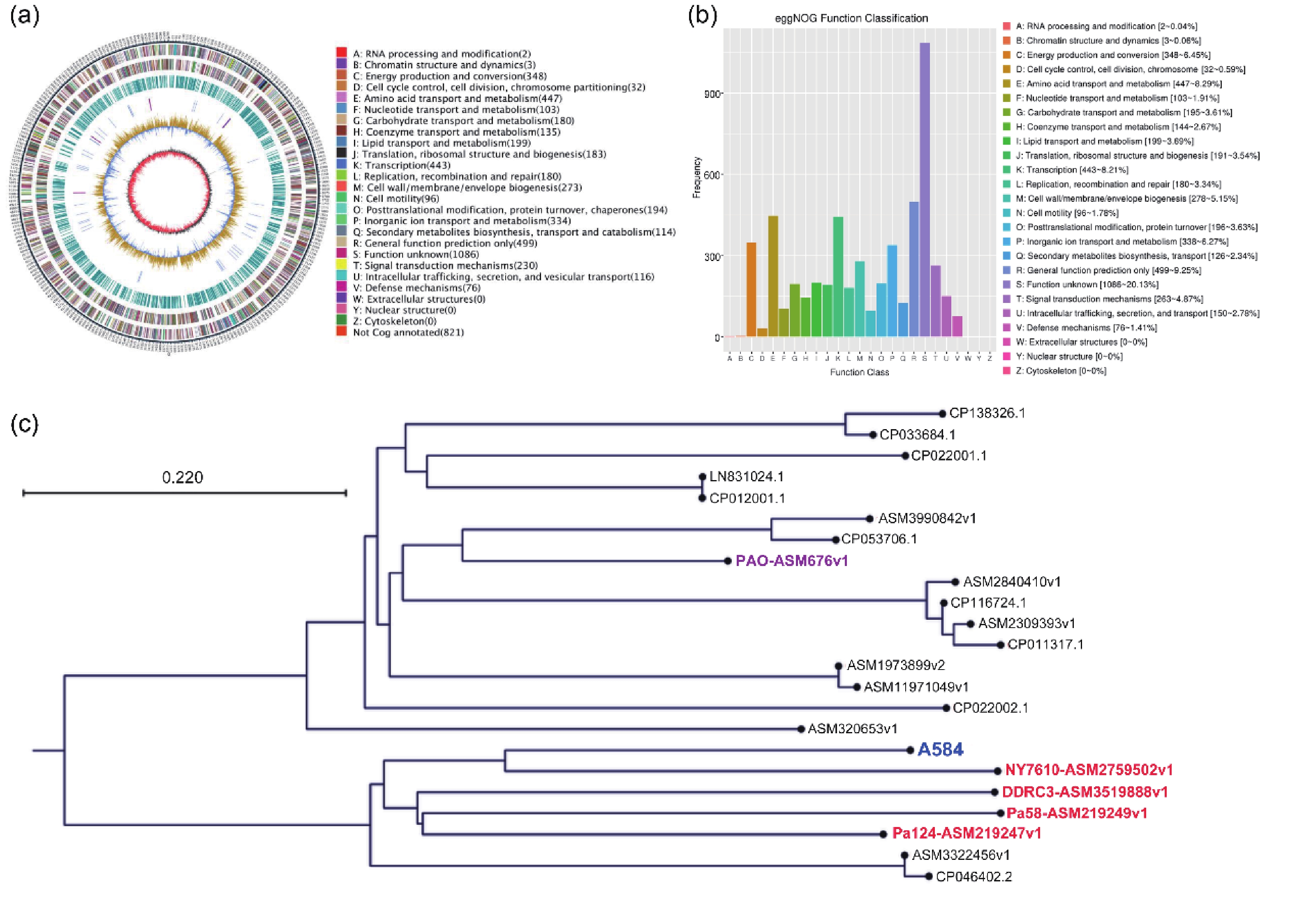
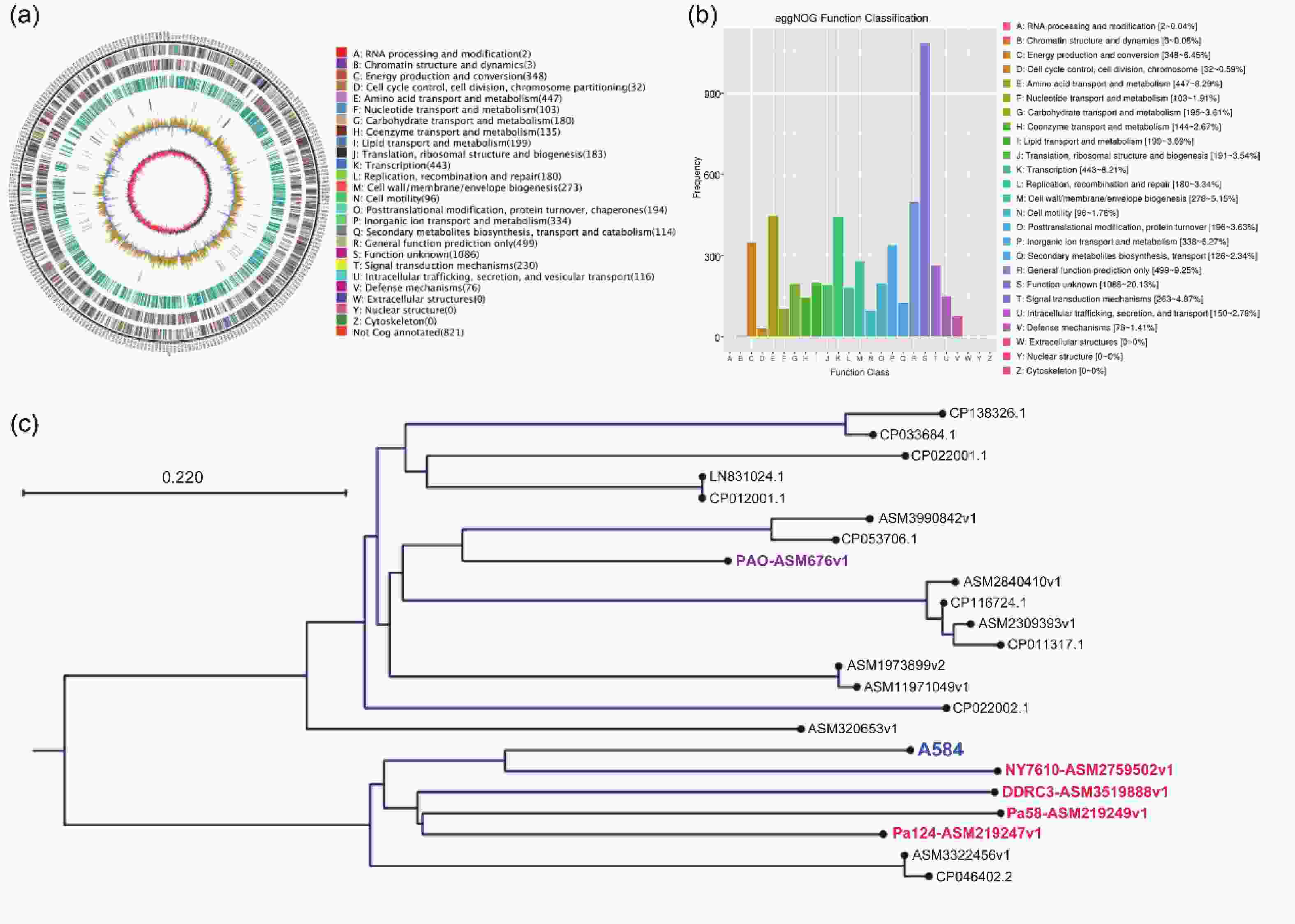
To investigate the relationship between P. aeruginosa strain A584, isolated from clinical meningitis tissues, and meningitis, we performed whole-genome sequencing of A584 to gain a preliminary understanding of the strain’s basic characteristics. Genome assembly results showed that strain A584 had a unique ring chromosome with a size of 5,974,167 bp and a GC content of 66.17% (Figure 1A). The A584 genes were predicted based on whole-genome sequence results, and the predicted coding sequences were annotated by protein homology alignment using the eggNOG-mapper functional annotation database. A total of 6,094 genes were predicted in the entire genome. Among them, 5,394 genes correspond to the eggNOG database. “General functional prediction only” had the highest number of genes (499), followed by “Amino acid transport and metabolism” (447), “Transcription” (443), “Energy production and conversion” (348), and “Inorganic ion transport and metabolism” (338), they are the most gene-rich class in the eggNOG group (Figure 1B). To determine the genomic variability and phylogenetic relationships among the different strains of P. aeruginosa, we performed a comparative genomic analysis of genomic assembly and constructed a genome-wide phylogenetic tree. The results showed that A584 clustered with NY7610-ASM2759502v1, DDRC3-asm3519888v1, Pa58-ASM219249v1, and Pa124-asm219247v1, suggesting that these five strains had similar biological characteristics (Figure 1C).
-
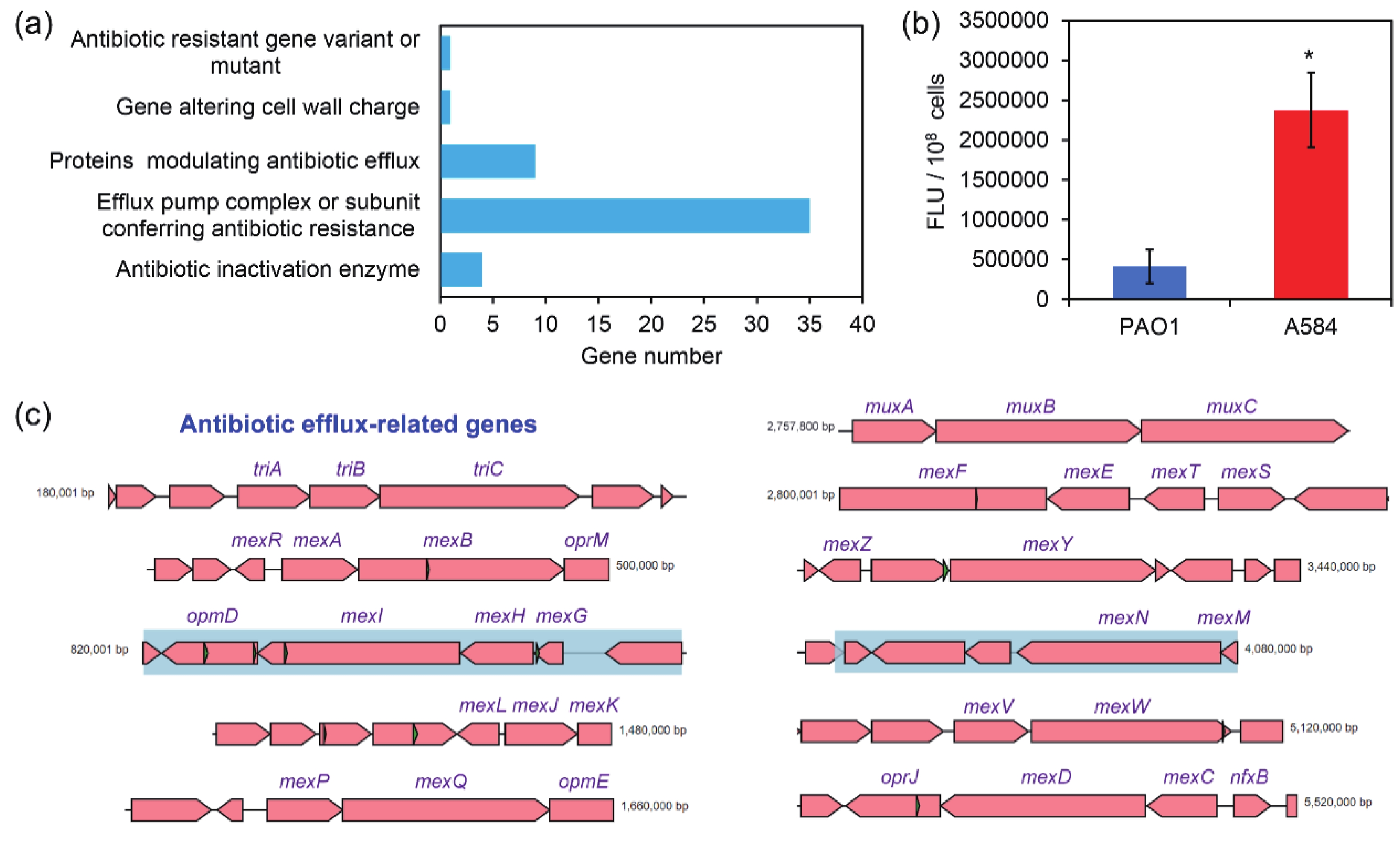
P. aeruginosa is resistant to various antibiotics, and its resistance mechanisms are mainly attributed to intrinsic, acquired, and adaptive resistance. Intrinsic resistance is primarily related to membrane permeability, efflux pump expression, and the production of antibiotic inactivation enzymes[21]. As revealed by drug sensitivity assays, A584 exhibited strong resistance to a wide range of antibiotics, including the aminoglycosides amikacin (≥ 64 mg/L) and tobramycin (≥ 16 mg/L), the β-lactams cefepime (≥ 32 mg/L), ceftazidime (≥ 64 mg/L), and imipenem (≥ 16 mg/L), the quinolones ciprofloxacin (≥ 4 mg/L) and levofloxacin (≥ 8 mg/L), together with β-lactamase inhibitors tazobactam (≥ 128 mg/L), clavulanic acid (≥ 128 mg/L) and sulbactam (≥ 64 mg/L) (Table 1).
Type Kind IC50 (mg/L) aminoglycoside amikacin ≥ 64 tobramycin ≥ 16 β-lactam cefepime ≥ 32 ceftazidime ≥ 64 imipenem ≥ 16 quinolone ciprofloxacin ≥ 4 levofloxacin ≥ 8 β-lactamase inhibitor tazobactam ≥ 128 clavulanic acid ≥ 128 sulbactam ≥ 64 Table 1. IC50 values of the P. aeruginosa strain A584 to different kinds of antibiotics
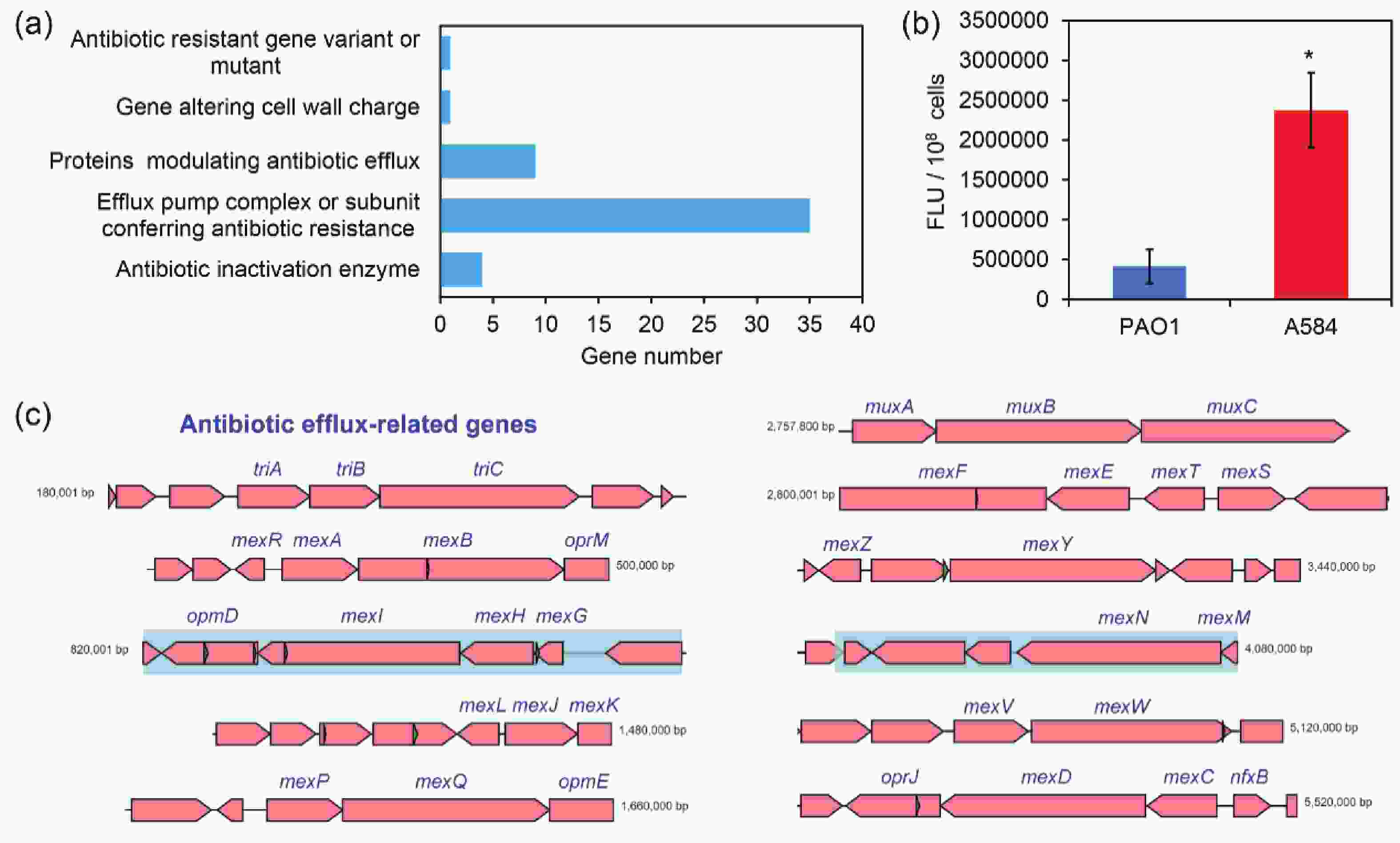
According to the CARD comparison, strain A584 harbored a series of antibiotic resistance genes. Among them, most belonged to genes involved in efflux pump complexes or subunits that confer antibiotic resistance. This indicates that the drug resistance of A584 cells is closely related to efflux pumps (Figure 2A and 2C). In addition, “antibiotic inactivation enzyme” genes ranked third. Consistent with this, we conducted a rhodamine 6G efflux experiment and found that the efflux capacity of A584 cells was significantly higher than that of PAO1 cells (Figure 2B). Therefore, A584 exhibits strong resistance to a wide range of antibiotics, which is associated with drug efflux. Interestingly, CARD analysis of known drug-resistance genes further revealed that A584 possessed a distinct OXA-488 gene (encoding an antibiotic inactivation enzyme) compared to PAO1 (Supplementary Table S1), highlighting the importance of this gene in A584’s drug resistance.
-
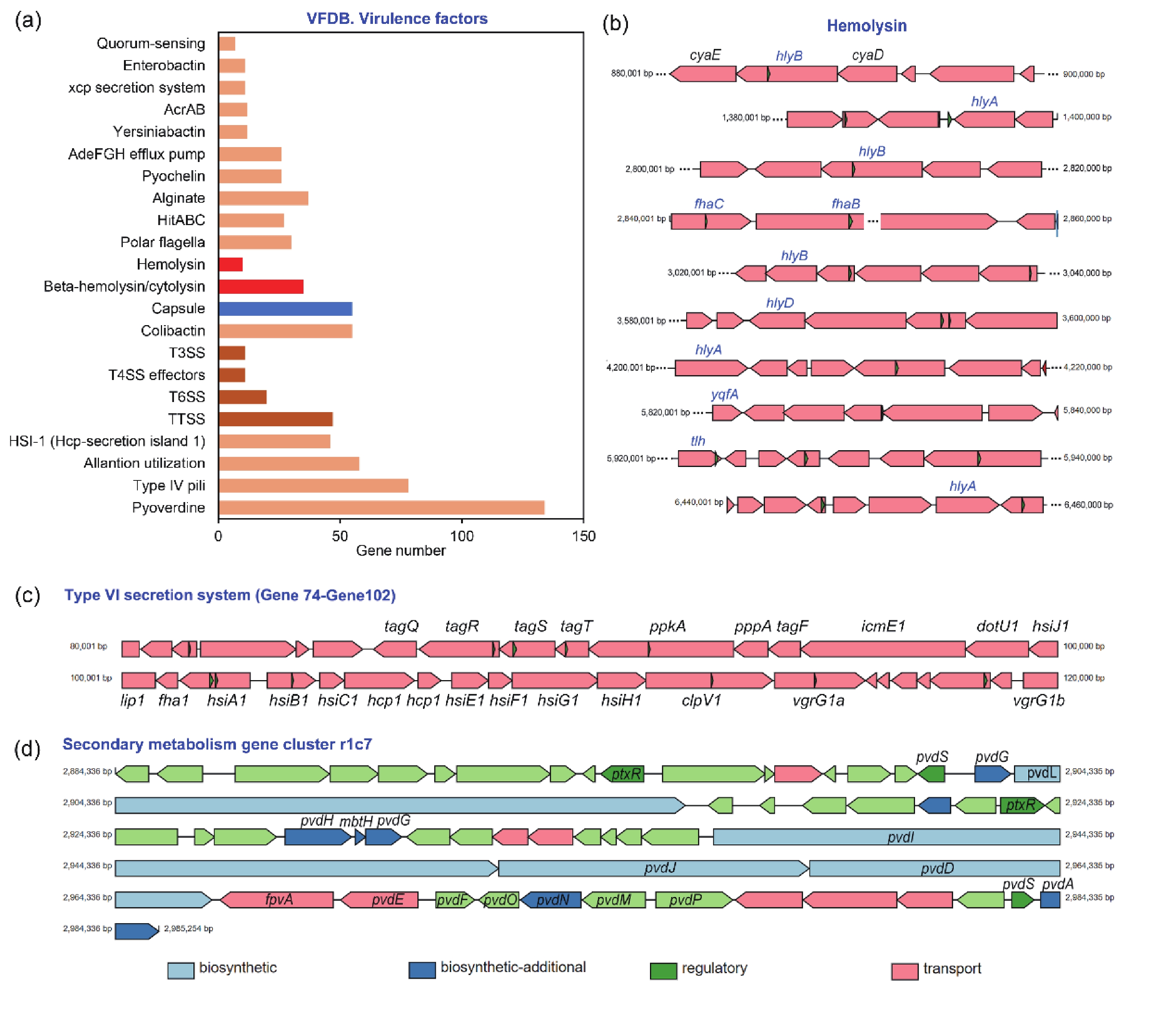
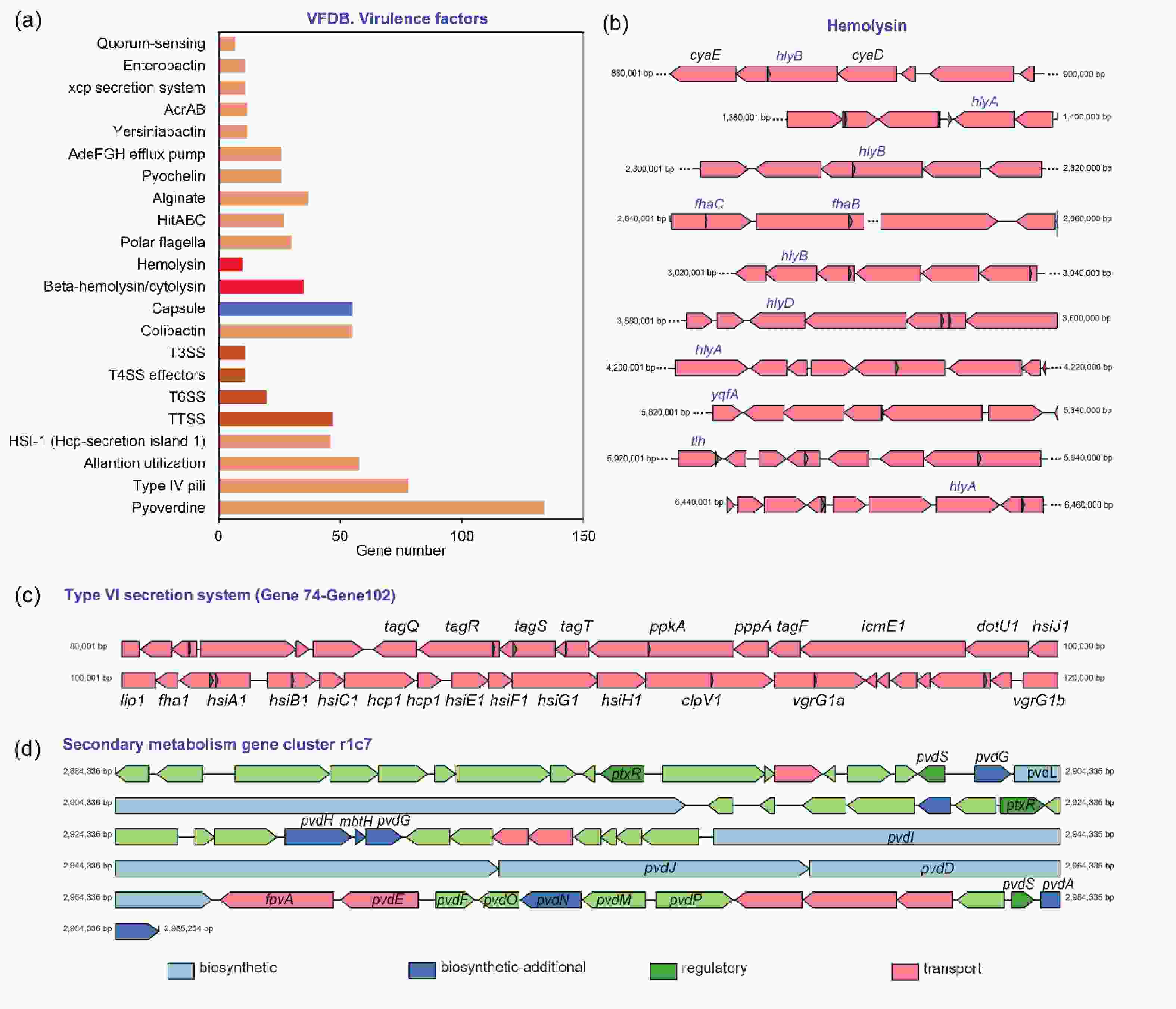
P. aeruginosa can adapt to adverse host environments by secreting a variety of virulence factors that contribute to successful infection and cause disease[22]. By comparing the virulence factors carried by A584 in the VFDB database (Figure 3A), we found that it carried more virulence genes related to pyoverdine, type IV pili, and capsules, which contribute to the growth and adhesion of A584. Additionally, A584 contained more genes related to ‘Hemolysin’ and ‘Beta hemolysin’ (Figure 3B), which lyse red blood cells and promote pathogen invasion, consistent with the results of our hemolysis experiment. Furthermore, secretory systems such as T3SS, T4SS, and T6SS are present in the strain (Figure 3C), and they express secreted virulence factors involved in protein secretion, colonization, and adhesion of A584. Moreover, this strain possessed the secondary metabolism gene cluster r1c7 (Figure 3D). Secondary metabolites play a role in host-pathogen survival, competition, and interaction with host tissues. The gene cluster r1c7 contains a series of genes related to the synthesis, secretion, and function of secondary metabolites, which may contribute to the virulence of A584. VFDB comparison also revealed that A584 had a distinct gene, exoU (encoding phospholipase A2), compared to PAO1 (Supplementary Table S2), suggesting that this gene may be involved in enhanced virulence in A584.
-
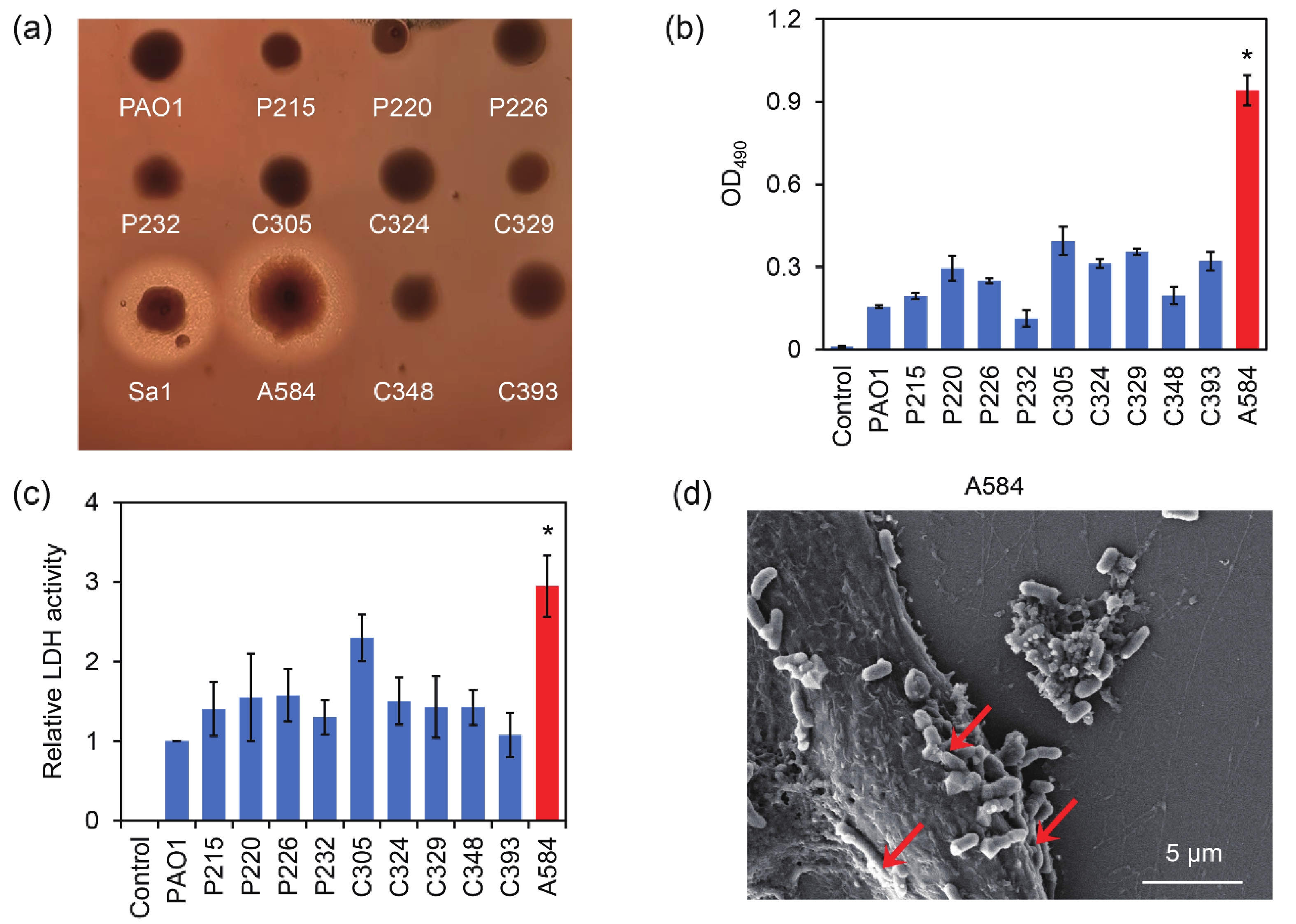
The strength of the hemolytic ability of pathogenic bacteria is often positively correlated with their virulence. These pathogens often produce hemolytic toxins that cause red blood cell lysis, causing damage to the body. To screen for hypervirulent P. aeruginosa, a series of the strains isolated from clinical cerebrospinal fluid samples, together with the standard strain PAO1 and the positive control Staphylococcus aureus strain Sa1, were used for hemolysis assays on BHI blood agar. The results showed that strain A584, together with S. aureus strain Sa1 (positive control), produced obvious hemolytic rings. In contrast, the other tested strains did not produce hemolytic rings (Figure 4A). The results indicated that A584 had a stronger hemolytic ability than the other P. aeruginosa strains. Consistently, as revealed by the measurement of the absorbance of the red blood cell supernatants, the strain A584 led to the highest absorbance of the supernatants among the tested P. aeruginosa strains, indicating that the highest levels of hemoglobin were released from red blood cells induced by A584 (Figure 4B). Therefore, A584 showed the strongest hemolytic ability among all P. aeruginosa strains.

Figure 4. Hemolysis and epithelial cell damage caused by the clinical isolated P. aeruginosa strains. (A) Image of the strains cultured in the BHI blood agar. (B) Hemolysis induced by the strains in the red blood cell suspensions. (C) LDH activity in the supernatants of the bEnd.3 endothelial cells after 12 h of treatment by the P. aeruginosa strains. (D) SEM image of the bEnd.3 cells treated by the A584 strain. The red arrows indicate the bacterial cells invading the epithelial cells.
During host cell invasion, pathogens commonly damage the host cell membrane, leading to the release of lactate dehydrogenase (LDH) from the host cells[23]. The LDH assay is commonly used to detect pathogen-induced cytotoxicity[24]. By measuring LDH release from cells, we found that LDH activity in the supernatant of A584-treated bEnd3 cells was significantly higher than in the other groups (Figure 4C). Additionally, the interaction between A584 and bEnd.3 was observed by scanning electron microscopy. The A584 strain strongly adhered to the surface of bEnd.3 and invaded host cells (Figure 4D). LD50 assays further revealed that the LD50 of PAO1 was 106 CFU/mL, while the LD50 of strain A584 was 105.167 CFU/mL. This suggests that the virulence of strain A584 in mice was greater than that of strain PAO1. Taken together, these results demonstrate that strain A584 has the highest virulence among the tested clinical P. aeruginosa strains.
-
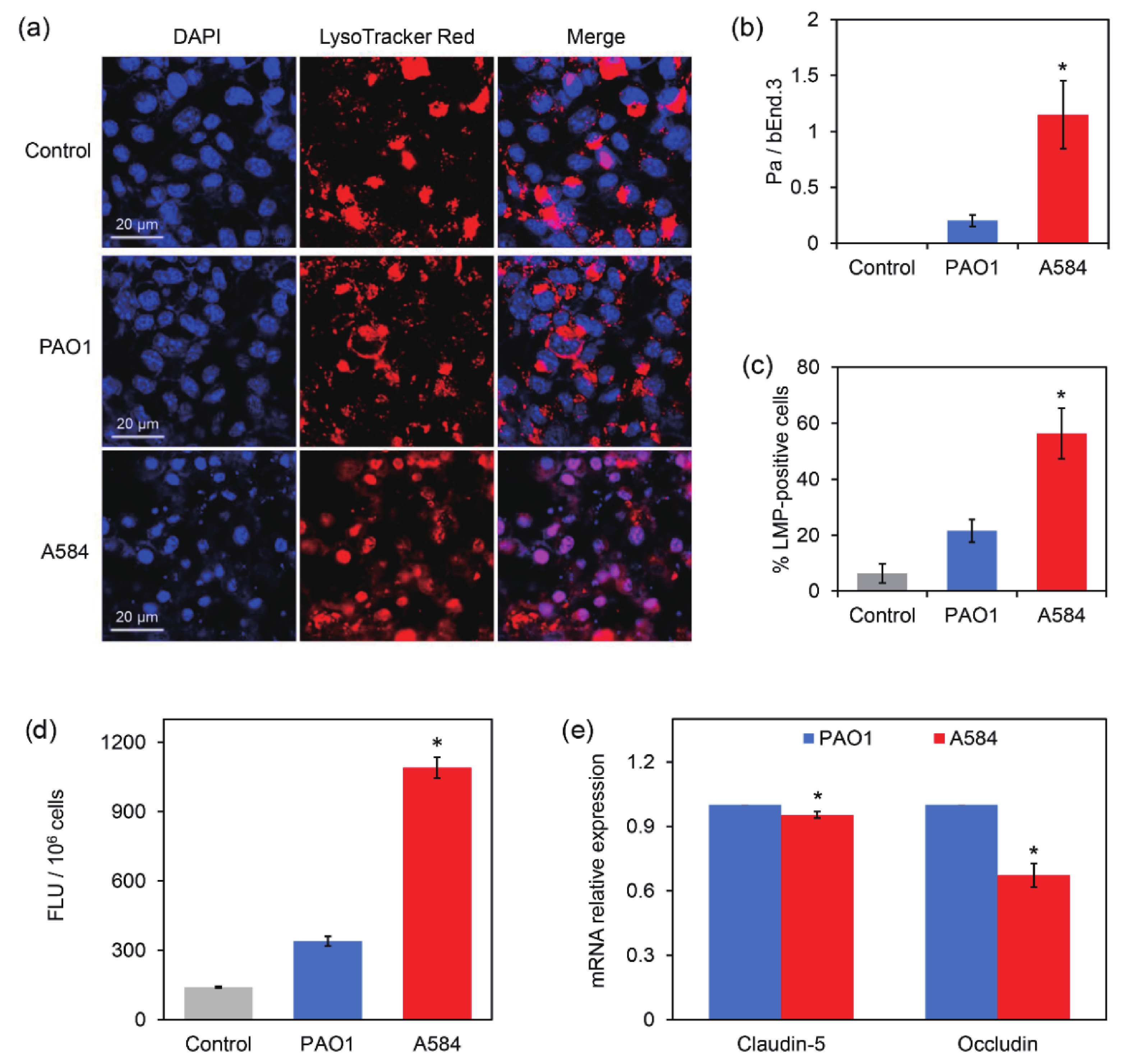
The BBB is a dynamic interface between the blood and brain tissue that selectively obstructs the passage of substances, playing an important role in maintaining homeostasis of the central nervous system and protecting the central nervous system from pathogen invasion[25]. Endothelial cells are key components of the BBB. A BBB model was constructed using bEnd.3 mouse endothelial cells. The P. aeruginosa strains were then added to the dense endothelial cell layer for 4 h. Lysosome staining and confocal microscopy revealed that the Control and PAO1-treated groups contained intact lysosomes localized in endothelial cells. However, most bEnd.3 cells treated with A584 had damaged lysosomes, widely distributed throughout the cells (Figure 5A). Statistical analysis showed a higher ratio of P. aeruginosa cells to bEnd.3 cells and a higher percentage of lysosome membrane permeabilization (LMP)-positive cells in the A584 group than in the Control and PAO1 groups (Figure 5B–5C). These results indicate that A584 causes significant damage to the BBB.
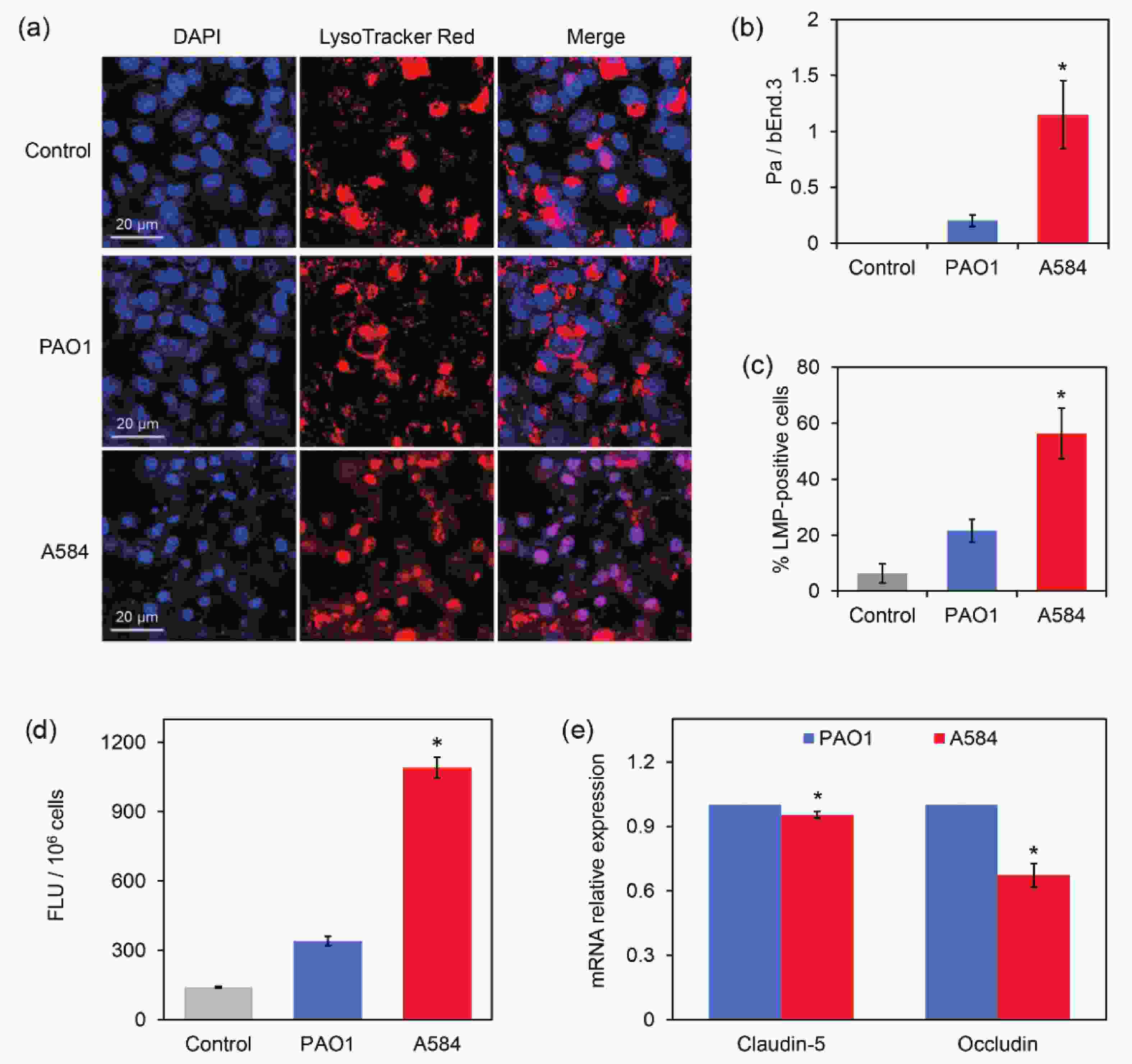
Figure 5. Damage of the bEnd.3-derived blood brain barrier model by the P. aeruginosa strains PAO1 and A584. (A) Confocal images of the blood brain barrier model. The cells were stained by DAPI and LysoTracker Red for 20 min before observation. (B) The ratio of Pa cells to bEnd3 cells. (C) The percentage of LMP-positive bEnd.3 cells. (D) Comparison of fluorescence intensity in blood-brain barrier model damaged by PAO1 and A584 (E) Relative expression of claudin-5 and occludin in the infected cells.
To further confirm the BBB damage, FITC-labeled dextran was added to the treated cell layer, followed by incubation in the dark for 1 h, and measurement of the FITC fluorescence intensity in the penetration liquid. Compared to the other groups, the A584-treated group exhibited a much higher fluorescence intensity signal in the penetration liquid than the PAO1 group (Figure 5D). This result confirmed the severe damage to the BBB caused by A584.
To further investigate the mechanisms by which A584 penetrates the BBB, bEnd.3 cells treated with P. aeruginosa strains were used for RNA extraction and qPCR analysis of Claudin-5 and Occludin expression. These two proteins are critical tight junction proteins that maintain the integrity of the BBB[26]. The results showed that the expression of these two genes were significantly downregulated in the A584-treated group compared to the PAO1-treated group (Figure 5E). Therefore, A584 significantly disrupted the BBB, which may be associated with the downregulation of tight junction proteins.
-
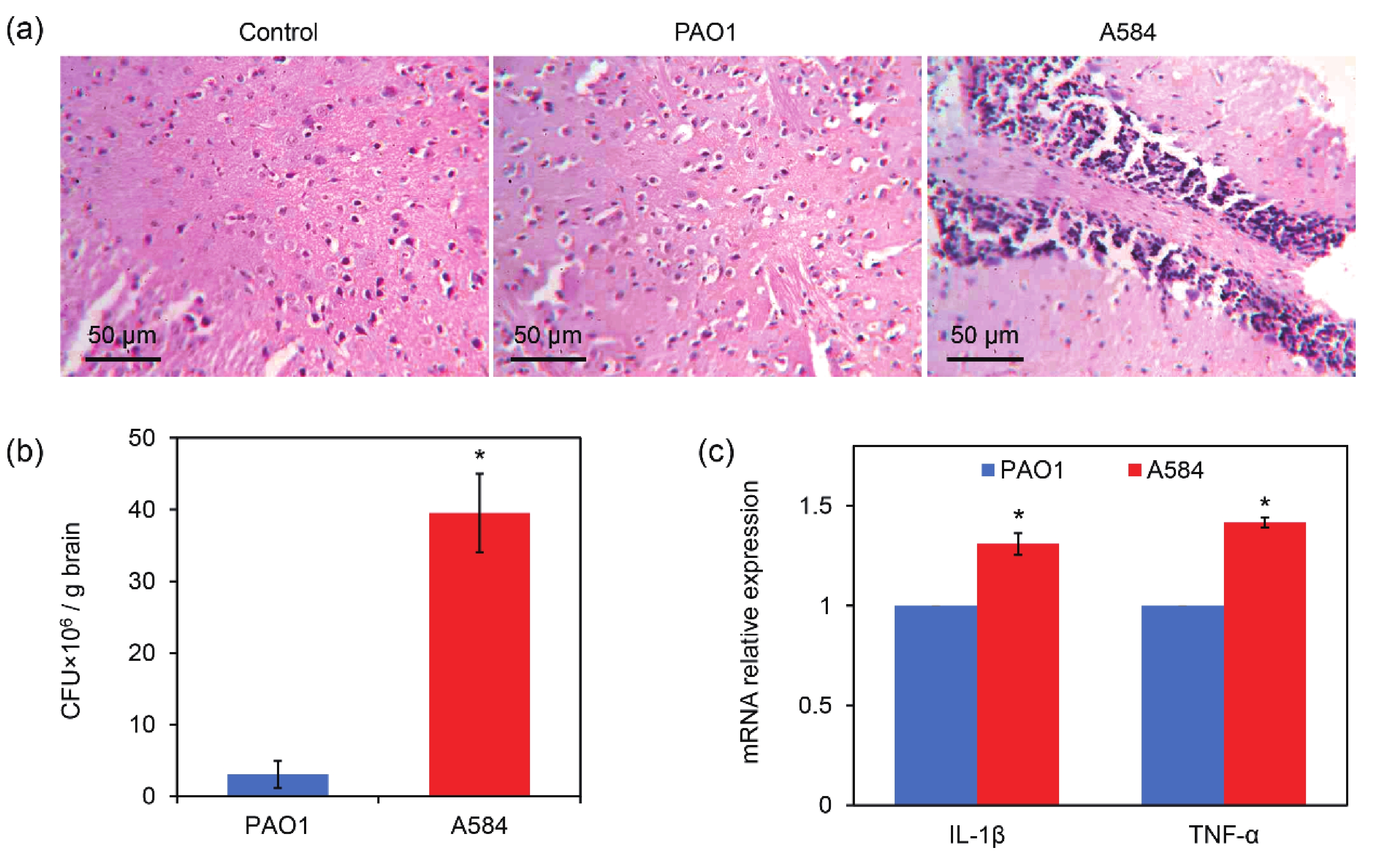
An animal model of A584 infection was established to observe the behavior of mice, and their brains and lungs were dissected for histopathological observation. The results showed that Control and PAO1-treated mice had intact brain tissue with no obvious inflammation (Figure 6A). In contrast, there was inflammatory cell infiltration and tissue damage in the brains of the A584-challenged group (Figure 6A). CFU assays of the tissue samples further showed that the number of P. aeruginosa cells in the brain tissues was much higher in the A584 group than in the PAO1 group (37.1 × 106 CFU/g brain versus 2.5 × 106 CFU/g brain, Figure 6B). Interestingly, we found that mice in the PAO1 challenge group died earlier than those in the A584 treatment group. In addition, the expression levels of IL-1β and TNF-α genes in mouse brain tissues were detected. The results showed that the expression levels of these two inflammatory cytokines were higher in the A584-challenged group than in the PAO1 group (Figure 6C). Hence, A584 can severely invade mouse brain tissue, resulting in inflammation and tissue damage.
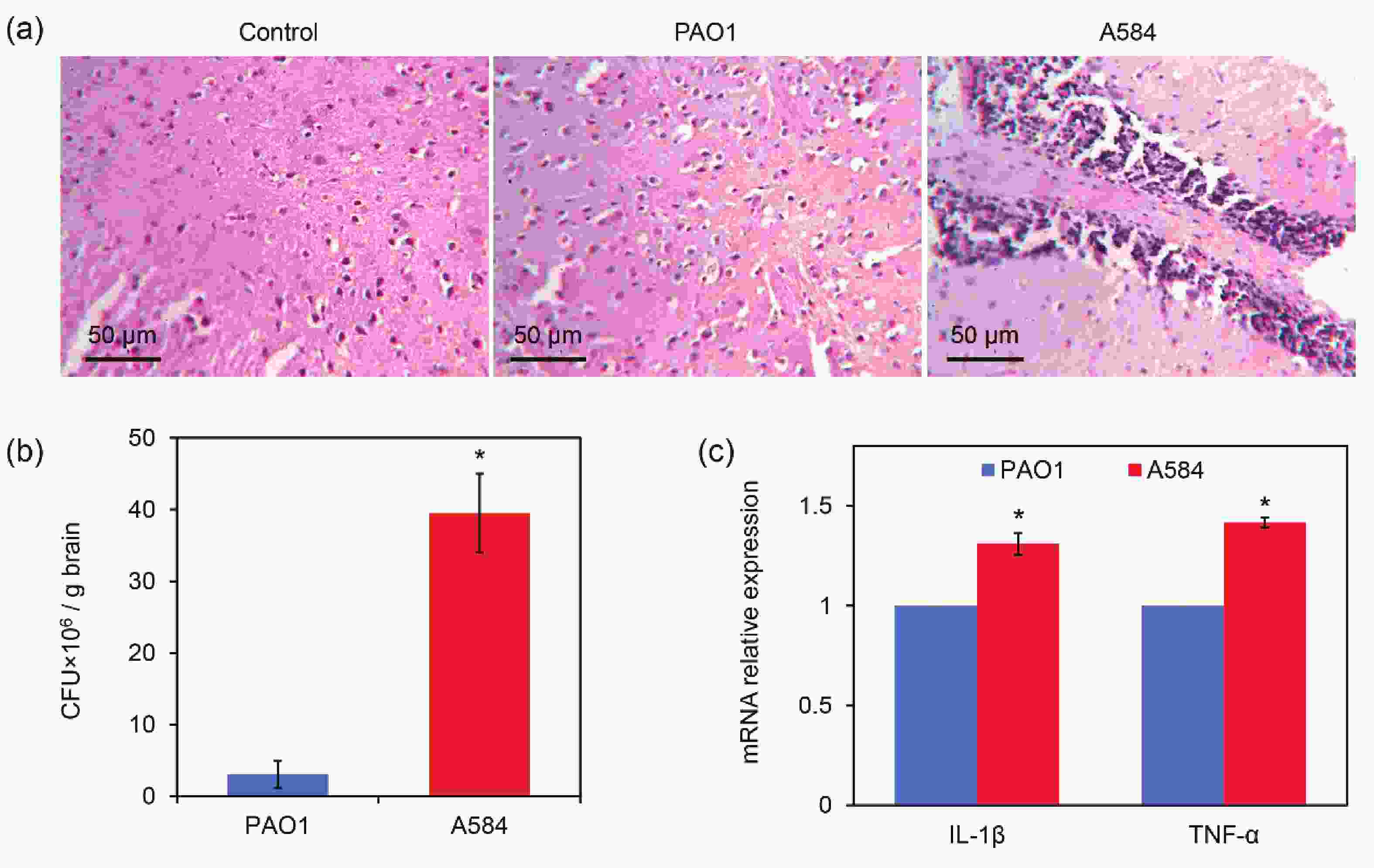
Figure 6. Invasion of A584 into the mouse brain tissues. (A) Histopathological images of the mouse brain tissue after 3 days of intravenous infection by the P. aeruginosa strains. (B) P. aeruginosa burden in the brain tissue after 3 days of infection. (D) Relative expressions of IL-1β and TNF-α in the infected tissues.
-
Meningitis is a widespread disease with a high mortality rate and long-term disabling sequelae, making its prevention critical for human health[12,27,28]. Understanding the impact of pathogens on the mechanisms underlying meningitis is of utmost importance. Cryptococcus neoformans is the most common pathogen in fungal meningitis. The host drives phenotypic drug resistance in C. neoformans, and glucose induces nuclear translocation of Mig1, which inhibits amphotericin B activity, leading to the development of drug tolerance[29]. Streptococcus pneumoniae, group B Streptococcus, and Escherichia coli (responsible for neonatal meningitis) cross the BBB by hijacking intracellular transport of iron transporter receptors[30]. These pathogens subsequently invade the brain and trigger inflammation. After reaching the meninges, both S. pneumoniae and Streptococcus agalactiae activate the pain neurons in the meninges, causing the release of calcitonin gene-related peptide (CGRP). CGRP then binds to the RAMP1 receptor on the surface of macrophages, rendering immune cells ineffective[31]. This leads to bacterial proliferation and widespread infection. Additionally, the inherent toxicity of these bacteria plays a crucial role.
In this study, we isolated a strain of P. aeruginosa, A584, from a clinical meningitis sample. The pathogenicity and virulence factors involved in meningitis were further studied, focusing on the toxicity, drug resistance, and the pathway by which the bacteria penetrate the BBB.
We found that the clinically isolated P. aeruginosa strains exhibited varying toxicities towards the host, with the A584 strain demonstrating the highest virulence. Cellular virulence includes both intrinsic and invasive factors. Traditionally, toxins produced by bacteria are substances toxic to host cells. From the perspective of P. aeruginosa, A584 has strong hemolytic ability, and its virulence is closely related to the secretion of hemolysin. A584 induces host cell rupture by secreting hemolysin, which facilitates host cell infection. Pyoverdine helps A584 replenish iron ions as nutrients and can precisely regulate the secretion of toxins such as exotoxins and exoproteases[32].
Bacterial invasiveness refers to the ability of bacteria to breach the body's defense mechanisms, colonize, multiply, spread, and disseminate within the body. In this study, when A584 was co-incubated with bEnd.3 cells, the results showed that A584 bacteria adhered tightly to the cells, which is crucial for bacterial colonization and subsequent infection. Furthermore, we found that A584 cells have a remarkable ability to enter host cells and traverse cell layers. In animals, bacterial invasiveness and toxin release occur simultaneously, more accurately reflecting the pathogenicity of bacteria. These results demonstrated that the pathogenicity of A584 was stronger than that of the control strain.
Many P. aeruginosa strains exhibit multidrug resistance, and the mechanisms of resistance are complex. In this study, the multidrug resistance of A584 cells was primarily dependent on the efflux pump system and antibiotic inactivation enzymes. The efflux pump system is the most rapid drug resistance mechanism in the bacterial stress response, expelling substances unfavorable for bacterial growth[33]. P. aeruginosa has 18 RND efflux systems[34], including MexAB-OprM, MexXY-OprM, MexCD-OprJ, MexEF-OprN, and other efflux systems[35-36], which are more common in clinical strains.
The key to meningitis caused by bacterial pathogens lies in the mechanism by which these bacteria penetrate the BBB. In fact, there are three main ways for pathogens to cross the BBB: tight junction crossing[37], transendothelial cell crossing, and receptor-mediated endocytosis[38]. This study revealed that A584 caused more severe damage to the BBB than PAO1, and the underlying mechanism merits further exploration. As shown in Figure 5, A584 may induce lysosomal dysfunction. Lysosomes, functioning as the “digestive workshops” within cells, are crucial for maintaining the stability of the intracellular environment. This strain impairs lysosomes in bEnd.3 cells, affecting normal cell functions and killing these epithelial cells, thereby increasing BBB permeability. At the molecular level, A584 disrupts the regulation of tight-junction proteins in bEnd.3 cells. Claudin-5 and Occludin expression was significantly downregulated in the A584-treated group, which may be part of complex molecular regulatory networks. Additionally, inflammatory factors may play a role. When A584 infects endothelial cells of the BBB, the release of inflammatory factors affects BBB integrity[39-42]. For example, TNF-α is abundantly secreted during inflammatory responses. It can degrade tight junction proteins by activating intracellular proteases and promoting endothelial cell contraction, thereby increasing intercellular spaces. As analyzed earlier, A584 cells expressed more potent virulence factors. These factors specifically bind to the surface receptors of endothelial cells, initiating intracellular toxic reactions that lead to cell damage and loss of barrier function.
Future research could focus on identifying the specific virulence factors secreted by A584 and their targets, conducting an in-depth analysis of the molecular mechanisms by which they interfere with cell signaling pathways and gene expression, and exploring the role of the inflammatory response in A584-induced BBB damage. This study provides a theoretical basis for developing treatment strategies for central nervous system diseases caused by P. aeruginosa infections.
-
In this study, we explored the virulence factors of meningitis-related P. aeruginosa strain A584. Whole-genome sequencing and analysis revealed that strain A584 possesses several virulence factors, including hemolysins, T3SS/T4SS/T6SS secretion systems, pyoverdine, and a secondary metabolism gene cluster. Additionally, A584 exhibited high multidrug resistance to various antibiotics, including aminoglycosides, β-lactams, quinolones, and β-lactamase inhibitors. Hemolysis, LDH release assays, and animal experiments demonstrated that the A584 strain caused more cell damage than the representative P. aeruginosa strain PAO1. Compared to PAO1, A584 caused much more severe damage to the BBB. This increased virulence is associated with lysosomal damage, downregulation of the endothelial tight junction proteins Claudin- 5 and Occludin, and excessive secretion of inflammatory factors. This study provides a foundation for developing antibacterial agents against bacterial meningitis.
Deciphering Virulence Factors of Hyper-Virulent Pseudomonas aeruginosa Associated with Meningitis
doi: 10.3967/bes2025.082
- Received Date: 2024-10-21
- Accepted Date: 2025-03-03
-
Key words:
- Pseudomonas aeruginosa /
- Virulence factor /
- Meningitis /
- Genome sequencing /
- Drug resistance
Abstract:
The authors declare no competing interests.
| Citation: | Liling Xie, Shuo Liu, Yufan Wang, Mingchun Li, Zhenhua Huang, Yue Ma, Qilin Yu. Deciphering Virulence Factors of Hyper-Virulent Pseudomonas aeruginosa Associated with Meningitis[J]. Biomedical and Environmental Sciences. doi: 10.3967/bes2025.082 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: