-
Organoids are three-dimensional (3D) micro-organ structures derived from pluripotent stem cells (PSCs) or tissue-specific progenitor cells in vitro. Their core features include: the presence of multiple, functionally interdependent cell types, self-organization to form spatial structures, and physiological functions similar to those of their in vivo counterparts. In 2009, Clevers et al. successfully constructed the first intestinal organoid with a crypt-villus structure by isolating a single Lgr5+ stem cell in the crypts of the small intestine of a mouse, marking a breakthrough in the development of organoid technology[1]. Since then, organoid models based on PSCs, including embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), and adult stem cells (ASCs), have rapidly expanded. Organoid systems of different embryonic origins have been formed, including ectodermal (such as retina, brain), mesodermal (including blood vessels, heart, and kidney), and endodermal (including intestinal tract, stomach, and liver) organoids[2].
Traditional research models, such as two-dimensional (2D) cell cultures and animal models, exhibit critical limitations in recapitulating the complex physiological and pathological mechanisms of human organs. 2D cell cultures fail to emulate 3D cellular interactions and tissue microenvironments, and animal models suffer from interspecific genetic and physiological disparities that compromise clinical translatability. To address these challenges, 3D organoid technology has emerged as a transformative approach, faithfully replicating cellular heterogeneity, spatial structure, and dynamic functions of developing organs, thereby overcoming the limitations of traditional models.
Organoids can be categorized into two major types based on their origin and function: healthy-tissue- and tumor-derived organoids[3]. Healthy tissue-derived organoids recapitulate the developmental and homeostatic regulatory mechanisms of normal organs and provide high-fidelity models for studying genetic disorders. In contrast, tumor-derived organoids—constructed from patient-derived tumor stem cells—retain the heritable heterogeneity and drug-response profiles of the primary tumor, making them essential tools for personalized cancer therapy[4]. Current organoid construction relies on multiple key technologies, including 3D culture within matrices (Matrigel), bioreactor-driven dynamic culture systems, and spatiotemporal regulation of growth factors. Self-organization is achieved through directed differentiation of stem cells (guided by extrinsic signals), coordinated interactions among multilineage precursor cells, or integration via assembloid engineering.
Yamanaka et al. achieved a landmark breakthrough in adult cell reprogramming by establishing the iPSC system[5,6]. Given the similarities between iPSCs and ESCs in morphology, gene expression, pluripotency maintenance, and self-renewal, researchers have successfully differentiated human iPSCs (hiPSCs) into cardiomyocytes, which are crucial for patient-specific cardiac organoid (CO) modeling. Notably, the integration of organoids with CRISPR/Cas9-mediated genome editing has revolutionized mechanistic studies. For instance, targeted editing of cardiac transcription factors (such as NKX2-5 and TBX5) in heart organoids enables real-time investigation of the impact of gene mutations on cardiomorphogenesis and their associations with congenital heart defects (CHD). Unlike zebrafish or murine models, organoid platforms circumvent interspecies signaling pathway discrepancies while permitting high-throughput screening. Currently, organoids serve not only as versatile platforms for developmental biology, disease modeling, and drug discovery but also as ethically superior alternatives to traditional animal models, with accelerating advancements in automation and standardization.
Cardiovascular disease, the leading cause of mortality globally, has long been hindered by the physiological irrelevance of traditional models[7]. For instance, the 2D differentiation of human PSCs (hPSCs) can generate cardiomyocytes; however, it fails to recapitulate key dynamic cell-matrix interactions, electromechanical coupling, and morphogenetic events during cardiac development. Critically, such static monocultures lack cellular heterogeneity, predominantly producing cardiomyocytes, while neglecting endothelial cells (ECs), fibroblasts (FBs), and epicardial cells that are essential for cardiac tissue function[8]. Animal models also suffer from species-specific disparities in electrophysiology, drug metabolism, and disease microenvironments, limiting their translational value in human cardiac research. However, neither model can replicate the complex cellular composition and dynamic biomechanical microenvironment of the native heart[9]. Recent advances in human heart organoids have addressed these limitations by modeling key cellular diversity and functional complexities observed in native tissues. Notably, disease-specific CO models (such as myocardial infarction (MI) and ischemia/reperfusion (I/R) injury) demonstrate remarkable potential for mimicking in vivo cardiac damage, offering high-throughput platforms for investigating metabolic dysfunction or drug-induced cardiotoxicity. Furthermore, the integration of organoid technology with microfluidic systems in heart-on-a-chip platforms promotes CO maturation through precise modulation of fluid shear stress and signal regulation. Compared with static culture models, dynamically loaded COs achieve enhanced functional maturation[10]. Furthermore, these platforms enable real-time monitoring of electrophysiological activity and perfusion under physiologically relevant conditions, providing a robust framework for drug toxicity evaluation and environmental cardiotoxicity research. The evolution and characteristics of the current mainstream cardiac models are summarized in Table 1.
Model Type Advantages Disadvantages Applications
2D cell cultureHigh throughput;
High genetic similarity;
Ease of probing cell-specific effects.Lack of 3D architecture and cell-cell/matrix interactions;
Poor mimicry of myocardial tissue mechanics and electrophysiology.Drug screening;
Basic cardiomyocyte research for studying cellular responses and signaling pathways.
Animal modelsCan simulate complex in vivo microenvironment and physiological processes;
Enables holistic disease mechanism studies.Genetic relevance varies depending on the animal model;
Lower throughput compared to in vitro models;
High cost and long experimental cycles.Disease mechanism studies;
Whole-organ disease modeling;
Drug development.
Cardiac spheroidsMedium/high throughput;
High genetic similarity;
3D microenvironment closer to in vivo conditions;
Customizability.Low contractility;
Limited long-term culture stability and functional complexity.Tissue engineering;
Drug screening and testing.
COsHighly organized: features a more complex 3D structure and cellular composition;
In vivo-like function;
Customizability.Lack of vascular/innervation networks;
Technical complexity;
Standardization challenges.Disease modeling;
Drug development;
Regenerative medicine.
CO-on-a-ChipMicrofluidics-enabled hemodynamic/mechanical stress simulation;
Multi-organ interaction studies;
Real-time monitoring;
High customizability.Technical complexity (requires advanced microfluidic technology and specialized equipment) and high cost;
Medium/low heart architecture (similar to a segment of working myocardial tissue, not fully representative);
Standardization challenges.Drug screening and testing;
Personalized medicine.Note. CO: cardiac organoid. Table 1. Evolution and characteristics of cardiac models
This paradigm shift has gained institutional validation: the United States Food and Drug Administration has announced plans to replace traditional animal models—used for almost a century—in favor of advanced technologies such as organoids and organ-on-a-chip systems for drug safety testing. These human-mimetic systems not only provide more accurate pharmacological data but also minimize animal experimentation while accelerating research efficiency.
Despite their potential in cardiovascular drug screening and disease modeling, COs face three critical challenges: 1) incomplete standardization protocols, 2) immature functional phenotypes (including contractile and electrophysiological properties), and 3) insufficient integration of vascular, neural, and immune networks. In addition, multisystem organ-on-a-chip platforms remain underdeveloped. To address these issues, bioengineering advancements, such as spatiotemporal transcriptomics-guided induction protocols and modular assembloid designs, hold substantial potential for advancing the study of cardiac development, environmentally induced cardiomyopathy, and personalized therapeutic prediction.
-
Cardiac research is rapidly transitioning from traditional 2D culture systems to advanced 3D CO models. Major advances include multicellular COs, vascularized COs, chambered COs, and cardiac organoid-on-a-chip (COoC) technologies. By regulating the spatiotemporal differentiation of hPSCs, these models effectively recapitulate the development of specific chambers in the human embryonic heart, providing an important tool for investigating the mechanisms of cardiac development and pathogenesis of chronic and systemic diseases. This section reviews the evolution of 3D CO models and construction strategies, as well as their potential for use in disease modeling, drug screening, and related applications. Cardiac spheroids are explicitly defined as simplified 3D aggregates formed through scaffold-based or scaffold-free assembly, primarily composed of cardiomyocytes, lacking vascular networks and complex spatial organization, and are suitable for high-throughput screening. In contrast, cardiac organoids are self-organizing structures with a multilayered architecture (such as myocardium and endothelium) and physiological functions that mimic aspects of organ development or disease pathophysiology. Understanding these distinctions is crucial to understanding their unique roles and applications in cardiac research.
-
With the rapid development of 3D cell culture technology, methods for constructing myocardial spheroids based on the properties of cell aggregates have been gradually established and continuously improved. In general, spheroids are obtained from one or multiple cell types that spontaneously form adherent cell populations with nonuniform sizes. Construction strategies are mainly based on (1) cellular aggregation and self-assembly in a scaffold-free cell culture framework and (2) co-culture systems that integrate multiple cell types with biomaterials[11]. The former approach focuses on regulating cardiac progenitor cell differentiation pathways to generate chamber-specific cardiomyocytes, forming atrial or ventricular spheroids with distinct electrophysiological properties for basic research, such as lineage specification and electrophysiological screening[12]. The latter approach involves co-culturing pre-differentiated cardiomyocytes with cardiac microenvironment cells (including ECs and FBs) at physiological ratios to form multicellular cardiac spheroids, specifically emphasizing the natural tissue architecture and cell-extracellular matrix (ECM) interactions to promote cardiomyocyte maturation. The resulting functional complexity offers significant advantages in studies requiring high-fidelity electrophysiology and predictive pharmacology[13].
Initially, in vitro cultures of mammalian embryos or human cardiac cells often led to the loss of myofibrillar proteins and organized morphology in cardiomyocytes, thus resulting in diminished cardiac differentiation characteristics. To address this, Maltsev et al. used long-term in vitro culture of ESCs to study cardiomyocyte differentiation. Using 3D embryoid bodies formed via hanging drop culture to mimic early embryonic conditions, they promoted cell-cell interactions and natural differentiation into mature cardiomyocytes. These cells exhibited spontaneous contractions and electrophysiological properties similar to those of native cardiac cells[14]. Despite variability in spheroid size and functional limitations, this approach laid the groundwork for subsequent 3D culture techniques[15]. Many studies have built upon this method, optimizing it to improve spheroid generation efficiency and function[16]. Lai et al. developed a novel “hanging heart” chip integrated with a microchannel flow-driven system, which reduced manual handling, simplified operations, increased throughput, and maintained contraction frequencies suitable for high-throughput toxicity testing[17]. In addition, low-attachment-well plates effectively inhibited cellular apposition and promoted the aggregation and self-assembly of cardiomyocytes to form spheroids. This technique also enhances the maturation of cardiomyocytes to a certain extent, and has been widely used to study developmental pathways and construct transplantable myocardial patches[18,19]. Suspension culture techniques have demonstrated considerable advantages in cardiomyocyte research by simulating physiological conditions using nutrient and gas gradients. This enhanced cardiomyocyte maturation and functionality, making it valuable for large-scale production and pathological simulation. Kempf et al. established a strategy to efficiently generate cardiomyocytes from hPSCs using suspension culture and achieved high-purity differentiation (> 80%) by modulating the WNT signaling pathway[20]. Sebastiao et al. developed a 3D suspension culture spheroid model to simulate myocardial I/R injury using hiPSC-cardiomyocytes cultured in a stirred-tank bioreactor, which successfully mimicked the disease characteristics of MI[21]. In addition, scaffold-based fabrication has emerged as a preferred strategy for generating multicellular cardiac spheroids, demonstrating enhanced cellular viability and functional maturation within 3D microenvironments[13,22].
Cardiac spheroids serve as physiologically relevant models for studying cardiac physiology and pathology, recapitulating critical myocardial tissue characteristics, including autonomous rhythmic contractions, synchronized calcium oscillations, and electrophysiological signal propagation[22]. Their ability to exhibit dose-dependent drug responses allows the evaluation of pharmacological effects on myocardial contractility, calcium homeostasis, and electrophysiological parameters, offering insights into cardiotoxicity mechanisms[23]. However, achieving precise dynamic regulation through internal self-assembly remains challenging, often leading to uneven cell distribution, undefined spatial organization, and mixed marker expression, which disrupt intercellular communication and signaling. In addition, the absence of vascular networks limits functional maturation, as tissue size is constrained by diffusion limits (≈ 100-150 μm), beyond which central cell apoptosis increases significantly[24]. Therefore, strategies are needed to enhance microvascular network formation, including adding angiogenic factors, integrating human umbilical vein endothelial cells (HUVEC), developing proangiogenic hydrogel matrices, and constructing EC-cardiomyocyte co-culture systems. When combined with 3D bioprinting technology, these approaches aim to induce neovascularization, enhance functional vascular network formation, and improve metabolic support, thereby ensuring effective metabolic exchange and spheroid viability of cardiac spheroids[11,13].
-
Human cardiac tissue is composed of 25‒35% cardiomyocytes and 65‒75% non-cardiomyocytes by cell number[25]. These cellular populations are embedded within a dynamic ECM containing interstitial collagens, elastin, fibrillin, laminin, fibronectin, proteoglycans, and regulatory molecules, including growth factors, cytokines, and proteases[26]. The ECM provides biomechanical support while establishing anisotropic topographical cues and biochemical signaling gradients that collectively enable electromechanical coupling, synchronized contraction, and efficient force transmission across myocardial tissue[26,27]. The native heart also features an extensive vascular network that meets the high metabolic demands of contractile cardiomyocytes. Farah et al. identified 39 major cell populations and 75 subgroups in the human heart using single-cell RNA sequencing (scRNA-seq). Cardiac cells form specific communities through spatial coordination rather than random arrangement, comprising diverse cell types that collectively maintain normal cardiac function[27]. The spatial organization of the heart is closely linked to its function, with specific cell distribution and organization enabling efficient pumping and electrical conduction.
Complex CO structures involve self-organizing processes such as cell fate specification, spatial patterning, and morphogenesis. This presents core challenges in their construction, such as how signaling guides cell self-organization, regulates spatial heterogeneity, and drives chamber morphogenesis. General principles of cardiac development, including the roles of signaling, cell types, lineage structure, and function, are conserved. To establish a controlled in vitro system that mimics human heart development, these in vivo principles must be applied to coordinate and validate CO constructs[28,29].
In 2002, Zimmermann et al. introduced the concept of “CO” and constructed an engineered cardiac tissue of macroscopic cell origin with organoid properties. This tissue exhibited highly differentiated characteristics and cardiac tissue-like functions but lacked full maturation[30]. Since 2021, considerable breakthroughs have been achieved in the development of COs. Compared to 2D-cultured cardiomyocytes and foundational cardiac spheroids, COs offer superior cellular composition, with greater complexity and functionality. Recent studies have identified diverse cardiomyocyte subtypes expressing specific genes for atrial and ventricular cardiomyocytes and cardiac progenitor cells, as well as smooth muscle cells, ECs, and even epicardial cells within the tissue microenvironment[31]. Advances in spatial organization have enabled COs to form multilayered structures, including myocardial, endothelial, and epicardial layers, which facilitate cell-cell and cell-matrix interactions. In addition, COs can form microchambers, such as ventricle- and atrium-like structures, and exhibit electrophysiological synchrony, such as calcium transients and action potential propagation, which are crucial for mimicking cardiac electrical activity. They also demonstrate mechanical contractility, simulating the pumping function and hemodynamic properties of the heart. Metabolically, they exhibit tissue-specific characteristics such as oxygen and nutrient utilization, and can be used to model pathological processes such as hypoxia-induced cell death in MI[21]. Furthermore, their responsiveness to drugs makes them ideal for the assessment of toxicity and efficacy. COs are typically constructed using self-organization and assembly-based methods. Self-organization approaches rely on endogenous signaling, whereas assembly strategies primarily depend on exogenous manipulation (Figure 1). Self-organization excels in developmental recapitulation, whereas assembly offers superior controllability in translational applications.
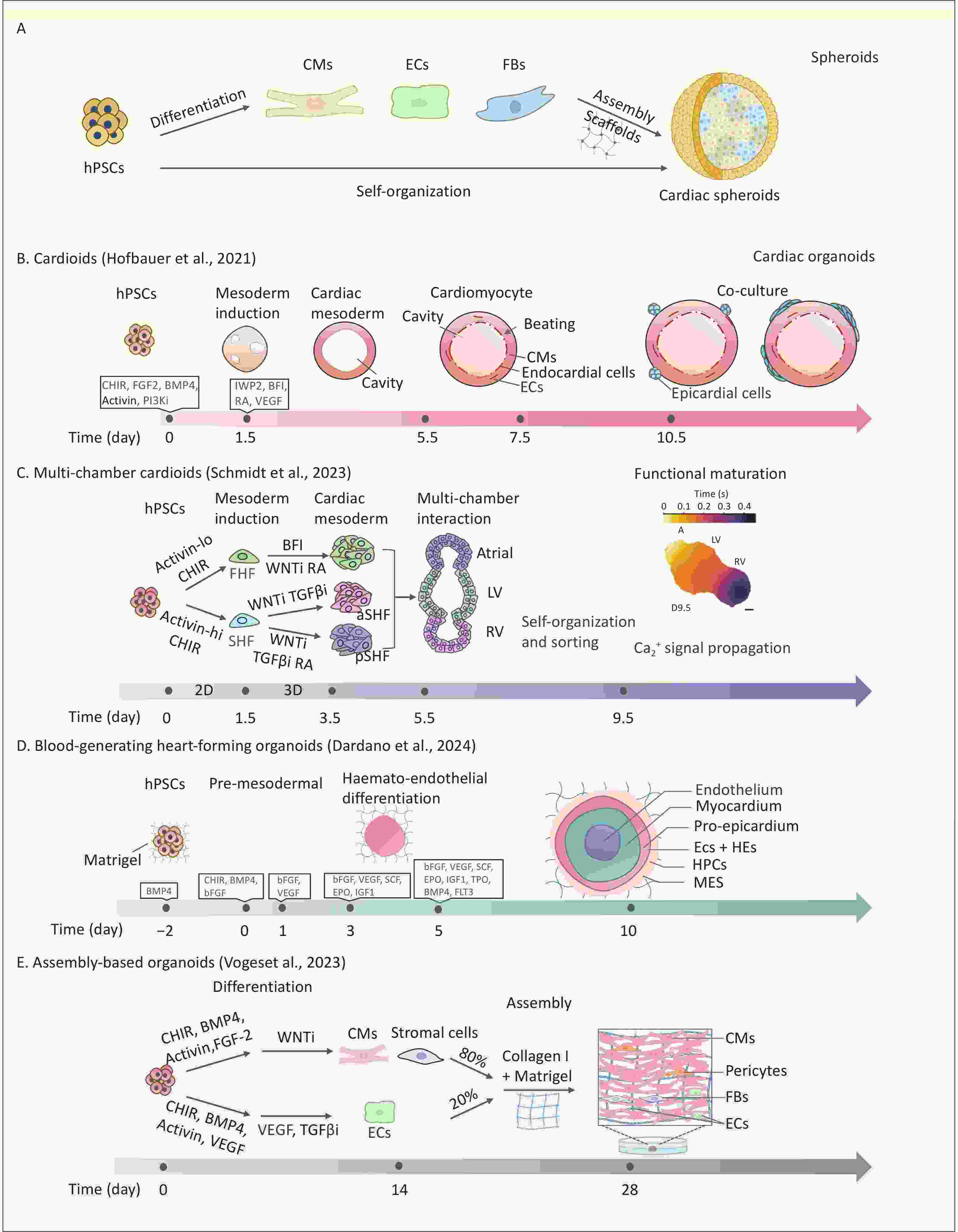
Figure 1. Comparison of different strategies for constructing cardiac spheroids and cardiac organoids in vitro. (A) hPSC-derived cardiac spheroid formation via direct differentiation or post-differentiation assembly; hPSCs: human pluripotent stem cells, CMs: cardiomyocytes, ECs: endothelial cells, FBs: fibroblasts. (B) Self organization of hPSCs into contractile single-chamber cardiac organoids via sequential induction steps[29]. (C) In vitro 2D and 3D culture of multi-chamber cardiac organoids including atrial, LV and RV via sequential induction steps[39]; LV: left ventricle,RV: right ventricle. (D) Blood-generating heart-forming organoids with definitive hematopoietic potential, generated via stage-specific BMP4/bFGF/VEGF induction, feature a trilaminar structure (inner core, myocardial layer, outer layer)[44]; HE: haemogenic endothelium, HPCs: haematopoietic progenitor cells, MES: mesenchymal cells. (E) Generation of vascularized complex organoids from hPSC-derived CMs and ECs in a Collagen I/Matrigel matrix[57].
-
Self-organization methods leverage the intrinsic differentiation potential of PSCs to construct COs via spatiotemporal signaling regulation, thereby recreating the biochemical and physical microenvironment of the embryonic heart. Cells form self-organizing networks through direct contact and paracrine signaling, which drives coordinated differentiation and spatial assembly. A stepwise induction strategy is typically employed to guide stem cells through the following key stages: mesoderm, cardiac mesoderm, cardiomyocyte specification, and maturation, ultimately generating complex COs. Biphasic WNT regulation is a central driver of cell differentiation and morphogenesis during organoid construction. In addition, Lewis-Israeli et al. developed a three-stage WNT signaling strategy (activation-inhibition-reactivation) by applying low-dose CHIR99021 on day 7 of differentiation to specifically induce WT1+ epicardial cells, achieving a spatially ordered assembly of epicardial, myocardial, and ECs[28], which has also been widely validated and applied to construct CO disease models[32,33].
Hofbauer et al. developed the first hiPSC-derived self-organizing CO model, termed “cardioids,” featuring 3D structures with enclosed cavities that contract rhythmically, mimicking pumping function (Figure 1B). They found that during mesoderm induction, WNT activation initiated mesoderm development, triggered BMP signaling, and regulated HAND1 to control cardiac mesoderm cell proliferation and polarity, driving chamber expansion[29]. In addition, the spatiotemporal synergy of WNT/activin/VEGF signaling accurately directs the hierarchical organization of cardiomyo-endothelial cells. This process mimics the bidirectional signaling between the myocardial and endocardial layers[29,34,35]. Physical geometric constraints and vascular structures are equally important in influencing the chamber structure of COs[36]. Although single-chamber cardioids exhibit robust beating, they have limitations in modeling complex cardiogenesis.
To create more precise models, controlling heart field specification, focusing on the spatiotemporal differentiation of the first heart field (FHF) and second heart field (SHF), is essential. The FHF contributes to the left ventricle and parts of the atria, whereas the SHF forms the outflow tract, right ventricle, and remaining atria. During cardiac development, the BMP/Activin A signaling gradient acts as a key morphogenetic switch, directing hPSCs toward specific cardiac mesodermal subtypes. Co-regulation of WNT and Nodal/Activin signaling spatiotemporally enables directed differentiation of hPSCs into progenitors of the FHF, anterior SHF (aSHF), and posterior SHF (pSHF)[37]. Notably, Retinoic acid (RA) signaling is crucial for constructing multi-chamber COs. This determines the fate of the aSHF/pSHF lineage via spatiotemporal concentration gradients. Refined RA gradients are essential for the precise patterning of atrial, ventricular, outflow tract, and atrioventricular canal substructures during chamber morphogenesis[33]. Several previous studies have used mouse PSCs to mimic the development of FHF and SHF to generate functional COs with atrial- and ventricular-like structures[38]. Yang et al. first modeled human FHF, aSHF, and pSHF lineage using hPSCs and created a multilineage developmental atlas of the human embryonic heart[31]. Schmidt et al. developed a human CO platform modeling all major embryonic heart regions, including the ventricles, atria, outflow tract, and atrioventricular canal, using combined 2D and 3D induction of lineage-specific progenitors[39]. These multi-chamber platforms are important for studying inter-chamber electrophysiological signaling and early cardiac development (Figure 1C).
Most CO models, which rely on predifferentiated cardiomyocytes or primary cardiac cells, face challenges in achieving sufficient spatiotemporal resolution of early developmental dynamics and reproducing key cardiac morphogenetic events. Cardiac development begins during gastrulation when visceral mesoderm cells migrate from the primitive streak to form the precardiac mesoderm. These progenitor cells establish bilateral heart fields near the embryonic midline and interact with the adjacent foregut endoderm to create an inductive microenvironment. Guided by BMP/FGF signaling, they form a primitive heart tube, which undergoes looping, regional expansion, and septation to develop distinct atrial, ventricular, and outflow regions. This process requires multi-germ layer coordination, including biomechanical and paracrine interactions crucial for cardiac looping and septation[40].
The multilineage organoid strategy has achieved groundbreaking innovations by reconstructing the synergistic microenvironment of germ-layer interactions. In 2021, Rossi et al. established mouse gastruloids that captured self-organized cardiogenesis. CHIR99021-mediated WNT activation induces anterior-posterior (A/P) axis formation, confining cardiac progenitors to the anterior domain. By day 6, these structures developed crescent-shaped cardiac fields morphologically aligned with E7.5, revealing the inductive role of the foregut endoderm in heart development. Crucially, the gastruloids exhibited compartmentalization where cardiac progenitors adjacent to Sox17+ primitive gut tube-like structures were separated by an endocardium-like layer, enabling paracrine crosstalk (endoderm-derived FGF2). Disruption of this signaling recapitulates endocardial cushion defects, thereby providing a congenital heart disease model that elucidates the supportive mechanisms of the foregut endoderm in cardiac morphogenesis[34]. Drakhlis et al. generated human heart-forming organoids (HFOs) through Matrigel-embedded hPSC aggregation and biphasic WNT modulation, recapitulating the early cardiac primordial features.
These HFOs exhibited a stratified architecture containing myocardial layers, endocardium-like cells, and A/P foregut endoderm layers with functional vascular networks. Notably, NKX2.5 knockout in this model successfully mimicked murine cardiac malformation phenotypes, validating its utility for the mechanistic dissection of congenital heart diseases[41]. Gastruloids are characterized by a symmetry-breaking event that induces organoid elongation and heart morphogenesis, making them suitable for studying global embryonic development. However, HFOs lack elongation and instead emphasize endoderm-heart interactions. Despite containing diverse cell types from different germ layers, both models lacked markers for cardiac and endodermal tissue maturation. Silva et al. achieved a considerable breakthrough by creating mesoendodermal organoids that facilitated symbiotic cardiac-gut co-development. These organoids spontaneously developed cardiac-gut structures, featuring myocardial layers, epicardium-like cells, smooth muscle tissues, and functional intestinal compartments with Paneth and goblet cells. Remarkably, the co-development of cardiac and gut tissues within these organoids, which maintained their structural integrity for over a year, significantly enhanced cardiomyocyte maturation. Mechanistic investigations have identified FGF10 and NPPB as gut-enriched paracrine factors that play critical roles in promoting mesodermal cell differentiation, supporting atrial morphogenesis, and contributing to cardiac structural properties. Cardiomyocyte maturation, sarcomere organization, and mitochondrial distribution mirrored those of late-stage human fetal cardiomyocytes, demonstrating that cross-germ layer interactions play a critical role in driving myocardial maturation[42].
Recapitulating functional hemogenic endothelial differentiation and endothelial-to-hematopoietic transition in vitro using conventional vascularization strategies remains challenging. Previous models, such as mouse embryonic stem cell-derived gastruloids[43], were limited to the generation of primitive hematopoietic lineages (erythroid/myeloid) analogous to embryonic yolk sac hematopoiesis. Dardano et al. addressed this limitation by developing blood-forming HFOs, an in vitro model of early human circulatory development[44] (Figure 1D). By temporally activating WNT signaling and using BMP4/VEGF, this model maintains the myocardium and mesenchyme while inducing CD31+CD34+ HE, producing multipotential hematopoietic progenitors (CD45+CD43+). Single-cell transcriptomics revealed that they contained arterial/venous endothelial subpopulations and RUNX1 (ALDH1A1) hematopoietic niches, and their spatial localization highly mimicked that of the embryonic aortic-gonadal-mesonephric region. This innovation addresses in vitro hemogenic microenvironment reconstruction and offers a cross-germ layer platform for studying congenital heart disease with hematologic abnormalities. Notably, the modeling capabilities of HFOs are currently limited to very early developmental stages. Nevertheless, it opens up new perspectives for studying cardiac HFOs for multifunctional applications, such as human embryonic developmental mechanisms, disease modeling, and drug discovery.
-
Current self-organization of CO construction faces challenges such as imprecise control of cell subpopulation ratios, limited non-cardiomyocyte lineage differentiation, insufficient 3D tissue structure, and difficulty in achieving functional maturation at physiological levels. Emerging assembly strategies aim to develop functional organoid models with precise tissue hierarchy by regulating multilineage cell proportions, spatiotemporal distribution, and intercellular interactions, while promoting cell self-organization[45].
Assembly strategies for COs primarily employ bioengineering approaches, such as scaffold design, geometric constraints, and ECM modulation, to guide the differentiation of PSCs and organize them into functional tissues with chamber-like, spherical, or organoid structures. The physiological relevance of these models is highly dependent on the rational design and application of exogenous biomaterial scaffolds[46]. An ideal bioactive scaffold should provide mechanical support and structural guidance for stable 3D organoid growth, allow geometric design flexibility to meet diverse research and application needs, integrate cell adhesion sites (such as RGD peptides) and growth factor signaling, and support vascular network formation. The commonly used biomaterials and their respective properties are presented in Table 2.
Material Key Properties Functional Impact Challenges Engineering Strategies Matrigel Complex ECM composition and biocompatibility;
Tunable stiffness.Promotes cell differentiation. Tumor-derived origin;
Considerable batch variation;
Low mechanical strength.Hybrid scaffold development (e.g., photo-crosslinking with GelMA[56]); dECM Tissue-specific ECM composition and biomimetic properties with low immunogenicity. Enhances cell proliferation and differentiation. Heterogeneous composition and lack of standardization;
Challenges in recapitulating complex 3D architectures.3D bioprinting with spatial control[68,69];
Microfluidic-guided self-assembly[47].Collagen Main structural protein in the ECM, biocompatible and biodegradable. Promotes cardiomyocyte alignment. Rapid degradation;
Low initial modulus.Diverse hybridization or cross-links to meet varied biomedical needs[50]. PEG Mechanically tunable;
Spatiotemporal control via photo-crosslinking;
Biocompatible.High-throughput drug screening compatibility[70]. Lacks intrinsic bioactivity and requires modification with bioactive molecules;
Degradation fragments inhibit cell migration.Incorporate ECM-derived peptides (such as RGD) and growth factors to enhance bioactivity[71]. Fibrin Derived from fibrinogen, biodegradable and biocompatible. Supports cell adhesion and migration. Rapid degradation;
Poor mechanical stability.Use crosslinking agents to control the degradation rate. Alginate Natural polymer derived from seaweed;
Tunable properties and high biocompatibility.Excellent for cell encapsulation and 3D culture;
Mild gelation process.Lack of cell adhesion;
Rapid degradation;
Limited bioactivity.Incorporate bioactive molecules (such as growth factors and peptides) to enhance cell interactions. GelMA Photo-crosslinkable;
Tunable mechanical properties;
High biocompatibility.Supports cell adhesion, proliferation, and differentiation;
Supports vascular co-culture.Potential softening during long-term culture;
Rapid sol-gel transition at room temperature in 3D bioprinting[72];
Complex synthesis.Modular bioink customization[73,74];
Co-crosslinking with other materials to significantly enhance mechanical stability and long-term culture performance[75].Note. dECM: decellularized extracellular matrix. PEG: polyethylene glycol. GelMA: gelatin methacryloyl. Table 2. Characteristics and engineering strategies of key biomaterial scaffolds for COs
Hydrogels—3D networks formed by hydrophilic polymers via chemical or physical crosslinking—are widely used owing to their high water content, tunable mechanical properties, and biocompatibility, and have become a leading choice for CO scaffolds[47]. Natural hydrogels, including collagen, fibrin, decellularized ECM (dECM), and Matrigel, are considered the “gold standard” for 3D tissue construction because of their high biofidelity. Collagen, the predominant structural protein in the cardiac ECM, provides essential fibrous networks that guide cardiomyocyte alignment[48]. It has excellent biocompatibility, with collagen-based scaffolds proving superior to alternative biomaterials (such as gelatin, hyaluronic acid, and glucan)[49]. However, collagen was relatively unstable post-implantation. Cross-linking can enhance stability and longevity by strengthening the physicochemical interactions between polymer chains[50]. Prepared by removing cellular components from tissues, dECM retains the physical and biological properties of native tissues. It offers high bioactivity and tissue specificity, making it effective in promoting regeneration[51]. Matrigel, derived from mouse Engelbreth–Holm–Swarm sarcoma as a basement membrane extract, contains abundant laminin, collagen IV, and growth factors (such as TGF-β and FGF), and strongly supports stem cell differentiation and angiogenesis[52]. Its compositional complexity results in in-batch variability, necessitating combination with collagen, fibrin, or elastin-like polypeptides to balance bioactivity and mechanical properties[53].
Synthetic hydrogels (including PEG and gelatin methacryloyl (GelMA)) exhibit tunable mechanical properties, controllable degradation kinetics, and facile functionalization, rendering them highly reproducible scaffold materials. However, their intrinsic limitations include suboptimal biocompatibility, slow degradation kinetics, and limited cell adhesion[47,54]. To address these constraints, molecular engineering strategies, such as chemical backbone functionalization or the incorporation of cell-adhesive peptides (RGD), enhance cell-ECM interactions, enable therapeutic molecule conjugation, and promote vascular network formation in 3D constructs. For example, PEG hydrogels are biologically inert, hydrophilic, and cost-effective synthetic biomaterials. Strategic modifications via end-group derivatization or peptide conjugation can transform these platforms into tunable biomimetic microenvironments that support organoid maturation[47,55]. Hybrid systems combining natural matrices such as Matrigel with engineered polymers demonstrate enhanced utility for tissue engineering. For example, the Matrigel-GelMA composite system uses photo-crosslinking to modulate mechanical properties, supporting long-term cardiomyocyte survival and spontaneous contraction[56].
Hydrogels can encapsulate various cell types, such as cardiomyocytes, FBs, vascular cells, and mesenchymal stem cells, to improve co-culture outcomes. Voges et al. optimized the collagen I/Matrigel composite matrix ratio and co-assembled hPSC-derived cardiac cells (including CMs, FBs, and epicardial cells; 80%) with vascular cells (20%) to construct vascularized COs (Figure 1E). This model not only accurately reproduced the cellular composition of the fetal heart but also significantly enhanced the contractility and electrophysiological maturation of organoids through PDGFB-LAMAS paracrine signaling from vascular ECs[57]. Interestingly, this multicomponent assembly approach has also yielded novel functionalized models such as vascularized and chamber-like COs integrating a chamber-specific myocardium and possessing a vasculature system, which provide more advanced tools for functional testing and drug action studies[58]. Furthermore, some hydrogels can be used as bioinks in 3D bioprinting[59,60].
By leveraging digital modeling—such as computed tomography/magnetic resonance imaging-derived chamber geometry reconstruction—and multi-material deposition, 3D bioprinting enables precise spatial control of cellular architecture, including vascular branching, heterogeneous cell patterning, and biomimetic hierarchical organization[61]. Vascular networks with hierarchical branching from large vessels to microvessels require precise size and shape control to achieve effective perfusion. Although self-organization forms primary vessels through endothelial cell migration, the resulting vasculature is limited in both size and complexity. In contrast, bio-printing uses strategies such as sacrificial templating to print hollow vascular structures using bioinks containing ECs, and integrates multiple cell types to build cardiac parenchymal tissue. This approach enables the construction of third-level branching vascular networks within organoids, mimicking the cardiac vascular system to supply oxygen and nutrients and remove metabolic waste. Some studies have achieved 3D printing of large-scale cardiac tissues with complex structures by developing novel bioinks, combining cardiomyocytes and ECs, and optimizing structural design and printing parameters[61,62]. Bio-3D printing continues to encounter challenges for large-scale applications, including high costs and significant cellular requirements for the production of large tissues. Currently, printing both external geometries and internal microstructures simultaneously is not feasible. Most studies have focused on cardiac tissues such as ventricular models[63], patches[64], and multicellular cardiomyocyte spheroids[65]. To better simulate and study cardiac development and diseases, methodological improvements and solutions to current technical limitations are required[66].
Combining lineage-specific differentiation with ordered assembly strategies has demonstrated considerable benefits in the construction of COs. In developing an atrioventricular conduction axis model, Li et al. first differentiated hiPSC into atrioventricular canal myocardial cells (AVCMs) via signaling pathway modulation. Subsequently, they prepared 3D spheroids of pre-differentiated atrial cardiomyocytes (ACMs), AVCMs, and ventricular cardiomyocytes (VCMs), positioning AVCM spheroids between ACM and VCM spheroids in microwell plates, similar to neuronal organoid assembly methods. After 24 h of co-culture, the spheroids fused to form an atrioventricular conduction axis assembly that replicated the “fast-slow-fast” electrical conduction pattern of the embryo heart[67]. This technique provides an accurate model for studying the development of the cardiac conduction system.
-
Traditional static cultures of self-organizing COs lack simulation of hemodynamic microenvironments (such as endothelial shear stress and cardiomyocyte mechanical stretching) and dynamic regulation of key parameters, including oxygen gradients and metabolic delivery, thereby limiting their physiological relevance. Organ-on-a-chip technology integrates microfluidics, smart biomaterials (anisotropic hydrogels and conductive GelMA), and multifunctional sensors to construct a dynamic biomimetic cardiac microenvironment. Using soft lithography or 3D bioprinting, these systems design microchambers and microchannel networks to replicate cardiac electrophysiological pathways and mechanical responses. By combining dynamic electromechanical signals (cyclic stretching and electrical stimulation) with real-time monitoring (action potentials, calcium transients, and contraction frequency), standardized platforms can simulate disease-related mechanical stress (hypertensive remodeling), directly linking dynamic cardiac microenvironments to pathophysiological phenotypes. The COoC system, which integrates organ-on-a-chip and CO technologies, demonstrates potential for (1) establishing standardized microfluidic culture protocols with embedded sensing modules and (2) enabling multi-organ interaction studies via modular chip designs for systemic disease modeling.
-
Directed CO differentiation requires tightly controlled activation of morphogenetic signaling pathways through the time-dependent delivery of exogenous morphogens to drive PSC differentiation. Conventional 3D culture systems using manual medium exchange in Petri dishes often generate non-physiological concentration gradients of these morphogens and endogenous paracrine signals. This setup fails to replicate the spatiotemporally heterogeneous microenvironment essential for embryonic development and introduces inter-batch variability that compromises reproducibility.
Microfluidic systems address these limitations by enabling precise control of culture medium components and dynamically maintaining nutrient concentration homeostasis, thereby avoiding cytotoxicity from metabolic waste accumulation. They also allow for the programmable delivery of exogenous factors, enabling precise simulation of the timed activation of signaling pathways during development. In addition, these systems can create controllable solute gradients through a flow-field design, providing a technical platform for studying cell migration and differentiation. Jayne et al. designed a microfluidic platform in which adjusting the pressure differences in microchannels enabled precise control of drug delivery rates. The circular microchannel layout allowed targeted drug delivery to specific areas, forming concentration gradients that facilitated 3D cardiac tissue self-assembly and growth[76]. In another study, standard source/sink-configured through-holes were used to establish morphogen gradients within Matrigel-filled chambers. This setup provided mechanical support for cell growth while creating a viscous fluid dynamic barrier, ensuring that the chemical environment of the chamber remained diffusion-controlled for precise regulation of the cellular microenvironment[77].
During embryonic development, tissues are subjected to diverse mechanical forces—from single-cell traction to fluid shear stress and solid mechanical loading—which act synergistically with soluble morphogens and ECM signals to regulate organ development and maturation. Microfluidic technology offers a controllable biomechanical loading platform. In a dual-layer channel design, researchers seeded HUVECs in the lower layer and perfused the channel with culture medium via a peristaltic pump to mimic endothelial mechanical responses to blood flow. Under fluid shear stress, ECs aligned with the flow direction, forming intercellular connections that enhanced barrier function. Moreover, cardiomyocytes and FBs formed multilayered structures, improving myocardial tissue differentiation and function, with spontaneous cardiac contractions sustained until day 60[78]. This approach addresses key limitations of traditional organoids in terms of nutrient supply and longevity. Further establishment of the physiological fidelity of the model requires expanded drug response testing and quantitative characterization of drug permeability across the endothelial barrier. Other vascularized COoC devices have also been developed. Di Cio et al. implanted COs into a microfluidic chip, creating a beating cardiac model perfused via a vascular system[79], which was successfully used for vandetanib toxicity testing.
-
Systemic diseases often arise from dysregulated dynamic crosstalk between multiple organs. For instance, the heart engages in multidimensional interactions with distant organs via metabolic regulation (including cardiac-liver lipid metabolic exchange), mechanical coupling (such as renal hemodynamic load), and neuroendocrine signaling. Multi-OoC, also known as body-on-a-chip, provides physiologically relevant models of systemic diseases by generalizing the spatiotemporal characteristics of interorgan substance exchange, biomechanical forces, and signal transduction.
The cardiorenal axis exemplifies such bidirectional crosstalk. Clinical studies have revealed that cardiorenal syndrome progresses via mutually reinforcing pathological loops, where dysfunction in one organ exacerbates that in the other through multiple pathways[80]. To investigate this network, Gabbin et al. engineered a “cardiorenal-unit” chip featuring dual-layer microfluidic chambers housing hiPSC-derived cardiac microtissues and kidney organoids, which dynamically exchanged soluble factors under physiological flow[81]. This platform not only replicated drug-induced cardiotoxicity-renal toxicity cascades but also enabled real-time monitoring of bidirectional signaling. Similar principles can be applied to other organ systems[82]. Pires de Mello et al. developed a heart-liver-on-a-chip integrating induced iPSC-derived cardiac and hepatic modules with a Strat-M skin-mimetic barrier, enabling systemic evaluation of drug metabolism and transdermal toxicity[83]. For neurocardiovascular interactions, Peng et al. created an arrayed brain–heart chip with fibrin-based selectively permeable endothelial barriers. This system maintains organ-specific microenvironments while allowing biomolecular crosstalk via circulatory perfusion, providing a paradigm for studying central-peripheral organ communication[84]. Rajan et al. developed a photosensitive hydrogel platform integrating a three-organoid system (liver, heart, and lung) and a six-organoid system ( including endothelium, brain, and testis)[85]. With organ volumes and cell ratios approximating physiological levels, and simulating sequential blood flow between organs, the model demonstrated that hepatic prodrug metabolism (such as the conversion of ifosfamide) induces toxicity in downstream cardiac, pulmonary, and neural tissues, consistent with clinical reports of drug-induced multi-organ toxicity.
Although the physiological relevance of existing models needs further verification, multi-OoC systems have shown great potential as platforms for preclinical drug screening. These advances not only provide technical support for complex organ interactions but also lay the experimental foundation for developing targeted therapeutic strategies for multiorgan diseases. With continued optimization of organoid culture techniques and microfluidic systems, increasingly sophisticated in vitro human physiological simulation platforms are expected to emerge, thereby accelerating the mechanism analysis and therapeutic innovation of systemic diseases.
-
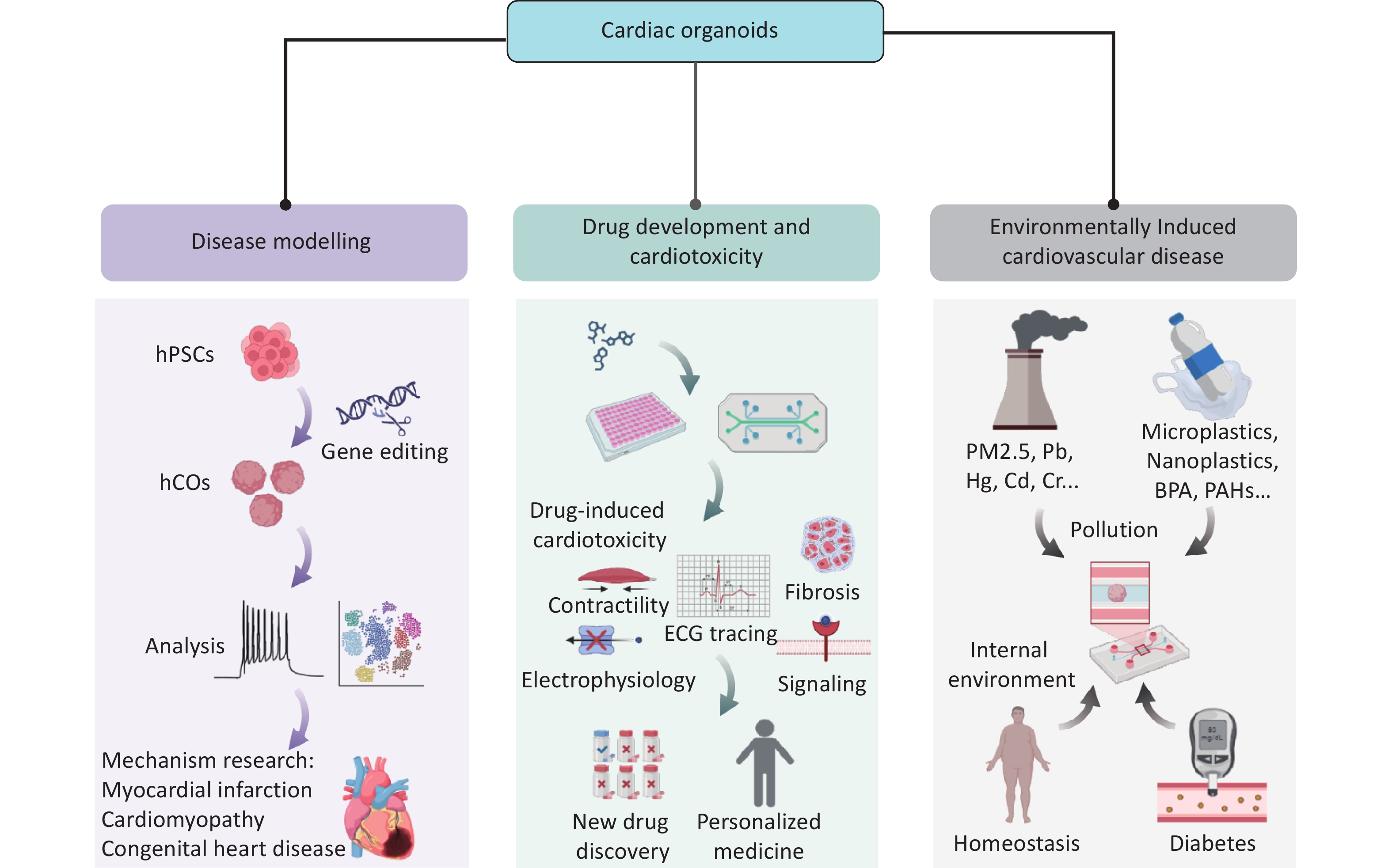
Owing to their highly bionic structures and functions, 3D COs have become important tools for disease and drug research. In this section, we systematically summarize the core applications of COs in disease modeling, drug screening, and environmental interactions, emphasizing their scientific and translational significance through the latest research (Figure 2).
-
Cardiac development is a coordinated and dynamic process involving the spatiotemporal differentiation of multiple cell lineages and the precise regulation of key signaling pathways (including Wnt, BMP, and Notch). Morphogens, such as BMP2/4 and FGF8, establish concentration gradients to direct mesoderm differentiation into cardiac progenitors. Core transcription factors (such as GATA4, TBX5, MEF2, and SRF) synergistically activate cardiac-specific gene expression (MYH6 and TNNT2) and drive heart tube formation, ventricular septation, and valve development. However, this complex network is vulnerable to disruptions due to drug exposure, environmental factors, and genetic mutations, which can lead to abnormal cell proliferation, migration, differentiation, and structural or functional heart defects. For example, maternal diabetes can adversely affect fetal heart development by increasing oxidative stress and altering the expression of genes crucial for cardiac development, potentially leading to CHD[86]. These factors can also contribute to acquired cardiomyopathies, including diabetic cardiomyopathy (DCM), hypertrophic cardiomyopathy (HCM), and ischemic cardiomyopathy[87-89]. Previous studies have primarily relied on animal models, such as mice, to investigate and uncover the underlying mechanisms of these conditions[90]. By modeling complex pathological microenvironments, COs offer a high-fidelity platform for studying genetic and acquired heart diseases (Table 3).
Disease Organoid Type Key Pathological Features Clinical Validation CHD NKX2.5-KO heart-forming organoids[41]. Structural abnormalities, cellular hypertrophy, and dysregulated gene expression. Atrial septal defects and cardiac conduction abnormalities caused by NKX2.5 mutations in patients. PDG-CHD PDG-induced human heart organoids[28,93]. Cardiac development abnormalities (impaired chamber formation, imbalanced cell subtypes) and electrophysiological abnormalities;
Metabolic dysfunction;
ER stress and VLCFA lipid imbalance[93].Macrosomia;
Electrophysiological abnormalities;
Metabolic disorders.MI Human cardiac organoids with oxygen-diffusion gradient and norepinephrine stimulation[96]. Structural remodeling (infarct-border-remote zones, fibrosis);
Metabolic shift;
Pathological calcium handling.Transcriptomic overlap with human ischemic cardiomyopathy and animal MI models;
Fibrosis and metabolic shifts align with those observed in patients with post-MI biopsies.Cryo-injured cardioids[29,58]. Oxidative stress;
Fibrosis;
Calcium handling abnormalities.Patient tissue match;
Captopril effectively mitigated cryoinjury-induced fibrosis and dysfunction in organoids[58].Myocardial I/R Injury Hypoxia/reoxygenation-induced ventricular cardiac organoids[97]. Increased cell death, oxidative stress, and inflammatory response;
Fibrosis;
Electrophysiological dysfunction.H/R-induced injury mimics myocardial I/R injury;
Anifrolumab attenuates inflammation and oxidative stress in the model.DCM Cardiac organoids mimicking high glucose and lipid conditions[113]. Cardiac injury and cell apoptosis;
Oxidative stress and inflammation;
Mitochondrial dysfunction.Metformin intervention reduced apoptosis, inflammation, fibrosis, and mitochondrial dysfunction;
Consistent drug response with traditional models.AV conduction block LMNAc.1477C>T hiPSC-derived cardiac assembloids[67]. Aberrant intracellular calcium handling and conduction block. The model recapitulates AV block manifestations, and S107 partially restores conduction. Note. CHD: congenital heart disease. PGD: pregestational diabetes. MI: myocardial infarction. I/R Injury: myocardial I/R injury. DCM: diabetic cardiomyopathy. VLCFA: very long-chain fatty acids. AV: atrioventricular Table 3. Disease-specific organoid models and validation
-
CHDs, caused by abnormal development of the heart or blood vessels, are the most common congenital disabilities in humans, affecting approximately 0.4-5% of newborns worldwide, making them the most common type of congenital disease[91]. Common CHDs, such as left ventricular maldevelopment syndrome, ventricular septal defects, and heart failure, often stem from abnormalities in cardiac primordia development during the embryonic stage. Although complex animal models such as zebrafish and mice are useful for identifying cardiac environmental risk factors, they are limited by species-to-species variability and low throughput.
Gene editing of transcription factors in COs enhances our understanding of early cardiac morphogenesis defects. hPSC-derived HFOs accurately mimic the spatiotemporal organization of early human cardiac primordia, uncovering structural and molecular causes of CHD. These models not only reproduce the multi-germ-layer co-development of the myocardium, endocardium, and foregut endoderm but also integrate gene-editing technologies to dynamically analyze key CHD-causing genes. Drakhlis et al. developed NKX2.5-KO HFOs that recapitulate CHD phenotypes linked to NKX2.5 mutations[41], showing reduced myocardial layer density, adhesive defects, and enlarged cell surface areas, aligning with cardiac malformations in transgenic mice[92]. Single-cell transcriptomic analysis suggests that loss of NKX2.5 causes structural defects by disrupting cardiomyocyte maturation and lineage-specific differentiation. Hofbauer et al.[29] knocked out HAND1 and NKX2.5 in human ESCs to model hypoplastic left heart syndrome. Li et al.[67] introduced LMNA mutations into cardiomyocyte assemblies, effectively recapitulating atrioventricular conduction blocks observed in vertebrate hearts. They revealed that intracellular calcium mishandling underlies the pathology and proposed treatment strategies. These insights have not been achieved using conventional 2D cardiomyocyte differentiation systems, underscoring the irreplaceability of organoids in simulating tissue-microenvironment interactions.
Pregestational diabetes (PGD), which occurs before pregnancy and during the first trimester, is a leading nongenetic risk factor for CHD. Approximately 12% of infants born to mothers with diabetes develop CHD, which can cause macrosomia, arrhythmias, and structural defects (such as tetralogy of Fallot). Lewis-Israeli et al. developed functional heart organoids that mimic the PGD metabolic environment (11.1 mM glucose/1.14 nM insulin) to investigate its impact on CHD[28]. Diabetic organoids showed significant phenotypic abnormalities, including enlarged morphology (macrosomia-like features), arrhythmias, metabolic reprogramming, and lipid metabolism disruption, along with structural defects consistent with clinical CHD. Kostina et al. demonstrated that COs under PGD conditions exhibited marked ER stress, resulting in the degradation of FADS2 mRNA, a critical enzyme in lipid metabolism, and causing an imbalance of very long-chain fatty acids[93]. These insights highlight potential therapeutic targets for preventing and treating PGD-associated cardiac conditions.
-
The major pathological burden of cardiovascular diseases stems from environmental exposures. Ischemic heart disease is a predominantly acquired cardiovascular disease characterized by myocardial ischemia and ischemic cardiomyopathy. Epidemiological studies indicate MI accounts for 8.5% of cardiovascular disease-related deaths and is the leading cause of heart failure[94]. Current research is limited by the inability of animal models to replicate the heterogeneity of human myocardial injury. In addition, clinically available myocardial samples are typically derived from end-stage tissues, which limit our understanding of dynamic acute injury mechanisms such as FB phenotypic switching, ECM remodeling, and immune microenvironment modulation. This translational gap substantially hinders the development of innovative therapies for myocardial repair.Compared with animal models, organoids preserve human-specific ECM remodeling, which is critical for mimicking human myocardial injury.
Cryoinjury, which induces cardiomyocyte necrosis via localized cooling (using pre-cooled needles or brief contact with dry ice), mimics the pathological cascade of MI[95]. It triggers abnormal calcium transients, cell damage, and fibrotic remodeling in COs. Researchers have successfully recapitulated the fibrotic features of human ventricular injury, including the directional migration of FBs around the injury foci and abnormal deposition of fibronectin and collagen I, matching in vivo pathology and offering a controllable platform for studying fibrosis initiation[29]. Yang's model of cryoinjury in vascularized COs demonstrated that captopril significantly inhibited fibrosis and improved contractile function, highlighting its potential for MI treatment and the translational value of organoids in drug efficacy assessments[58].
Richards et al. integrated oxygen diffusion gradients and chronic noradrenaline stimulation to create a spatially heterogeneous MI model in a 3D COs, mimicking the “infarct core-border zone-remote” pathological gradient[96]. This model revealed hypoxia-induced metabolic dysfunction—including glycolysis upregulation and mitochondrial dysfunction—as well as fibrotic features (such as FB activation and increased matrix stiffness). Co-culture with THP-1 monocytes confirmed inflammatory activation in damaged regions, offering a tunable humanized model for acute myocardial injury research. Recent studies using human ventricular organoids have established hypoxia/reoxygenation (H/R) injury models that characterize multidimensional I/R injury features[97], including cardiomyocyte apoptosis, oxidative stress, structural damage, and arrhythmias. THP-1 immune cell co-cultures showed significant inflammatory crosstalk between H/R-injured organoids and monocytes/macrophages, further validating the use of THP-1 cells to model immune response in cardiac injury.
Notably, organoids across genetic backgrounds exhibit human-like pathological phenotypes under diverse environmental stimuli, underscoring the significance of nongenetic factors in myocardial injury.
-
According to data from the United States Department of Health and Human Services (2014), nearly one million patients are hospitalized annually because of adverse drug reactions, with proarrhythmic toxicity identified as a leading causative factor[98]. hiPSC-derived cardiomyocytes are widely used for personalized drug screening. Techniques such as patch-clamp and microelectrode arrays enable the quantitative analysis of ion channel kinetics, including hERG inhibition. However, 2D monolayer models fail to simulate cardiac electromechanical coupling because of the lack of FB-cardiomyocyte interactions and 3D tissue structures, limiting their clinical translation value[99,100]. COs have overcome these limitations by offering high-throughput screening, cost-effectiveness, and greater physiological relevance to humans. They provide a precise, robust, and human-relevant platform for drug discovery and cardiac toxicity testing. For example, Mills et al. used hiPSC-derived COs to screen for promyocardial regenerative compounds[101]. Metabolomics analysis revealed that mevalonate pathway inhibitors promoted cell proliferation while avoiding arrhythmia risks. This dual assessment of efficacy and toxicity is unachievable in traditional models. Recent studies have demonstrated that COs can accurately recapitulate doxorubicin-induced cardiotoxic phenotypes, including contractile inhibition, calcium handling dysfunction, and apoptosis activation, consistent with clinical observations. These findings highlight their potential as ideal platforms for toxicity screening[58].
Current drug development processes remain limited by insufficient integration across stages. Early-stage optimization often focuses on target activity, whereas metabolism-dependent cardiotoxicity (including hepatic enzyme-mediated proarrhythmic risks) is frequently identified only in later clinical phases. This sequential approach carries high risk, as drug candidates with potential cardiotoxicity may lead to substantial financial losses due to late-stage failures[102]. Moreover, the stage-based research model may fragment the holistic understanding of drug mechanisms.
To evaluate potential side effects, chip-based models of the heart, gut, liver, kidneys, and pancreas have been studied[103] to understand drug absorption, metabolism, clearance, and organ-specific toxicity, as well as to evaluate interorgan interactions. Multi-OoC can integrate these models to reproduce drug metabolism-toxicity effects in vitro. Yin et al. demonstrated the metabolism-dependent cardiotoxicity of the antidepressant clomipramine using a liver-heart multi-organ chip[82]. When the drug was applied to the upper chamber containing hepatic organoids, its metabolites significantly reduced the viability of COs in the lower chamber and induced apoptosis of cardiomyocytes. Fluorescent calcium imaging further revealed reduced intracellular calcium flux in COs, underscoring the value of the chip in early, synchronized pharmacological and toxicological analysis. Recent studies have established a high-throughput Microphysiological System Chip Platform that integrates intestinal, hepatic, cardiac, and lung cancer spheroids to simulate the systemic effects of anti-lung cancer drugs. The platform demonstrated varying degrees of damage to the gut, liver, and heart, as well as reduced efficacy of four drugs against lung cancer owing to their absorption by normal spheroids. For example, pemetrexed exhibited diminished targeting efficacy after intestinal absorption and hepatic metabolism, while reducing liver spheroid viability[104]. This system also evaluates the true pharmacological effects of drug molecules reaching target lesions after absorption by normal organs through fluid-mediated physiological communication.
Other multi-OoCs, such as heart-kidney and heart-brain chips, can simultaneously evaluate drug efficacy and organ-specific toxicity in early experiments, recapitulating metabolism and cross-organ toxic effects in vitro[81,84]. These platforms, featuring miniaturized designs and dynamic fluidic systems, support a “single-experiment, multi-dimensional assessment” strategy. They enable concurrent monitoring of drug targeting, metabolic responses, and toxic phenotypes, considerably shortening drug discovery timelines and advancing an integrated drug development workflow.
-
Environmental pollution is an important but often overlooked risk factor for cardiovascular diseases such as heart failure. COs offer a substantial advantage owing to their ability to precisely mimic complex pathological microenvironments, including gestational diabetes, pathogenic infections, ionizing radiation, and exposure to drugs and other environmental toxins. They enable dynamic dissection of the regulatory mechanisms of environment-genetic interactions in cardiac development and homeostasis, emerging as a powerful tool for assessing the cardiac toxicity of environmental pollutants.
COs have been used to assess the cardiac toxicity of lead, mercury, thallium, and the pesticide glyphosate, demonstrating accurate responsiveness to external stimuli and providing powerful technical support for environmental toxin screening[105]. In addition, low-dose cadmium exposure affects cardiac development by inhibiting cardiomyocyte differentiation and reducing the beating frequency of mature COs, posing a risk of CHD. Notably, these effects were observed in organoids but not in undifferentiated cardiomyocyte cell lines[106].
The exponential increase in global plastic production and inefficient recycling systems have led to the accumulation of microplastics (< 5 mm) and nanoplastics (< 1 μm), collectively known as MNPs, in ecosystems. MNPs can enter the human body via the digestive and respiratory tracts, breach physiological barriers, and accumulate in tissues such as blood, placenta, and myocardium, raising international concerns about their potential cardiotoxicity[107-109]. Although traditional animal studies have confirmed that MNPs can accumulate in myocardial tissue and induce oxidative stress and inflammatory responses, their limited species specificity and insufficient spatiotemporal resolution in static exposure models hinder precise analysis of chronic damage mechanisms and dose-response relationships in human cardiomyocytes under pathological conditions[110]. COs, which functionally mimic the human heart (exhibit sustained contractile activity in vitro), enable the continuous monitoring of MNP-induced chronic cardiac injury. In addition, integration with microfluidic systems addresses gravity-driven sedimentation in static exposure models and enhances cellular uptake efficiency. Zhang et al. developed a functional COoC model featuring myocardial spheroids (approximately 600 nm in diameter) that matured and maintained stable contractility within the chip[111]. Short- and long-term exposures to polystyrene nanoplastics (PS-NPs) revealed dynamic cardiac injury, with long-term exposure significantly promoting collagen deposition and markers of myocardial fibrosis (such as α-SMA and COL1A1). Notably, under adrenaline-induced simulated MI conditions, subtoxic doses of PS-NPs synergistically exacerbated cardiomyocyte apoptosis, mitochondrial damage, and reduced contractility, indicating heightened susceptibility to MNPs in patients with cardiovascular disease.
In a recent study, Yang et al. utilized human self-organized COs to determine the cardiotoxicity of triclocarban (TCC) at environmental exposure doses[112]. TCC, a broad-spectrum antibacterial agent widely added to personal care products (such as antibacterial soaps and toothpaste), is among the top 10 global water pollutants. Its high lipophilicity and environmental persistence lead to continuous bioaccumulation in aquatic sediments, soil, and organisms, with human exposure occurring via dermal absorption, water intake, and food chain transfer. They found that low-dose TCC exposure induced cardiomyocyte hypertrophy, whereas high-dose TCC exposure triggered severe cardiotoxic effects, including cardiomyocyte apoptosis and heart failure. Single-cell transcriptomic analysis revealed that TCC disrupted EC metabolic homeostasis, causing microvascular dysfunction and activating hypertrophic signaling. Notably, TCC-induced reprogramming of amino acid metabolism (enhanced glutaminolysis and suppressed branched-chain amino acid metabolism) closely matched metabolic features observed in patients with HCM, underscoring the clinical relevance of organoid models in toxicity mechanism analysis. These models provide highly predictive tools for assessing the cardiotoxicity of environmental pollutants.
As human lifestyles change, DCM has emerged as a key contributor to secondary heart failure. Chronic exposure to a high-glucose, high-lipid microenvironment induces progressive cardiomyocyte injury through mechanisms such as advanced glycation end product accumulation, mitochondrial oxidative stress, and calcium dysregulation. However, the cell-specific response networks remain poorly defined. Recent in vitro studies have modeled DCM using human COs[113]. The high-lipid environment induced apoptosis in COs, with significant upregulation of pro-apoptotic markers (including BAX and APAF1). In addition, mRNA levels of cardiac injury markers were aberrantly altered, indicating severe disruption of normal cardiac function and structural integrity, accompanied by inflammation and fibrosis. Notably, metformin intervention reversed these phenotypes, validating the reliability of this model for toxicity assessment and drug screening. This study highlights the utility of COs in elucidating the pathogenesis of DCM and evaluating therapeutic interventions.
Furthermore, the systematic integration of COs with microfluidic chips provides a highly bionic assessment platform for subsequent cardiac developmental toxicity studies of a wider range of environmental pollutants (including PM2.5 and BPA)[105]. For example, BPA disrupts the directed differentiation of cardiac progenitor cells via an ERα-dependent oxidative stress pathway, leading to congenital structural abnormalities such as ventricular septal defects. This mechanism has been validated through a bioinformatics-driven Adverse Outcome Pathway framework[114]. In the future, the application of COs and COoC systems will enable precise simulation of vascular endothelial transport processes. This advancement will substantially enhance the controllability of pollutant-tissue interactions and support high-throughput exposure experiments, offering a standardized and scalable new paradigm for environmental toxicology research.
-
Despite considerable progress in modeling cardiac development, disease mechanisms, and drug screening, COs face substantial technical and biological challenges. Current models primarily reflect embryonic or fetal cardiac features and lack the structural and functional maturity of adult hearts. For instance, they lack key anatomical features, such as perfusable vascular networks, valvular structures, and defined outflow tracts, which limit their ability to model ischemic injury dynamics or long-term viability[115]. In addition, the absence of tissue-resident immune cells, neural crest-derived autonomic neurons, and endocardial-myocardial interface barriers severely restricts the modeling of complex pathologies such as immune-cardiac interactions or neuro-cardiac regulation[116,117]. Current protocols for COs fabrication lack standardization, leading to unpredictable cardiomyocyte maturation within 3D differentiation systems. Furthermore, the spherical geometry of 3D organoids causes uneven circumferential stress distribution between peripheral and central regions, which affects cell differentiation, maturation, and chamber formation via mechanotransduction pathways[118]. Although geometric constraints have been employed to induce multi-chambered structures, considerable variability in chamber number and overall morphology persists across iPSC-derived organoids[28,36]. Research on COoC and multi-OoC platforms remains in its early stages. Several challenges must be addressed, including achieving synchronous maturation of organoids within the chip, accurately simulating inter-organ physiological coupling (such as liver-heart metabolism), and enabling medium transfer. Collectively, these limitations restrict the potential application of COs in disease modeling, personalized therapy, and regenerative medicine.
Given these challenges, new technologies should be rapidly integrated into CO research to drive translational progress. Single-cell atlas analysis is a vital tool for unraveling multicellular COs. The analysis of cellular heterogeneity provides insights into cell fate transitions and deepens our understanding of cardiac development and disease mechanisms. Zhang et al. constructed a single-cell resolution map of 3D-cultured multilineage cardiac organoids, enabling in-depth dissection of cell fate transitions, state changes, and underlying gene regulatory networks. Transplantation studies further validated the reparative capacity of multilineage COs in MI models[119]. Feng et al. established the first chamber-specific disease organoid model using integrated scRNA-seq and machine learning-driven cross-species annotation transfer. This platform provides a novel tool for screening structural heart disease drugs. Their single-cell pseudo-temporal trajectory analysis demonstrated that NKX2-5 deficiency redirects ventricular progenitors toward atrial lineage by dysregulating WNT/β-catenin signaling[120]. Spatial transcriptomics enables 3D structural investigation of organoids by preserving in situ spatial information[121,122]. Although spatially resolved omics is well established for native cardiac tissues, its application in COs remains in its nascent phase. In the future, the integration of single-cell and spatial omics will advance the precise interrogation of cardiac organoid heterogeneity. By mapping gene/protein expression profiles to 3D coordinates, these technologies can resolve cell type distributions, niche formation mechanisms, and intercellular interactions within organoids, systematically elucidating cellular diversity and spatial patterns[123].
The integration of CRISPR technology with COs is driving considerable advancements in cardiac research. In monogenic disorder studies, CRISPR/Cas9-mediated genome editing in iPSC-derived cardiomyocytes has been achieved for pathological modeling. For example, Liang et al. demonstrated that TRPM4-mutant models exhibit hyperactivation of PDGFRB signaling, which triggers calcium overload, thereby elucidating the pathogenesis of BrS[124]. Extending genetic editing to functional COs, such as constructing SCN5A-mutant models, may overcome the limitations of 2D systems in simulating conduction abnormalities, owing to their deficient structural microenvironments, thereby enhancing their clinical relevance in pathophysiological studies. Integrating CRISPR-based screening platforms with multichambered COs enables high-throughput analysis of drug toxicity mechanisms. The use of CRISPR interference or activation libraries, particularly genome-wide collections or those targeting specific pathways, within mature organoids facilitates unbiased mechanistic discovery. Wang et al. have developed a double-strand break-free genome-wide CRISPR screening method called BARBEKO. This approach integrates the iBAR molecular barcoding system, substantially reducing cellular input required for screening, while mitigating the impact of DNA-cleavage-induced cytotoxicity[125]. Critically, this strategy demonstrates considerable potential for application in complex physiological models such as organoids.
Emerging machine learning and artificial intelligence (AI) technologies are revolutionizing organoid research by directly addressing core challenges ranging from structural design to functional characterization. Kowalczewski et al. constructed a high-throughput geometric organoid library and integrated an AI-driven workflow for unsupervised bias-free analysis, revealing that hexagonal designs optimized calcium transients in COs and established standards for organ-on-chip systems[126]. Similarly, Lin et al. developed a multimodal AI framework integrating electrophysiological signals, cytokine profiling, and volumetric imaging to precisely quantify immune cell infiltration and fibrosis within inflammatory COs; this platform successfully validated the efficacy of IL-1β inhibitor and accelerated translational exploration for immune-mediated cardiomyopathies[127].
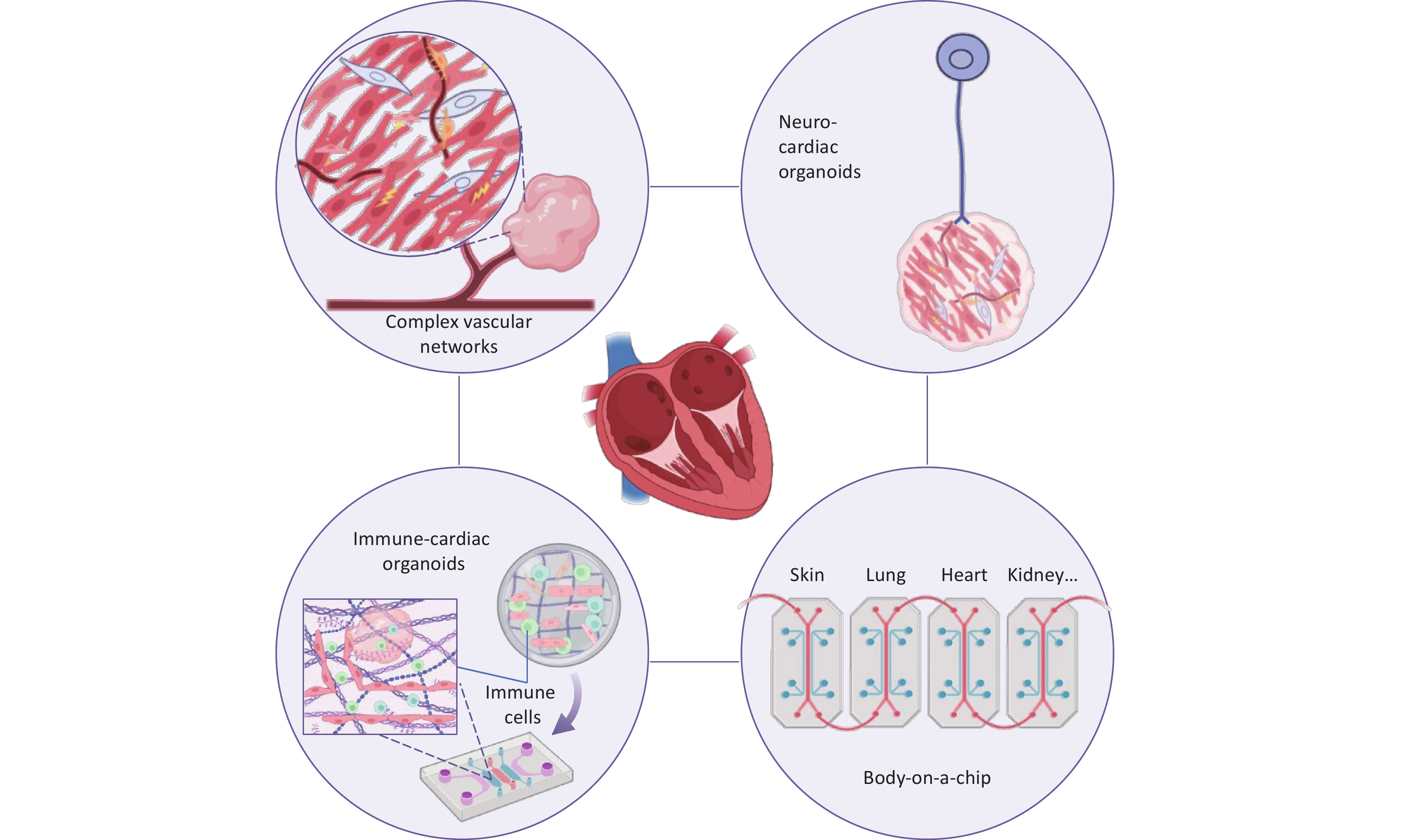
In addition to these advances in technology integration, we propose that future organoid research should focus on several key directions (Figure 3).
a. Human vascular-COs (hVCOs): Currently, promising vascularization strategies include directed activation of endogenous developmental signals[28,42], temporal regulation of exogenous factors[44,128], and engineering approaches[58], among others, collectively providing the basis for future vascularization of COs. Modified pro-angiogenic scaffolds, such as peptide-functionalized hydrogels[129], are also being explored. In addition, combining organoid technology with bio-3D printing and microfluidic perfusion systems offers a promising avenue to establish mature, perfusable models[62]. The InVADE platform proposed by Lai et al. offers an innovative approach to constructing integrated vascularized systems[130]. In this setup, HUVECs are seeded into the lumen of the AngioTube scaffold, forming a continuous barrier that mimics the inner wall of blood vessels. A gravity-driven perfusion system simulates the physiological blood-flow environment within the body. Cardiomyocytes encapsulated in hydrogel remodel along the vascular structure, forming a functional 3D cardiac tissue that exhibits spontaneous contraction and electrical signal conduction. This platform enables dynamic simulation of drug permeation and metabolism, facilitating accurate cardiac toxicity assessment. Future advancements in hVCOs platforms could enhance physiological relevance by precisely mimicking the microvasculature, for example, by reducing scaffold wall thickness. Furthermore, integrating hVCOs into multi-OoC to simulate systemic drug metabolism-cardiotoxicity cascades will provide a more powerful tool for cardiovascular disease modeling[10,131].
b. Immune-COs: Cardiac development and homeostasis rely on dynamic immune regulation; however, conventional COs lack tissue-resident immune cells such as macrophages, limiting their ability to model this process[132]. Recent studies have addressed this by integrating hPSC-derived primitive or autologous heart tissue-resident macrophages to develop organoid models with partial immune functions. O'Hern et al.[133] developed a human heart-macrophage assembloid platform, revealing that macrophages maintain cardiac homeostasis via pro-inflammatory apoptosis, integrate into cardiomyocyte electrophysiological systems, and support catabolism. Chronic exposure to proinflammatory factors on this platform replicated key features of inflammasome-mediated atrial fibrillation. Another study incorporated hPSC-derived primitive macrophages into vascularized cardiac chips, showing enhanced cardiomyocyte electromechanical coupling and prolonged chip perfusability, associated with the upregulation of cardiac maturation genes and angiogenic factors[134]. These findings highlight that macrophages influence cardiac function through immune-metabolic-electrophysiological coupling. Currently, immune-integrated models face several challenges. First, controlling immune cell heterogeneity is difficult, as precise regulation of macrophage polarization (M1/M2) and subset ratios remains elusive, which affects the fidelity of inflammation simulations. Second, dynamically modeling the spatiotemporally dependent functions of immune cells during cardiac development—such as embryonic macrophages guiding endocardial cushion remodeling—requires new strategies. Immune-integrated CO models offer a valuable platform for investigating cardiac anomalies, inflammatory heart diseases (such as myocarditis and heart failure), and immunomodulatory drug screening, thereby accelerating clinical translation.
c. Neuro-COs: Cardiac physiological functions are highly dependent on dynamic regulation by the autonomic nervous system (ANS)[135]. Studies have shown that excessive activation of the sympathetic nervous system can enhance calcium cycling in cardiomyocytes and shorten the duration of action potentials, thereby inducing adrenergic arrhythmias. In contrast, dysfunction of the parasympathetic nervous system is closely associated with pathological phenotypes, such as vagally mediated atrial fibrillation and Brugada syndrome[116]. The molecular mechanisms underlying ANS regulation of cardiac electrophysiology and its relationship to specific arrhythmia types remain incompletely understood, highlighting an urgent need to develop in vitro systems that can simulate dynamic neural-cardiac interactions. Cardiac neural crest cells, a migratory cell population derived from the cranial neural tube, serve as the developmental origin of cardiac autonomic nerves and play essential roles in maintaining electrophysiological homeostasis, organogenesis, and post-injury cardiac repair[136]. hPSC-based in vitro models offer new approaches for studying these neural-cardiac interactions. For instance, Oh et al. established a co-culture system combining hPSC-derived sympathetic neurons with neonatal mouse ventricular myocytes[137]. Ultrastructural imaging revealed vesicle-like structures resembling neuromuscular synapses at sympathetic neuron terminals, physically connecting to cardiomyocytes. However, this model is limited by the immaturity of cardiomyocytes and inherent constraints of co-culture systems. Notably, studies on human neuromusculoskeletal organoids have shown that suppressing SMAD signaling (along with providing other regulatory factors) directs neuroectodermal differentiation. This process forms neuronal networks (Tuj1+) whose axons penetrate muscle domains (TITIN+) and transmit electrical signals via synaptic vesicles to induce target tissue contraction[136]. This approach provides valuable insights for studying neural innervation in COs. Future studies should integrate cutting-edge technologies such as transcriptomics, optogenetics, and microfluidic chips to address the inefficiency of neural innervation in COs, recapitulate the dynamics of ANS regulation, and elucidate the molecular mechanisms of neural-cardiac interactions. Such innovations will offer powerful platforms for uncovering ANS imbalance-driven arrhythmia pathways, developing personalized anti-arrhythmic drugs, and advancing cardiac regenerative therapies.
d. Multi-OoC: The Multi-OoC platform aims to construct a micro-physiological system, often referred to as a “body-on-a-chip.” Future development should focus on creating integrated multi-system platforms that connect individual organ chambers to precisely recapitulate in vivo organ interactions, physiological relationships, metabolic pathways, key biological barriers, and drug responses[138]. The primary objective is to replicate the continuous interorgan medium exchange process present in the body, thereby substantially enhancing the dynamic associations that traditional single-organ models fail to capture. By simulating complex physiological environments, the multi-OoC platform enables precise disease modeling and drug screening, providing a superior platform for studying systemic and body-wide diseases.
COs have provided high-fidelity models for decoding cardiac pathogenesis. The convergence of bioengineering, developmental biology, computational modeling, and AI offers immense hope for advancing CO technology. Innovations such as 3D bioprinting, optogenetics, CRISPR-based editing, and AI-driven analysis/modulation are poised to enable precise spatiotemporal control over multicellular crosstalk within tri-lineage neuro-immune-cardiac assembloids. Integrating vascularized COs with parenchymal tissues may bridge the gap between single-organ models and systemic physiology. Standardized maturation protocols and scalable production pipelines are crucial for clinical translation, particularly in cell therapy and personalized medicine. Addressing these challenges could enhance our understanding of heart disease and accelerate the development of targeted therapies, bringing in vitro models closer to reflecting human pathophysiology.
Cardiac Organoids: Emerging Tools for Investigating Environmental Roles in Cardiomyopathy Pathogenesis and Therapeutic Development
doi: 10.3967/bes2025.104
- Received Date: 2025-04-19
- Accepted Date: 2025-08-01
-
Key words:
- Cardiac organoids /
- 3D cell culture /
- Disease modeling /
- Drug discovery /
- Environmental cardiotoxicity /
- Organoid-on-a-chip /
- Self-organization /
- Vascularization
Abstract: Human cardiac organoids have revolutionized the study of cardiac development, disease modeling, drug discovery, and regenerative therapies. This review systematically discusses strategies and progress in the construction of cardiac organoids, categorizing them into three main types: cardiac spheroids, self-organizing/assembloid organoids, and organoid-on-a-chip systems. This review uniquely integrates the advances in vascularization, organ-on-chip design, and environmental cardiotoxicity modeling within cardiac organoid platforms, offering a critical synthesis that is absent in the literature. In the context of escalating environmental threats to cardiovascular health, there is an urgent need for physiologically relevant models to accurately identify cardiac toxicants and elucidate their underlying mechanisms of action. This review highlights advances in cardiac organoid applications for disease modeling—including congenital heart defects and acquired cardiovascular diseases—drug development, toxicity screening, and the study of environmentally induced cardiovascular pathogenesis. In addition, it critically examines ongoing challenges and underscores opportunities brought by bioengineering approaches. Finally, we propose future directions for developing standardized cardiac organoid platforms with clinical predictability, aiming to expand the utility of this technology across broader research applications.
The authors declare that they have no competing interests.Not applicable.
| Citation: | Yaoyao Xu, Zhimin Wang. Cardiac Organoids: Emerging Tools for Investigating Environmental Roles in Cardiomyopathy Pathogenesis and Therapeutic Development[J]. Biomedical and Environmental Sciences. doi: 10.3967/bes2025.104 |





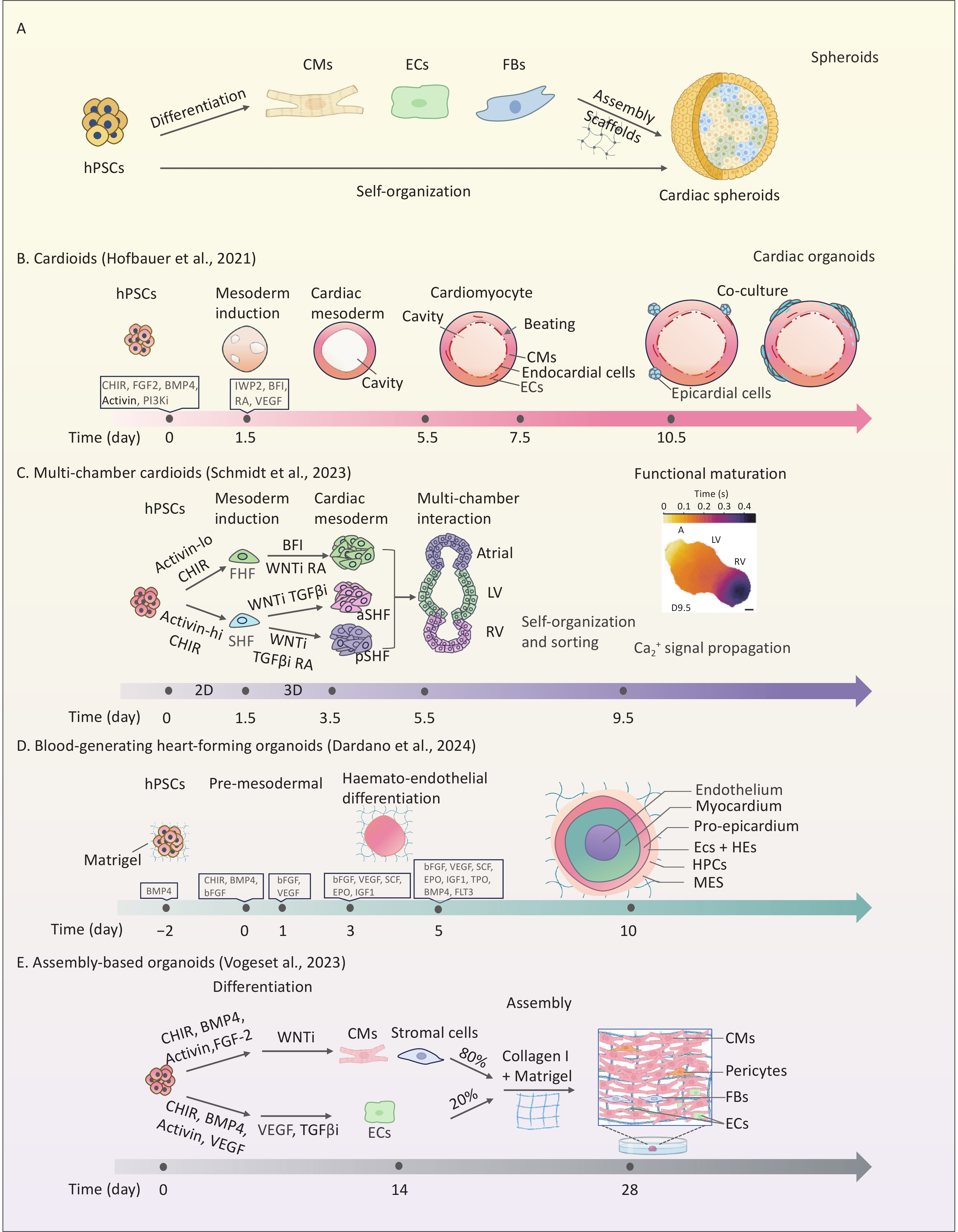

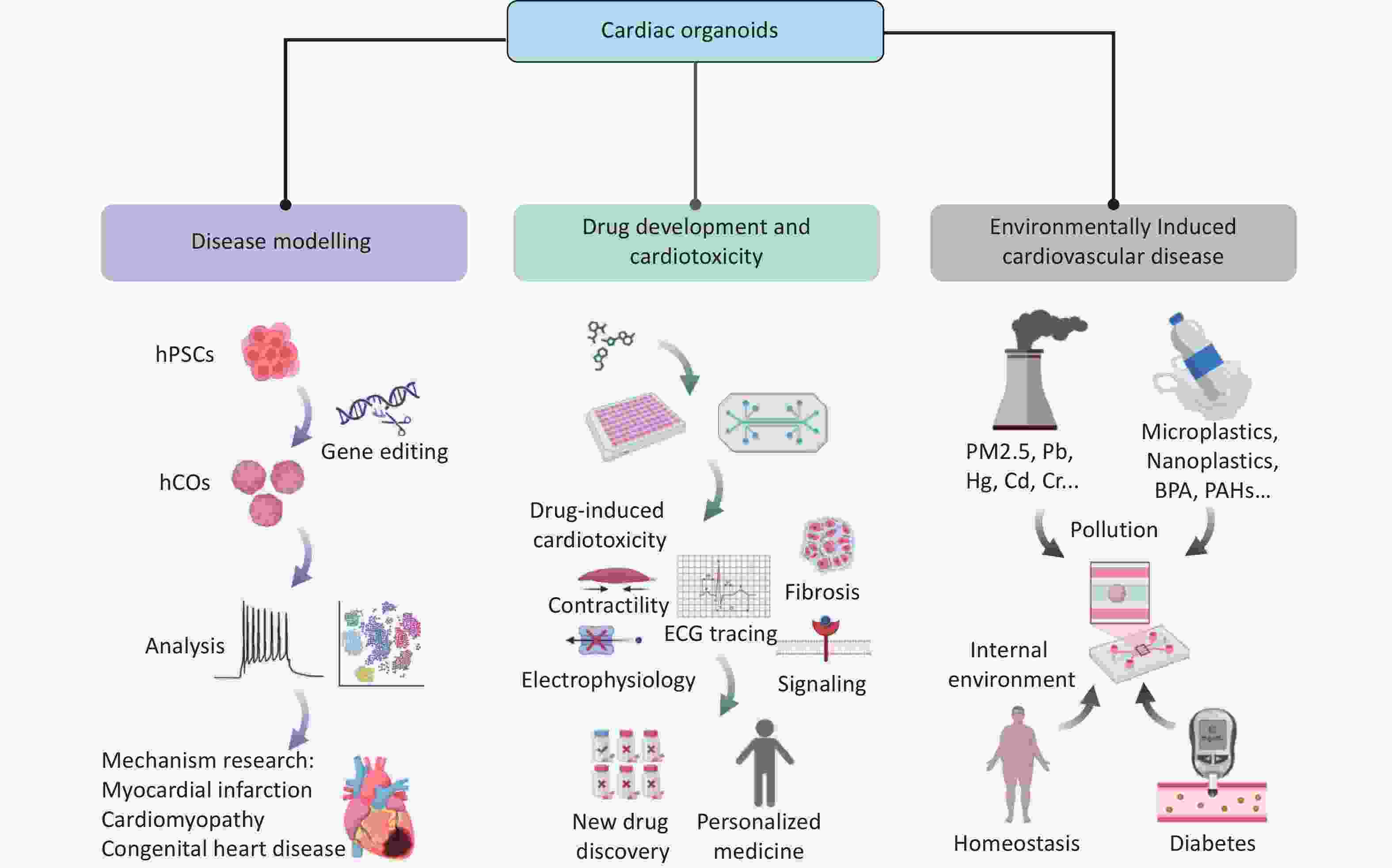
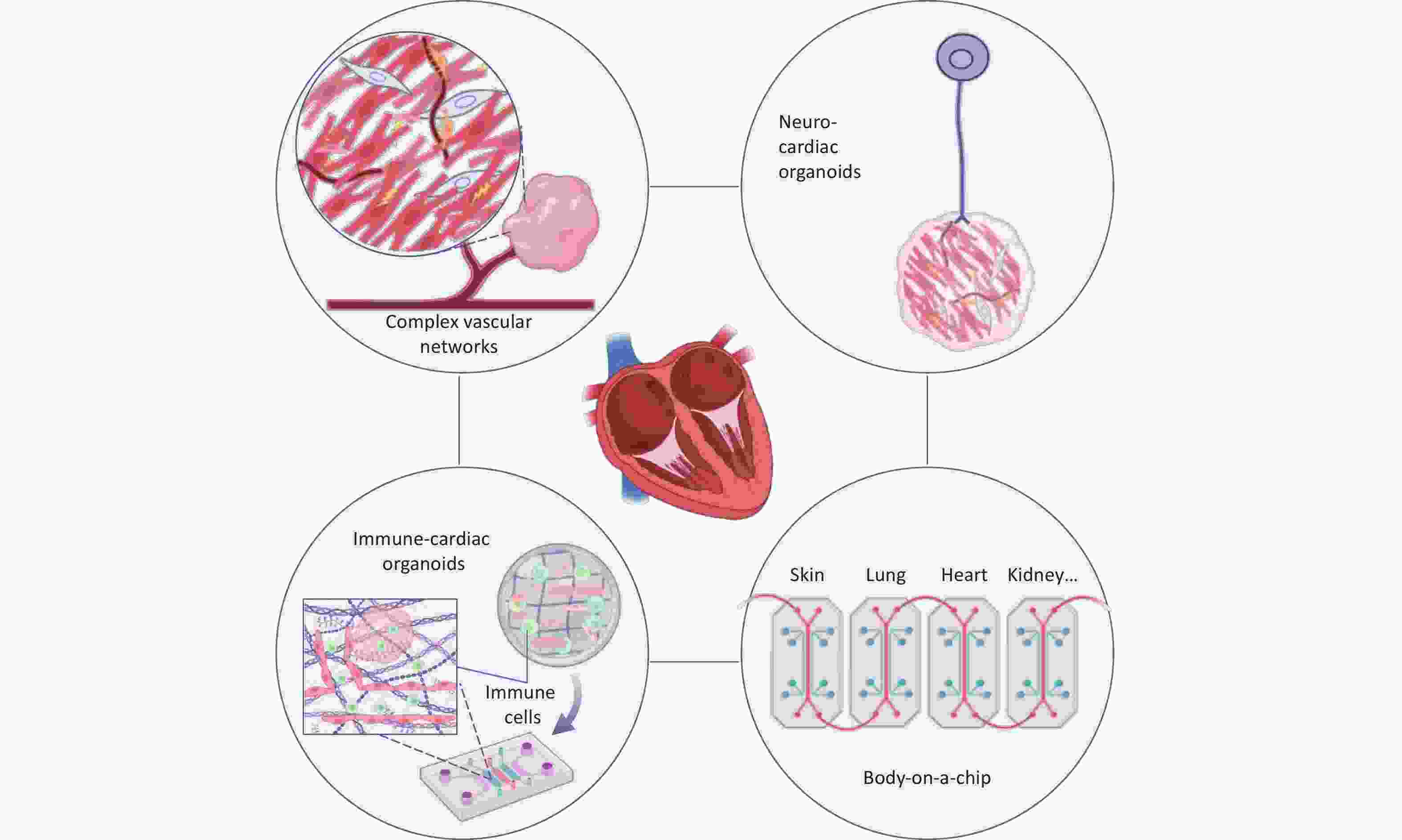

 Quick Links
Quick Links
 DownLoad:
DownLoad: