-
Legionella is a Gram-negative bacterium. Legionella is widely found in freshwater environments, moist soil and compost, showers, hot tubs, plumbing networks, air-conditioning systems, and other natural and artificial water environments close to areas inhabited by humans. Legionella spreads from these environments to humans via aerosols and infects lung macrophages, causing Legionnaires' disease[1,2]. More than 70 Legionella species have been identified, 25 of which can cause disease. Legionella pneumophila is one of the most pathogenic Legionella species and has 16 serotypes; serotype 1 accounts for 70%–90% of human Legionella infections[3,4] The antibiotics used to treat Legionnaires' disease mainly include macrolides and fluoroquinolones[5,6].
CRISPR-Cas is an adaptive immune system that protects bacteria and Archaea from invasive mobile genetic agents, particularly phages. The CRISPR-Cas systems are broadly categorized into two classes, six types, and 44 subtypes according to the Cas protein content and locus arrangement [7-10]. Class 1 systems are divided into types I, II, and IV, all of which employ multiprotein effector complexes to interfere with the invasion mechanisms of mobile genetic elements. Class 2 systems are classified into types II, V, and VI, all of which use single effector proteins for interfering with invasive mobile genetic agents[11]. The CRISPR-Cas system enables adaptive immunity through a three-stage process: adaptation, expression, maturation, and targeted interference[12-14]. CRISPR-Cas systems widely occur in nature, being found in approximately 50% and 90% of bacterial and archaeal genomes, respectively[15,16], and their occurrence does not correlate with ecological niche or phylogenetic branch. The type I CRISPR-Cas system is the most common, which uses a CRISPR RNA (crRNA)-bound multiprotein complex called the CRISPR-associated complex in antiviral defense to identify the target. This multiprotein complex uses nuclease Cas3 to cleave the target[12,17]. However, only a few type I CRISPR-Cas system systems have been characterized, such as type I-E in Escherichia coli[18,19]. Many Vibrio species have miniature I-F systems comprising tniQcas5cas7cas6f, which is associated with TN7-like transposons. Miniature I-F CRISPR-Cas systems identify Tn7-like transposons and target them at their insertion sites[20-22]. The type I-Fv CRISPR-Cas system from S. putrefaciens provides E. coli with heterogeneous protection against lambda phage infection[21].
The CRISPR-Cas system has been thoroughly investigated in several bacterial species, including numerous human pathogens, such as Staphylococcus epidermidis, Streptococcus pneumoniae, Yersinia pestis, Klebsiella pneumoniae, and Salmonella spp.[23-29] The CRISPR-Cas system is the core adaptive immune mechanism through which microorganisms resist phage invasion. The type I-C, I-F, and II-B CRISPR-Cas systems protect against exogenous genetic elements through sequence-specific cleavage (DNA or RNA targets)[30]. The understanding of the functions of the CRISPR-Cas have expanded from classical genome defense to nonclassical regulatory fields, such as gene expression regulation and epigenetic modifications. The II-B-type CRISPR-Cas system is dominant in Legionella pneumophila[31], and the nuclease activity of Cas2 is crucial for L. pneumophila growth within amoeba hosts[32,33]. The heterogeneous distribution, dynamic evolution, and the type I-F CRISPR-Cas system enable L. pneumophila to adapt to the environment[34]. The CRISPR-Cas spacer sequence library suggests that the CRISPR-Cas system enables continuous defense against Microviridae phages[35]. However, the biological functions of the type I-F CRISPR-Cas system in L. pneumophila have not been explored.
We assessed the functional phenotypes of the type I-F CRISPR-Cas system in L. pneumophila to explore the relationship between L. pneumophila and CRISPR-Cas. Our results lay the foundation for discovering the biological functions of the CRISPR-Cas system in L. pneumophila.
-
The type I-F environmental strain, L. pneumophila WX48, was used as the wild-type (WT) strain. All Legionella strains were cultured on buffered charcoal yeast extract (BCYE) agar plates (Oxoid) at 37 °C under 5% CO2 or in N-(2-acetamido)-2-aminoethanesulfonic acid-buffered yeast extract broth at 37 °C. E. coli DH5α was used for cloning via culturing in Luria-Bertani medium containing 100 µg/mL ampicillin and 25 µg/mL chloramphenicol at 37 °C.
-
Legionella strains (WX48) from aquatic environments with complete I-F type CRISPR-Cas system were screened and identified using whole-genome sequencing data. CRISPR-Cas system loci were annotated using CRISPRCasFinder, a web-based tool for computationally predicting CRISPR arrays and associated Cas proteins, with the default parameters.
The L. pneumophila deletion mutants were generate by separately amplifying the upstream and downstream fragments of Cas1, Cas2-Cas3, Csy1, Csy2, Csy3, Cas6f, and CRISPR arrays with homologous arms from L. pneumophila WX48 genomic DNA using polymerase chain reaction (PCR) with the appropriate primers. The primers are listed in Supplementary Table S1. Overlapping PCR was used to fuse the upstream and downstream fragments; the product was ligated to the pGEM-T Easy vector via TA cloning to generate pT-Cas/CRISPR constructs. The constructs were enzymatically digested using SmaI.
A kanamycin resistance (Km) cassette generated from pUT-mini-Tn5 Km was inserted into the upstream and downstream fusion fragments to generate pT-Cas/CRISPR-Km via enzymatic digestion and ligation. These pT-Cas/CRISPR-Km constructs were digested with NotI. Fragments carrying the disrupted Cas/CRISPR region were cloned into the NotI site of the allelic exchange vector PLAW344, generating PLAW344-Cas/CRISPR-Km. PCR amplification and sequencing were conducted using specific primers for each gene to confirm the recombinant plasmid was successfully constructed.
Recombinant plasmids carrying Cas and CRISPR arrays were introduced into competent cells of the original strain through electroporation. Transformants were selected on BCYE agar supplemented with 50 µg/mL kanamycin. Subsequently, the transformants were cultured on BCYE agar containing kanamycin and 5% sucrose at 37 °C under 5% CO2 for approximately 5 days to screen for ΔCas1, ΔCas2-3, ΔCsy1, ΔCsy2, ΔCsy3, ΔCas6f, and ΔCRISPR mutant strains. PCR amplification and sequencing were performed using specific primers for each gene, and successful construction of the mutants was confirmed.
We constructed a plasmid for the complementation of the Cas and CRISPR mutant strains by amplifying the target Cas genes and CRISPR array from the genomic DNA extracted from the WT strain. The product was digested with the corresponding enzymes and cloned into a PMMB207 plasmid. Finally, the plasmids were transferred into the corresponding competent cells via electroporation, and the transformants were cultured on BCYE agar containing chloramphenicol and sucrose to screen for ΔCas1, ΔCas2-3, ΔCsy1, ΔCsy2, ΔCsy3, ΔCas6f, and ΔCRISPR deletion strains. Positive colonies were confirmed by PCR and sequencing. The results of whole-genome sequencing (Illumina NovaSeq 6000 platform, 150-bp paired-end reads) with comparative genomic analysis revealed no detectable single-nucleotide polymorphisms (SNPs) or indels between the mutant and WT genomes, confirming the absence of secondary mutations.
-
We monitored extracellular growth by inoculating L. pneumophila into BYE broth, and incubating the culture at 37 °C overnight with shaking at 200 rpm. The culture was then quantitatively transferred to 50 mL of fresh broth, and the optical density at 600 nm (OD600) was measured using a spectrophotometer (NanoPhotometer NP80 Touch, Germany). The initial OD600 was 0.2. The cultures were incubated again at 37 °C under shaking at 200 rpm, and the OD600 was measured every 6 h until the growth stabilized. Each experiment was performed in triplicate.
-
Antimicrobial susceptibility was evaluated following the EUCAST recommendations and manufacturer’s instructionsthe. The minimum inhibitory concentration (MIC) of the antibiotic (Liofilchem, Teramo, Italy) against the complement, deletion, and WT strains was evaluated using the E test strip. Briefly, we prepared a bacterial suspension and adjusted its concentration to 0.5 McFarland standard. A sterile cotton swab was dipped into the bacterial solution, which was then evenly applied to the BCYE agar surface, and an E test strip was placed at the center. The agar was incubated at 37 °C for 48 h, after which the MIC of the E test strip was determined. L. pneumophila ATCC 33152 was used as the control in all tests. Each experiment was conducted in triplicate.
-
Fresh Legionella colonies on BCYE agar were suspended in BYE broth, and the OD600 of the suspension was adjusted to 0.3. Next, 200 µL of this suspension was added to each well of a 96-well microplate. The plate was then incubated at 37 °C under 5% CO2 for 3 days. We then added 40 µL of 0.25% crystal violet or 1.25% safranin to each well. The plate was allowed to stand at 25 °C for 15 min, after which the plate was washed three time with 300 µL of ddH2O and dried in air. L. pneumophila biofilms were visible as purple rings after crystal violet staining or red rings after safranin staining around the walls of the inoculated wells. We then added of 300 µL of 30% acetic acid to each well, and the plate was incubated for 15 min until the crystal violet or safranin dissolved. Finally, 200 µL of the sample was transferred to a new microtiter plate; the samples were then treated with crystal violet and read at OD600, or the samples treated with safranin were read at OD530 using a Synergy H1 plate reader (Biotek). Each experiment was conducted in triplicate.
-
A pure culture of fresh Legionella was resuspended in phosphate-buffered saline (PBS), and the Legionella concentration was adjusted to a McFarland standard of 2.0 (1 × 108 CFU/mL). The bacterial suspension was diluted 10-fold in DMEM-H and used to infect the mouse macrophage line J774 (1 × 10^5 cells/mL) at a multiplicity of infection of 100. The infected cells were incubated at 37 °C for 1 h under 5% CO2. The cells were then washed twice with PBS to remove extracellular bacteria, and 1 mL of sterile water was added. The cell suspension was then transferred to a 1.5 mL centrifuge tube. A series of 10-fold dilutions of the cell suspension were plated on BCYE agar, and the bacterial colonies that developed were counted. We measured the invasive strength of the bacteria by removing the extracellular bacteria via washing the cells with PBS three times, and 1 mL of the medium with 100 µg/mL of gentamicin was added to each well. The wells were incubated for 1 h to kill extracellular bacteria. The cells were washed again with PBS, and the number of intracellular bacteria was determined as described above in this section.
This infection process was repeated for an intracellular growth assay. The infected cells were incubated at 37 °C for 1 h under 5% CO2 and then washed three times with PBS to remove extracellular bacteria. Next, 1 mL of DMEM-H was added to each well, and the bacterial colonies proliferating within the J774 cells were counted every 24 h for three consecutive days. The total number of colony-forming units (CFU) formed after coating the BCYE agar with the cell suspensions was also calculated.
-
Cell smears were fixed on glass slides with methanol, stained with carbonate fuchsin for 5 min, gently washed with water, stained with 8% malachite green for 1 min, rinsed again, and air-dried for 1 min. The slides were dried and then observed under a light microscope. We randomly selected 100 fields of view under a microscope, counted the total number of cells and the number of bacteria-infected cells in each field, and calculated the ratio of bacteria-infected cells to the total number of cells.
-
Total RNA was reverse-transcribed into complementary DNA (cDNA) using a SuperScript III First-Strand Synthesis System for qRT-PCR analysis. cDNA was amplified using quantitative real-time PCR (qRT-PCR) with SYBR Green Real-time PCR Master Mix (Takara, Japan). The gene expression levels were relatively quantified using the 2−∆∆Ct method. GyrB was used as the internal control. Supplementary Table S1 presents the primers used for qRT-PCR.
-
We assessed the statistical significance of our data using Student’s t-test and one-way analysis of variance with Dunnett’s multiple comparison tests. P < 0.05 was considered as indicating significance.
-
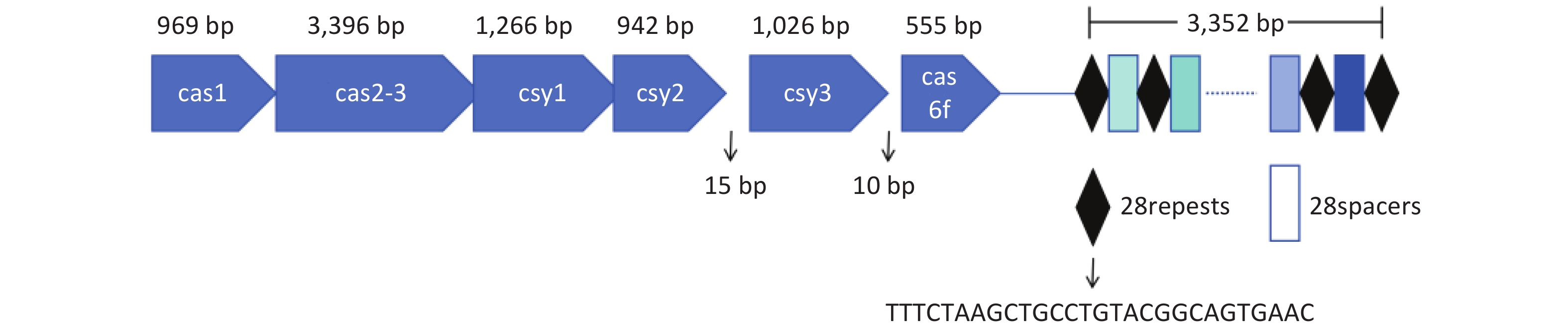
The type I-F CRISPR-Cas system of L. pneumophila WX48 was analyzed using CRISPRCasFinder[34]. Figure 1 shows that the type I-F system consists of six Cas genes, namely, Cas1, Cas2-Cas3, Csy1, Csy2, Csy3, and Cas6f, along with a downstream CRISPR array with a conserved repeat sequence (5’-TTTCTAAGCTGCCTGTACGGCAGTGAAC-3’).
-
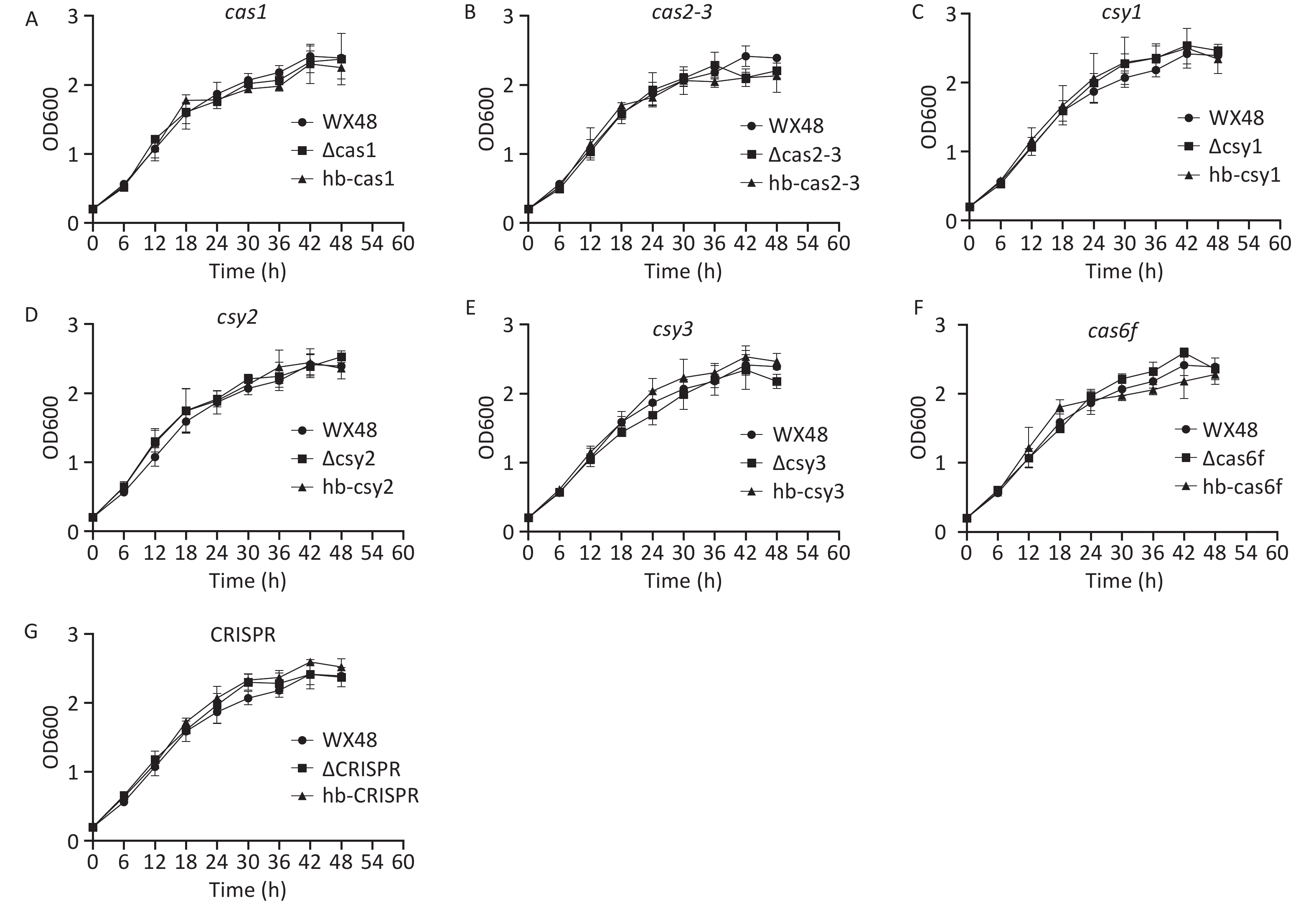
L. pneumophila WT as well as its ΔCas1, ΔCas3-Cas2, ΔCsy1, ΔCsy2, ΔCsy3, ΔCas6f, ΔCRISPR, and complementation mutant strains were grown in BYE broth. Their standard growth curves were plotted. The proliferation of the deletion mutants, complementation mutants, and WT strain was determined after their incubation at 37 °C for 48 h. The growth trends were consistent among all the strains, indicating that the CRISPR-Cas system did not affect the growth of L. pneumophila (Figure 2).
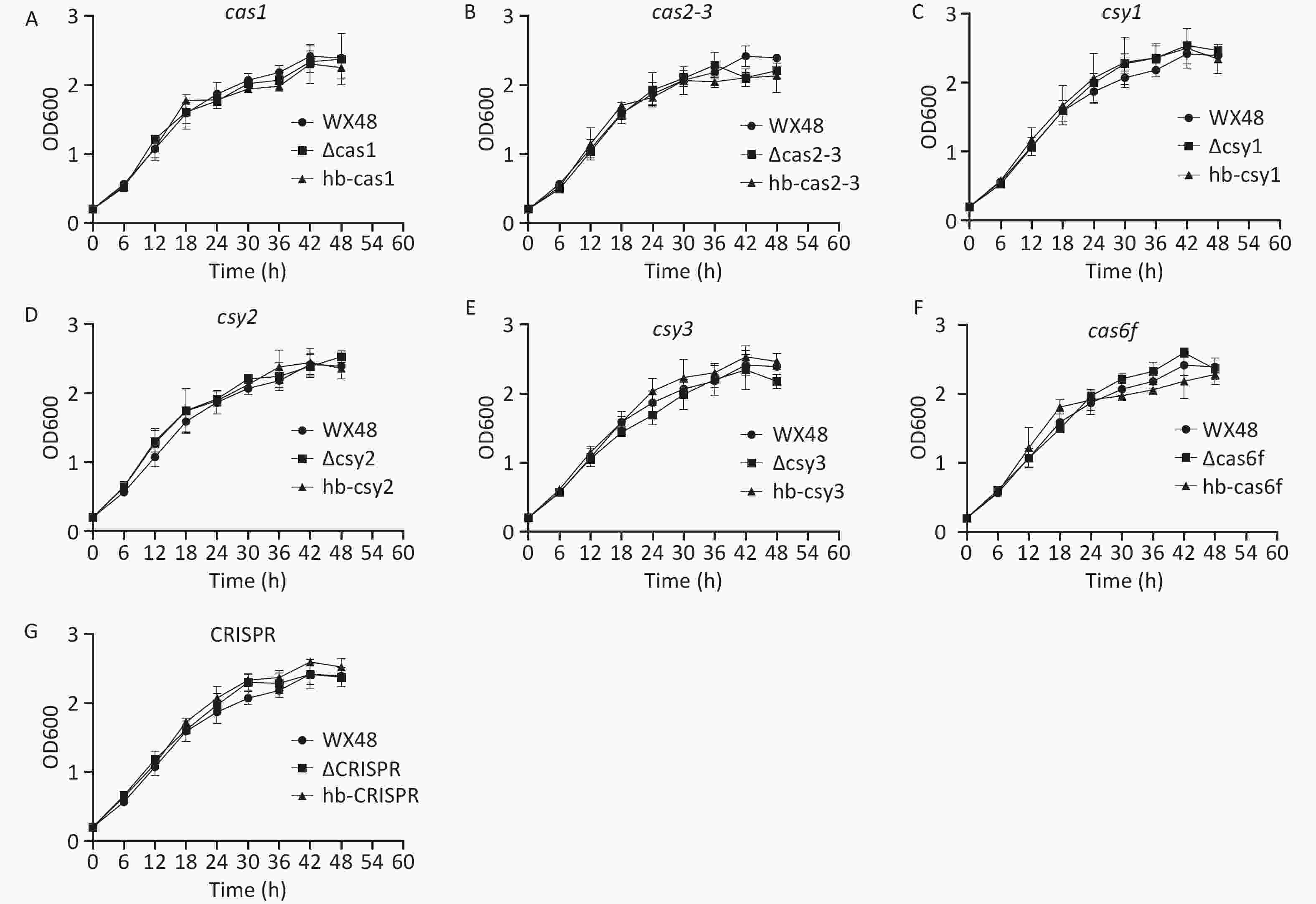
Figure 2. In vitro growth curves of various L. pneumophila strains. WX48: wild-type L. pneumophila strain; ∆Cas1, ∆Cas2-3, ∆Csy1, ∆Csy2, ∆Csy3, ∆Cas6f, and ∆CRISPR: mutant strains lacking Cas1, Cas2-3, Csy1, Csy2, Csy3, Cas6f, and the CRISPR gene, respectively. hb-Cas, hb-Cas2-31, hb-Csy1, hb-Csy2, hb-Csy3, hb-Cas6f, and hb-CRISPR: Cas1-, Cas2-3-, Csy1-, Csy2-, Csy3-. Cas6f-, and CRISPR-complemented strains, respectively. In vitro growth curves of (A) wx48, ∆Cas1, and hb-Cas1; (B) wx48, ∆Cas2-3, and hb-Cas2-3; (C) wx48, ∆Csy1, and hb-Csy1; (D) wx48, ∆Csy2, and hb-Csy2; (E) wx48, ∆Csy3, and hb-Csy3; (F) wx48, ∆Cas6f, and hb-Cas6f; (G) wx48, ∆CRISPR, and hb-CRISPR.
-
The antimicrobial susceptibilities of WT, deletion mutants, and complementation mutant L. pneumophila strains were evaluated. The deletion mutant strains were slightly less sensitive to erythromycin than the WT and complementation mutant strains, with their MICs decreasing from 0.75 in the WT 0.125 or 0.5. Although the MIC values fluctuated, the sensitivity remained within this range. The MIC of the deletion and complementation mutant strains did not substantially differ from those of the WT strain for the other antibiotics (Table 1).
Drug MIC range (mg/L) ECOFF (mg/L) ATCC
33152WT (WX48) Cas1 Cas2-3 Csy1 Csy2 Csy3 Cas6f CRISPR ∆Cas1 hb-Cas1 ∆Cas
2-3hb-
Cas2-3∆Csy1 hb-
Csy1∆Csy2 bb-
Csy2∆Csy3 hb-
Csy3∆Cas6f hb-
Cas6f∆CRISPR hb-CRISPR Erythromycin 0.032-2 1 0.5 0.75 0.5 0.75 0.125 0.75 0.5 0.75 0.5 0.75 0.5 0.75 0.5 0.75 0.5 0.75 Rifampicin 0.004-0.032 0.032 0.016 0.016 0.016 0.016 0.016 0.016 0.016 0.016 0.016 0.016 0.016 0.016 0.016 0.016 0.016 0.016 Levofloxacin 0.064-1 0.5 0.125 0.125 0.125 0.125 0.125 0.125 0.125 0.125 0.125 0.125 0.125 0.125 0.125 0.125 0.125 0.125 Moxifloxacin 0.25-1 1 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 Azithromycin 0.038-8 1 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 Tigecycline 1-16 16 0.5 0.5 0.75 0.5 0.75 0.5 0.5 0.5 0.75 0.5 0.5 0.5 0.5 0.5 0.5 0.5 Ciprofloxacin 0.25-2 1 0.5 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 0.75 Clarithromycin 0.064-1 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 Doxycycline 1-8 8 0.75 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 Note. MIC, minimum inhibitory concentration; ECOFF, epidemiological cutoff. Table 1. MICs of various antibiotics for WT and mutant strains of L. pneumophila.
-
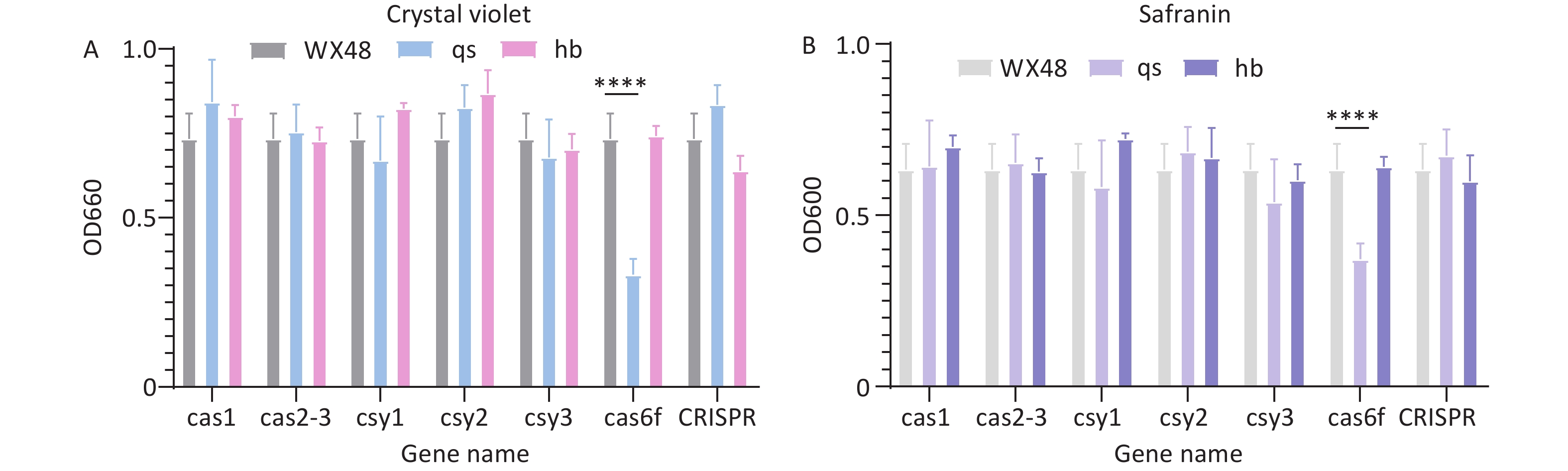
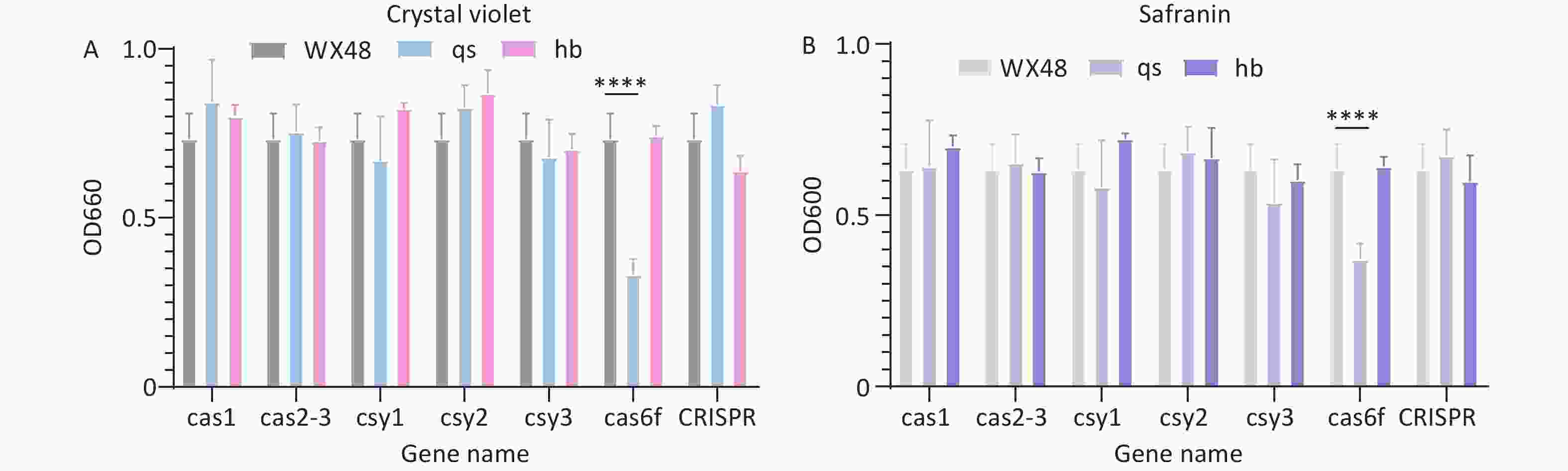
We used crystal violet staining to evaluate the impact of Cas gene deletion on the biofilm-forming ability of L. pneumophila mutants. The biofilm formation ability of the ∆Cas6f mutant was weaker than that of the WT. Introducing the complete Cas6f gene restored the biofilm formation ability of the mutant to that of the WT strain, indicating that ∆Cas6f impacts biofilm formation ability. The biofilm formation ability of the other deletion mutant strains remained similar to that of the WT strain (Figure 3A). We obtained similar results when we measured biofilm formation ability using safranin staining (Figure 3B).
-
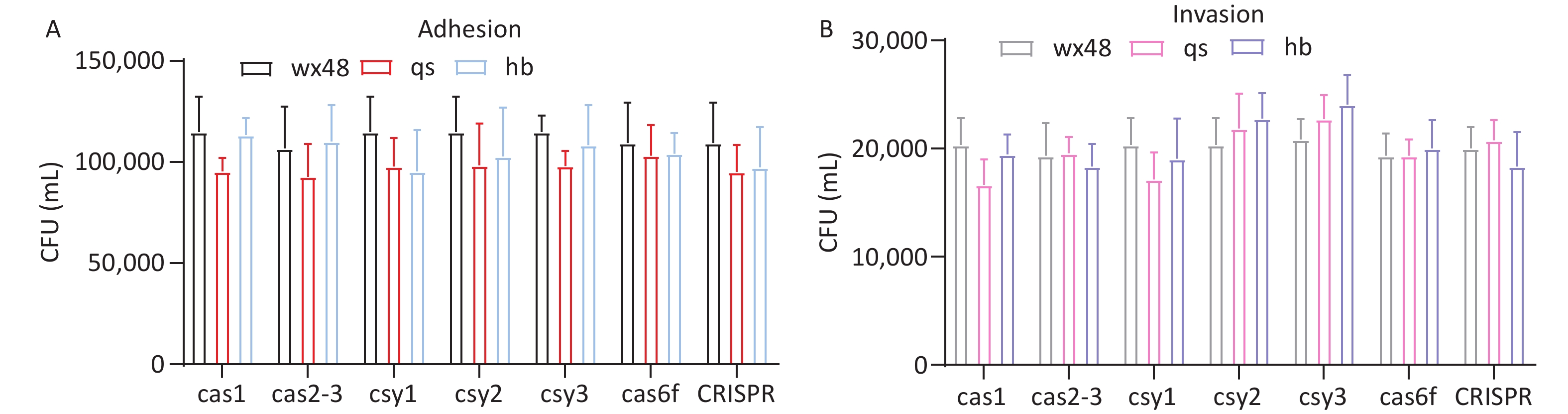
We assessed the adhesion and invasion abilities of the WT, deletion mutants, and complementation mutants of L. pneumophila using J774 cells. The adhesion or invasion abilities of these strains did not substantially differ (Figure 4). Therefore, the CRISPR-Cas system may not affect the adhesion and invasion abilities of L. pneumophila.
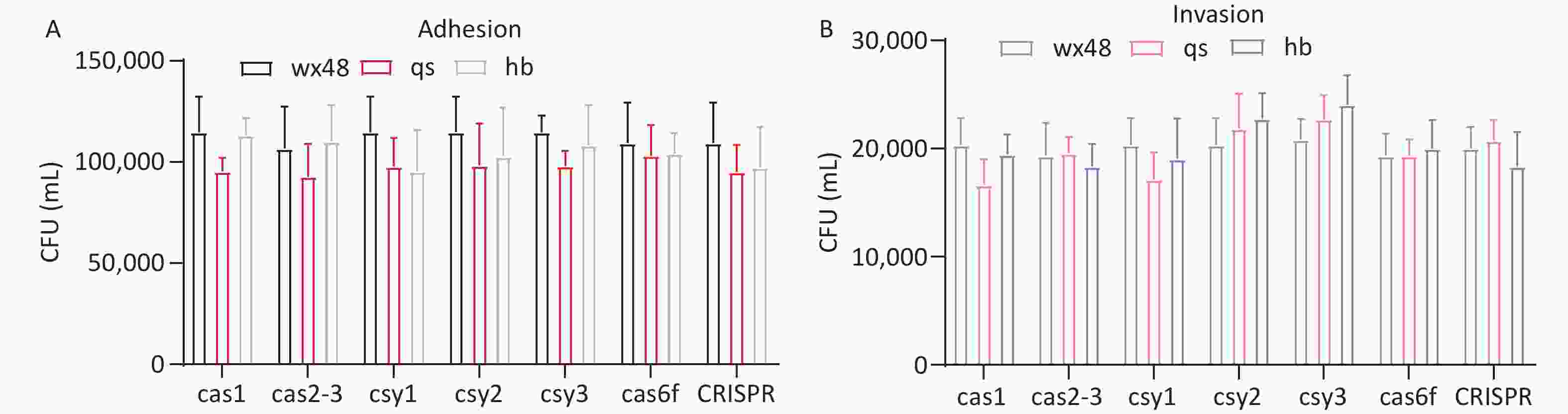
Figure 4. Adhesion and invasion of J774 cells by various L. pneumophila strains. (A) The adhesion ability of each strain. Black (WX48), red (qs), and blue (hb) representing the wild-type, deletion mutant, and complemented strains, respectively. (B) The adhesion ability of each strain. Gray (WX48), pink (qs), and purple (hb) represent the wild-type, deletion mutant, and complemented strains, respectively.
-
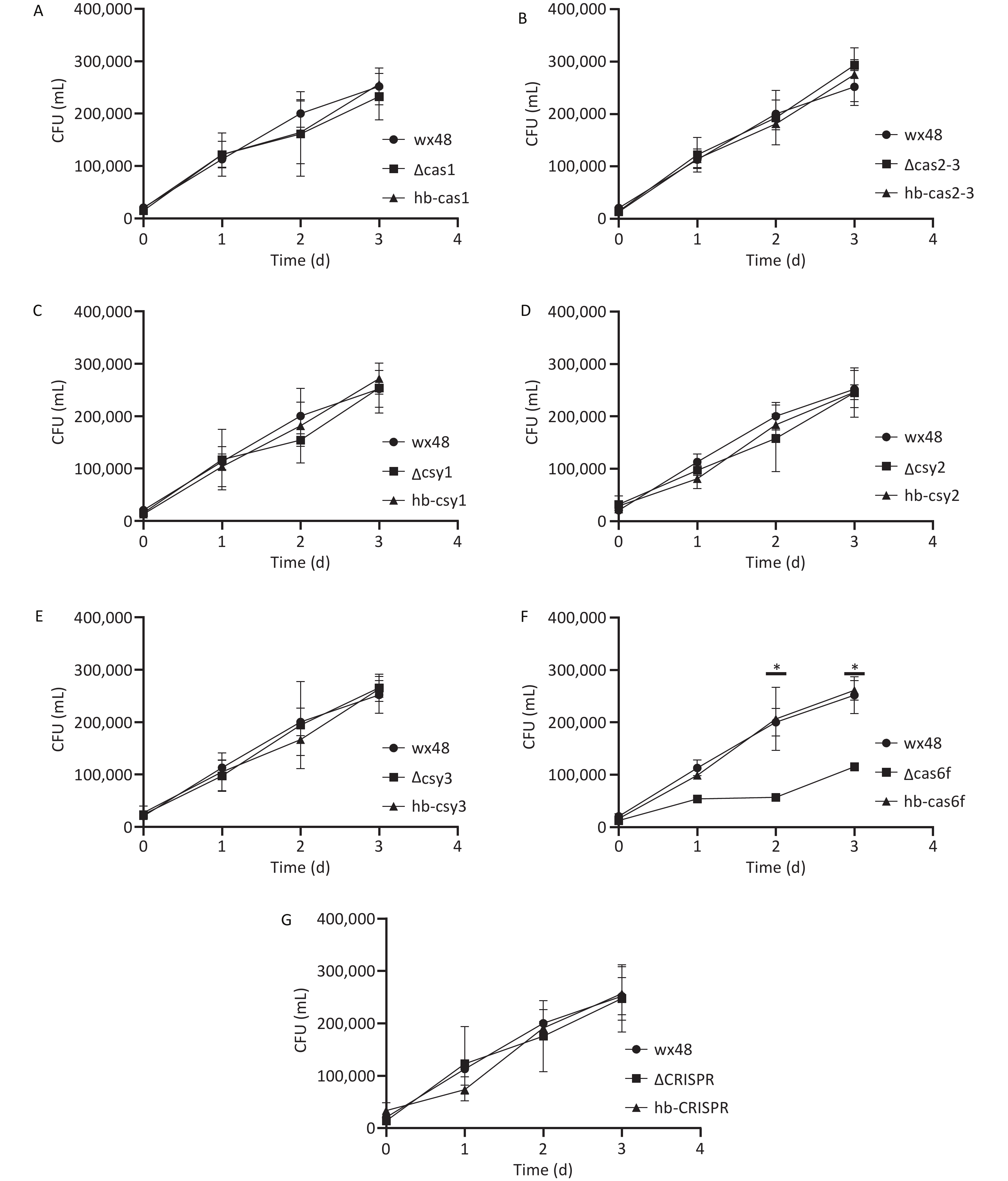
We evaluated the effect of the CRISPR-Cas system on L. pneumophila proliferation in human alveolar macrophages by infecting J774 cells with WT, deletion mutants, and complementation mutants of L. pneumophila in vitro and assessing their proliferative ability. The numbers of the WT strain, as well as those of the ΔCas1, ΔCas2-3, ΔCsy1, ΔCsy2, ΔCsy3, ΔCRISPR, and complementation mutants, within the macrophages increased gradually over time, indicating that Cas gene deletion did not affect the intracellular proliferation ability of L. pneumophila (Figure 5A–E,G). The intracellular survival ability of the ∆Cas6f strain was lower on the first day than that of the WT strain, and the intracellular proliferation ability of the ∆Cas6f strain was markedly lower than that of the WT strain on days 2 and 3. The complete Cas6f gene was introduced into the ∆Cas6f mutant strain, which restored its intracellular growth capacity to that of the WT strain (Figure 5F). Thus, Cas6f deletion may affect the intracellular proliferation of L. pneumophila.
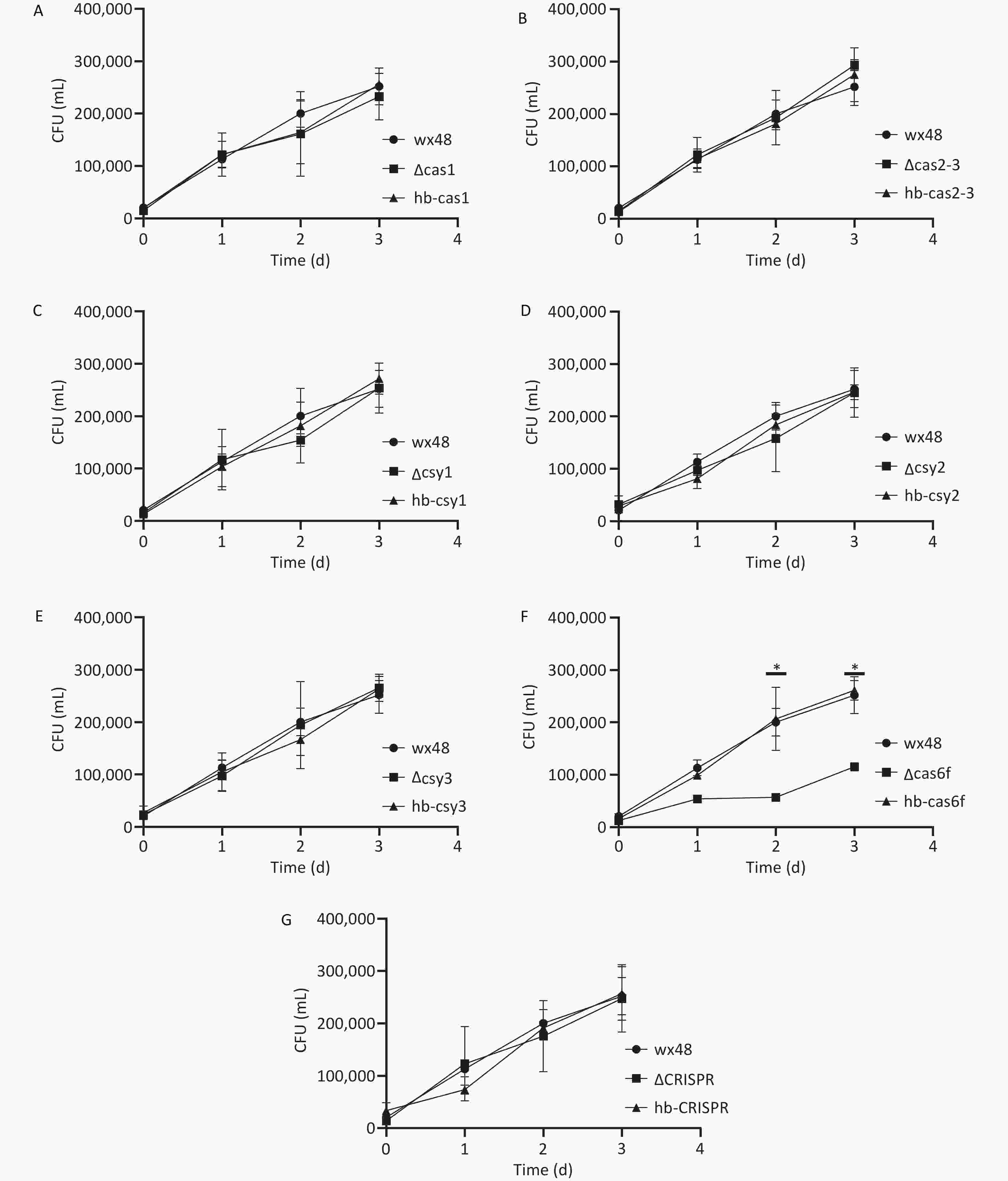
Figure 5. Proliferation ability of CRISPR-Cas deletion and complemented mutants of L. pneumophila in J774 mouse macrophages. *P < 0.05. The proliferation ability of (A) WT 48, ∆Cas1, and hb-Cas1; (B) WT 48, ∆Cas2-3, and hb-Cas2-3; (C) WT 48, ∆Csy1, and hb-Csy1; (D) WT 48, ∆Csy2, and hb-Csy2; (E) WT 48, ∆Csy3, and hb-Csy3; (F) WT 48, ∆Cas6f, and hb-Cas6f; (G) WT 48, ∆CRISPR, and hb-CRISPR.
-
We used Gimenez staining to visualize the bacterial growth in J774 cells infected with WT, deletion mutants, and complementation strains of L. pneumophila. The quantities of the WT, ΔCas1, ΔCas3-Cas2, ΔCsy1, ΔCsy2, ΔCsy3, ΔCRISPR, and complementation strains within the cells gradually increased over time, and their growth trends were consistent. Although the intracellular quantity of Cas6f also gradually increased over time, its intracellular bacterial load was markedly lower than that of the WT and complemented strains. This result is consistent with the results reported in Section 3.6, indicating that deleting Cas6f reduces the intracellular proliferative ability of L. pneumophila (Figure 6). We randomly selected 100 fields of view under a microscope and counted the number of cells infected with bacteria in these fields. The proportion of infected cells in the WT and mutant strains (ΔCas1, ΔCas3-Cas2, ΔCsy1, ΔCsy2, ΔCsy3, and ΔCRISPR), along with complemented mutants, consistently increased with infection time, similar to the results described above in this section (Figures 7A– J,M–O). However, the proportion of cells infected with the ∆Cas6f strain began to decrease after the first day compared with those in the WT and complemented strains. The curve depicting the infection ratio (infected/total cells) flattened for ∆Cas6f (Figure 7K,L,O).
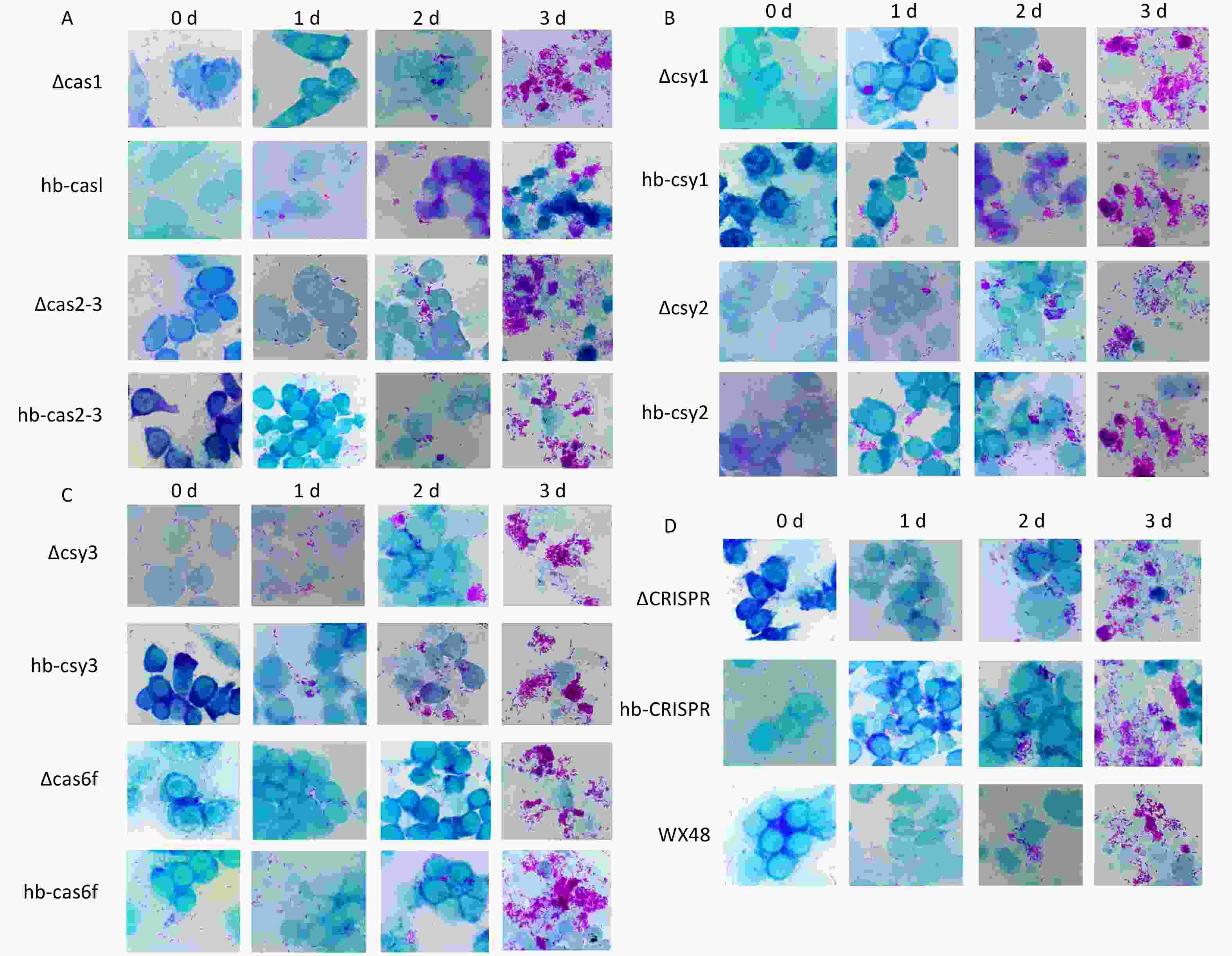
Figure 6. Micrographs of Gimenez-stained monolayers of J774 cells from 0 to 3 days after infection with various L. pneumophila strains. Scale bar: 25 μm. Magnification: 100×. Microscopic images of (A) ∆Cas1, hb-Cas1, ∆Cas2-3, and hb-Cas2-3; (B) ∆Csy1, hb-Csy1, ∆Csy-2, and hb-Csy2; (C) ∆Csy3, hb-Csy3, ∆Cas6f, and hb-Cas6f; (D) ∆CRISPR, hb-CRISPR, and WX48.
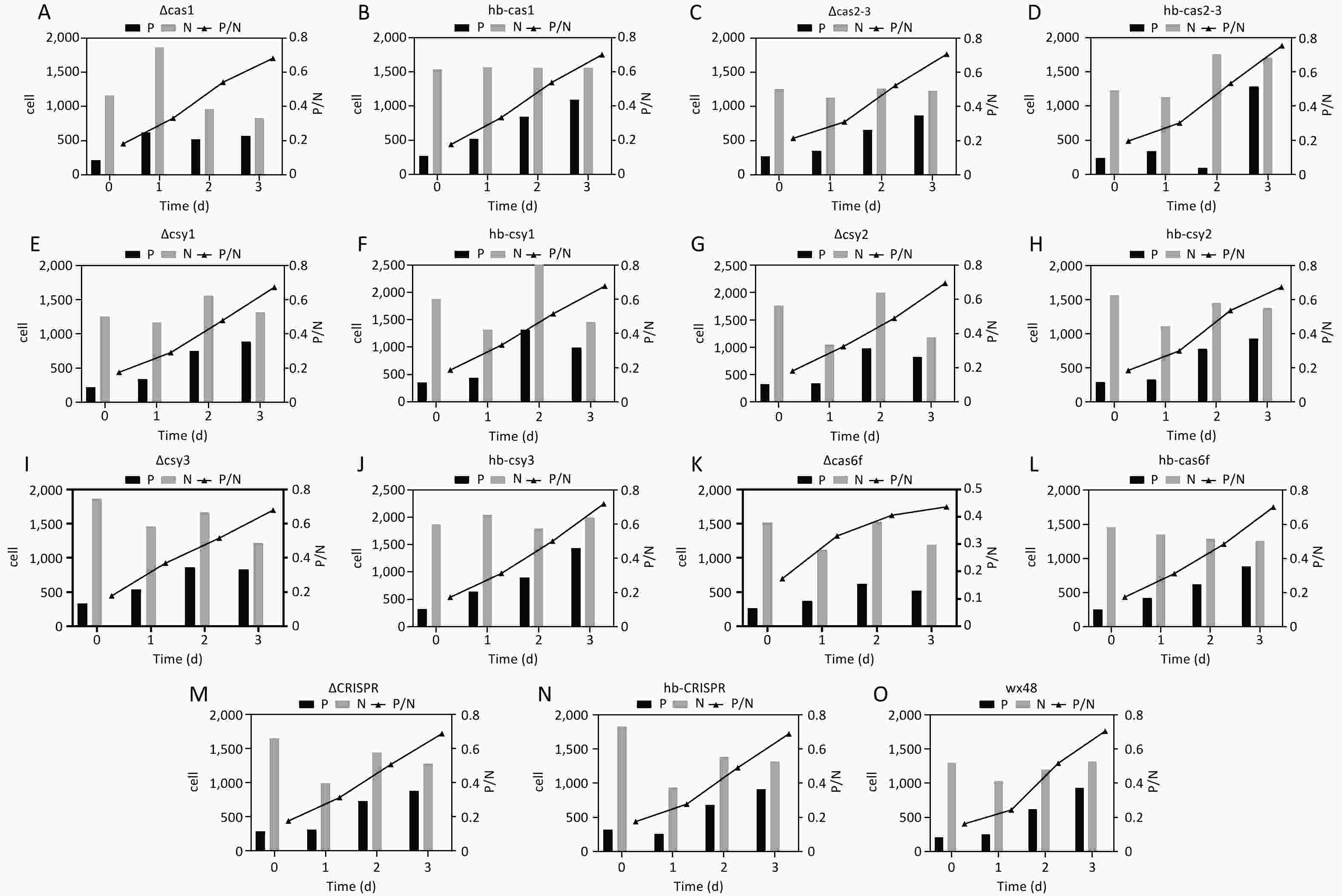
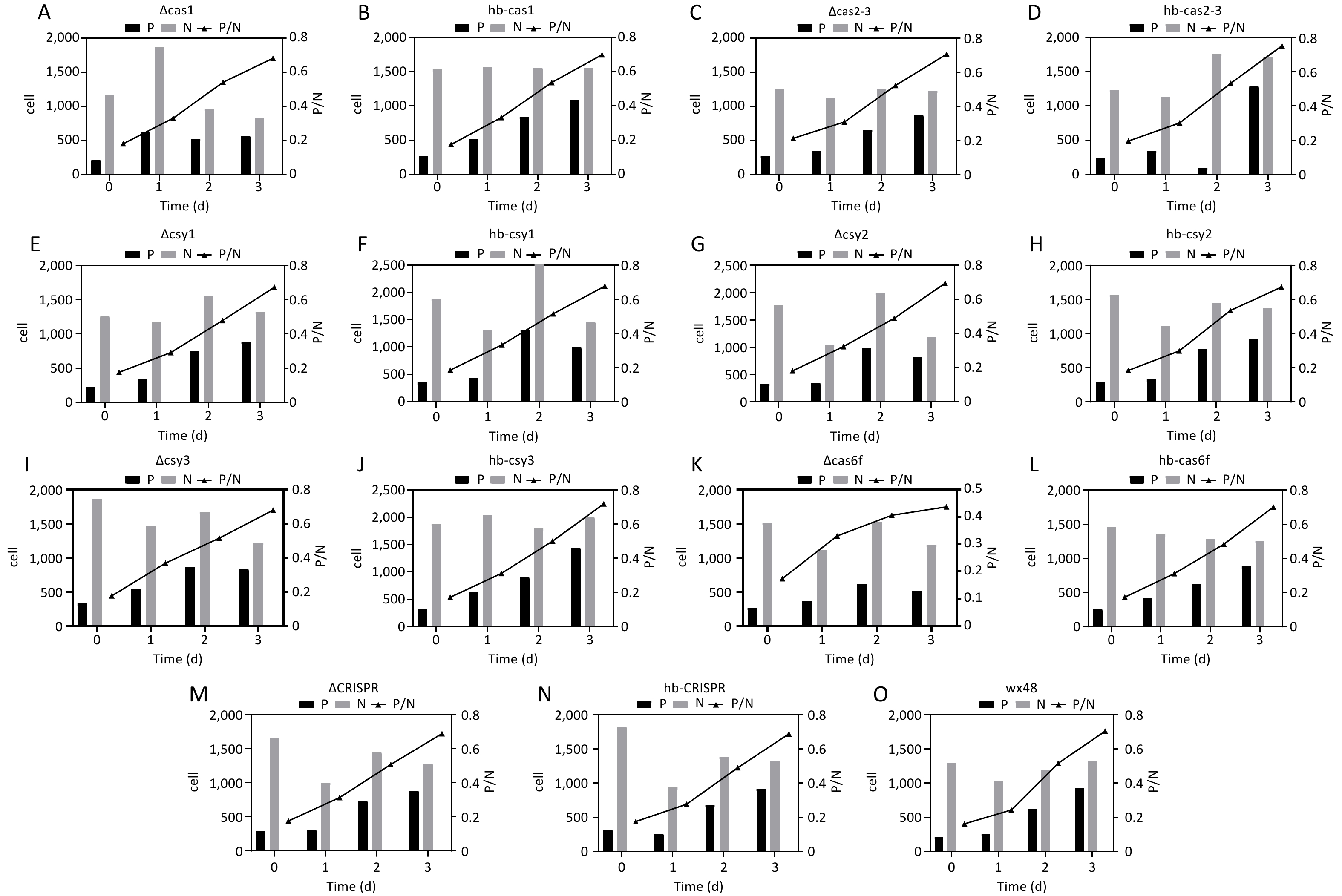
Figure 7. Cell infection status of various L. pneumophila strains from 100 microscopic fields. P: The number of bacteria-infected cells in a single field of view. N: The total number of cells in a single field of view. P/N: The ratio of cells infected with bacteria to the total number of cells in a single field of view. All values are presented as averages. The cell infection status of (A) ∆Cas1, (B) hb-Cas1, (C) ∆Cas2-3, (D) hb-Cas2-3, (E) ∆Csy1, (F) hb-Csy1, (G) ∆Csy2, (H) hb-Csy2, (I) ∆Csy3, (J) hb-Csy3, (K) ∆Cas6f, (L) hb-Cas6f, (M) ∆CRISPR, (N) hb-CRISPR, and (O) WX48.
-
We analyzed the expression levels of genes related to intracellular proliferation and biofilm formation, specifically the CRISPR-Cas, Dot/Icm secretion, and pili systems, using qRT-PCR.
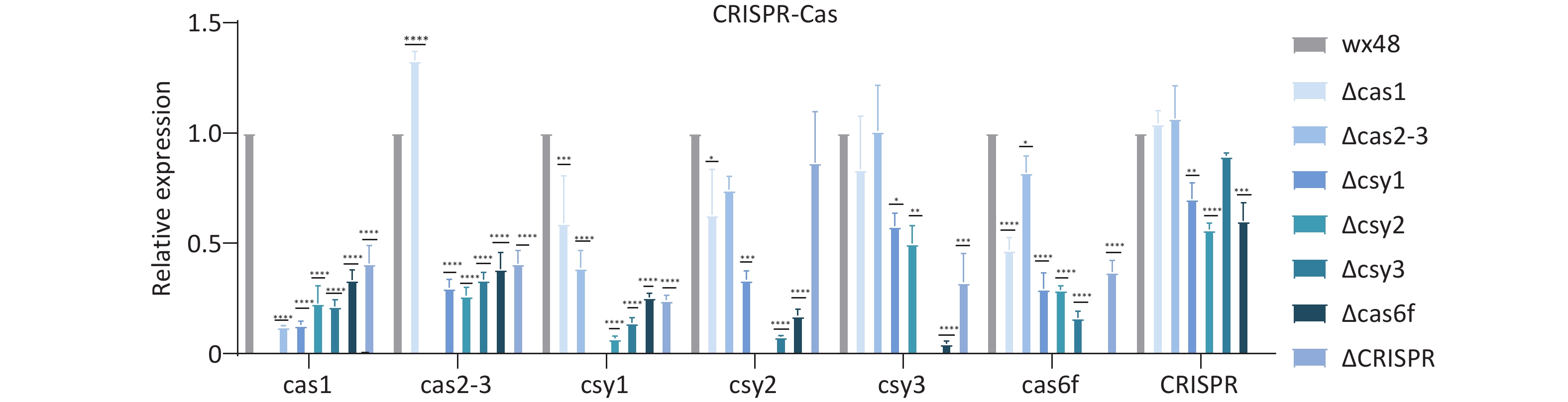
The type I-F CRISPR-Cas system consists of six Cas genes and a CRISPR array, with the Cas gene constituting an operon headed by Cas1. The CRISPR-Cas system genes of L. pneumophila regulated each other, and deleting any of these genes affected the expression levels of their downstream genes. Cas1, Csy1, and Cas6f were individually deleted; the expression levels of the upstream and downstream genes of these three deletion mutants were considerably lower compared with those of the WT. The expression level of the Cas1 gene was slightly higher in the Cas2-3 deletion mutant, whereas the expression levels of other genes were lower compared with those in the WT. The expression levels of the Cas2-3 and CRISPR genes of the Csy2 deletion mutant, whereas the expression levels of other genes were lower compared with the WT. The expression levels of Cas1 and Cas2-3 did not differ compared with the WT, but those of other genes were lower. CRISPR deletion did not affect the expression levels of Cas1, Cas2-3, and Csy3 in the mutant strain, whereas those of the other genes were substantially downregulated compared with the WT (Figure 8).
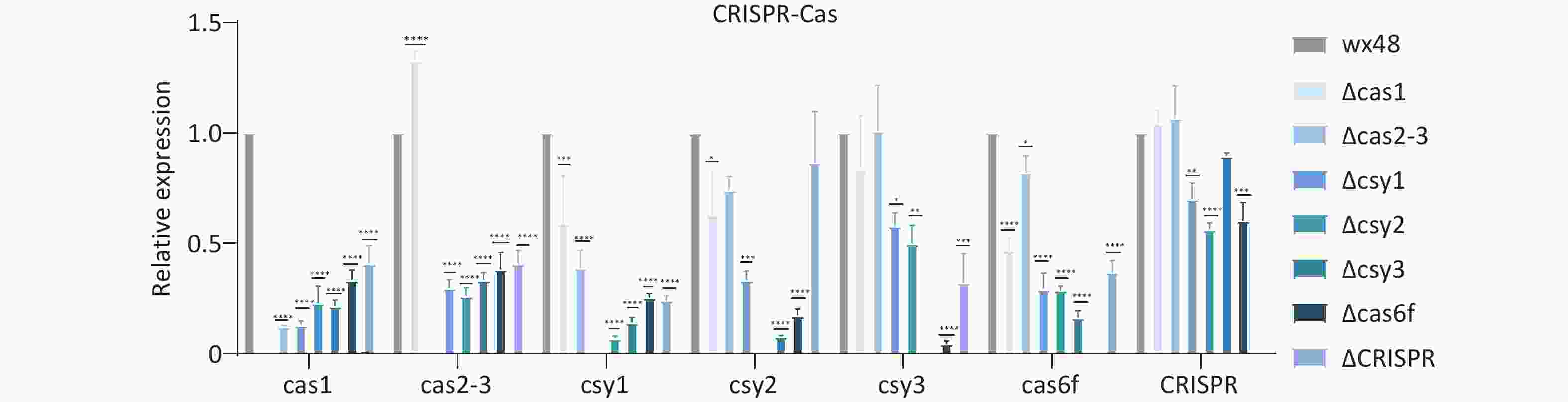
Figure 8. Expression levels of CRISPR-Cas system-related genes in L. pneumophila CRISPR-Cas deletion strains. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. WX48 served the control group. Cas1, Cas2-3, Csy1, Csy2, Csy3, Cas6f, and CRISPR: the expression levels of the Cas1, Cas2-3, Csy1, Csy2, Csy3, Cas6f, and CRISPR genes in the deletion mutants, respectively.
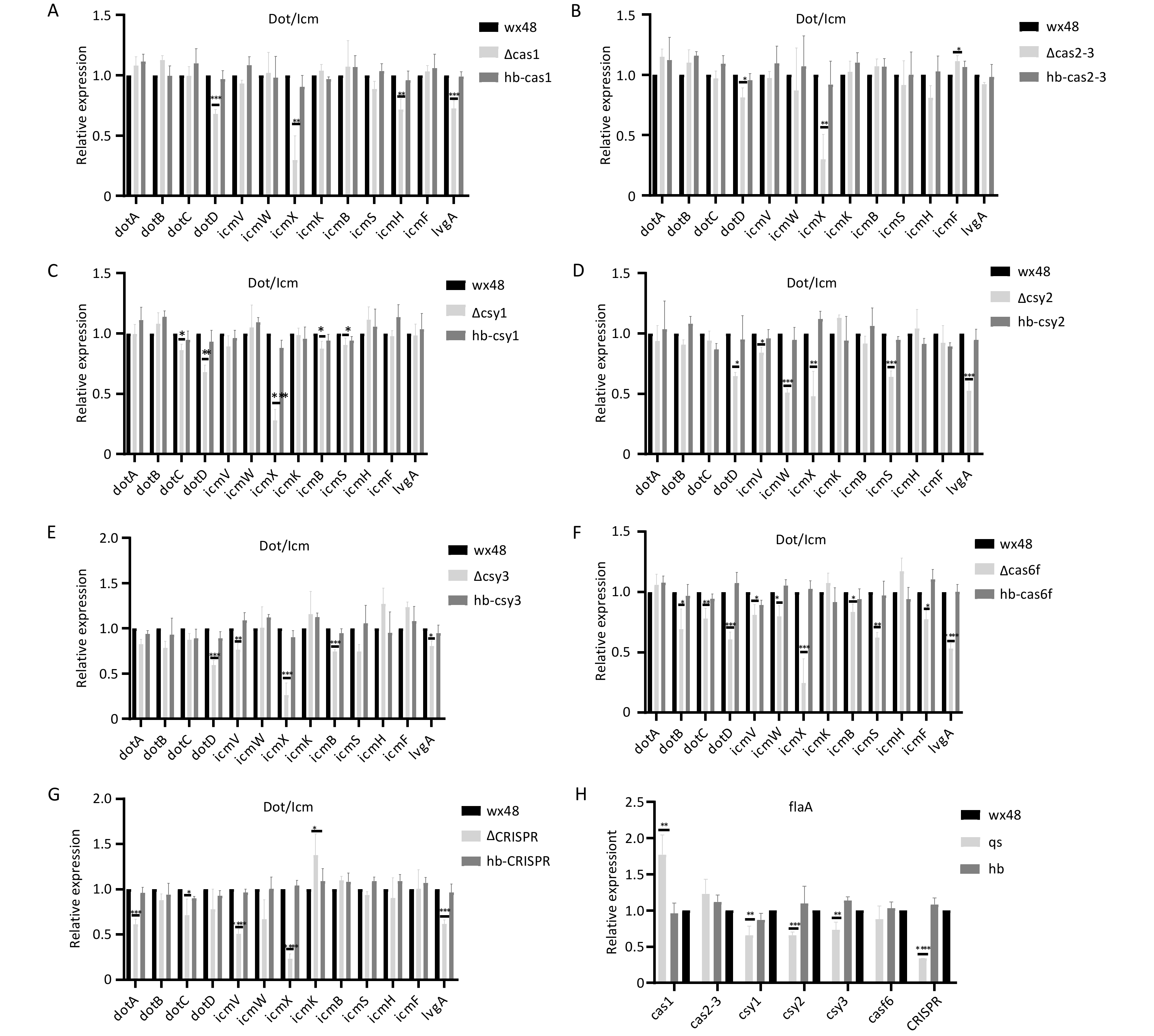
We analyzed the structural gene expression levels of the Dot/Icm type IV secretion system (T4SS) in Cas6f because its intracellular proliferation was notably lower than that in the WT. The expression levels of DotB, DotC, DotD, DotV, DotW, IcmX, IcmB, IcmS, IcmF, and LvgA were considerably lower in the Cas6f strain than in the WT strain. Therefore, Cas6f deletion may reduce the expression levels of the most structural genes of Dot/Icm T4SS, reducing the intracellular ∆Cas6f proliferation (Figure 9F). In addition, although the intracellular proliferation of ∆Cas1, ∆Cas2-3, ∆Csy1, ∆Csy2, ∆Csy3, and ∆CRISPR was not affected compared with that of the WT, the expression levels of the individual structural genes of the Dot/Icm T4SS were slightly lower (Figures 9A–E,G), which may have been due to the mutual regulation among the Cas genes in the CRISPR-Cas system.
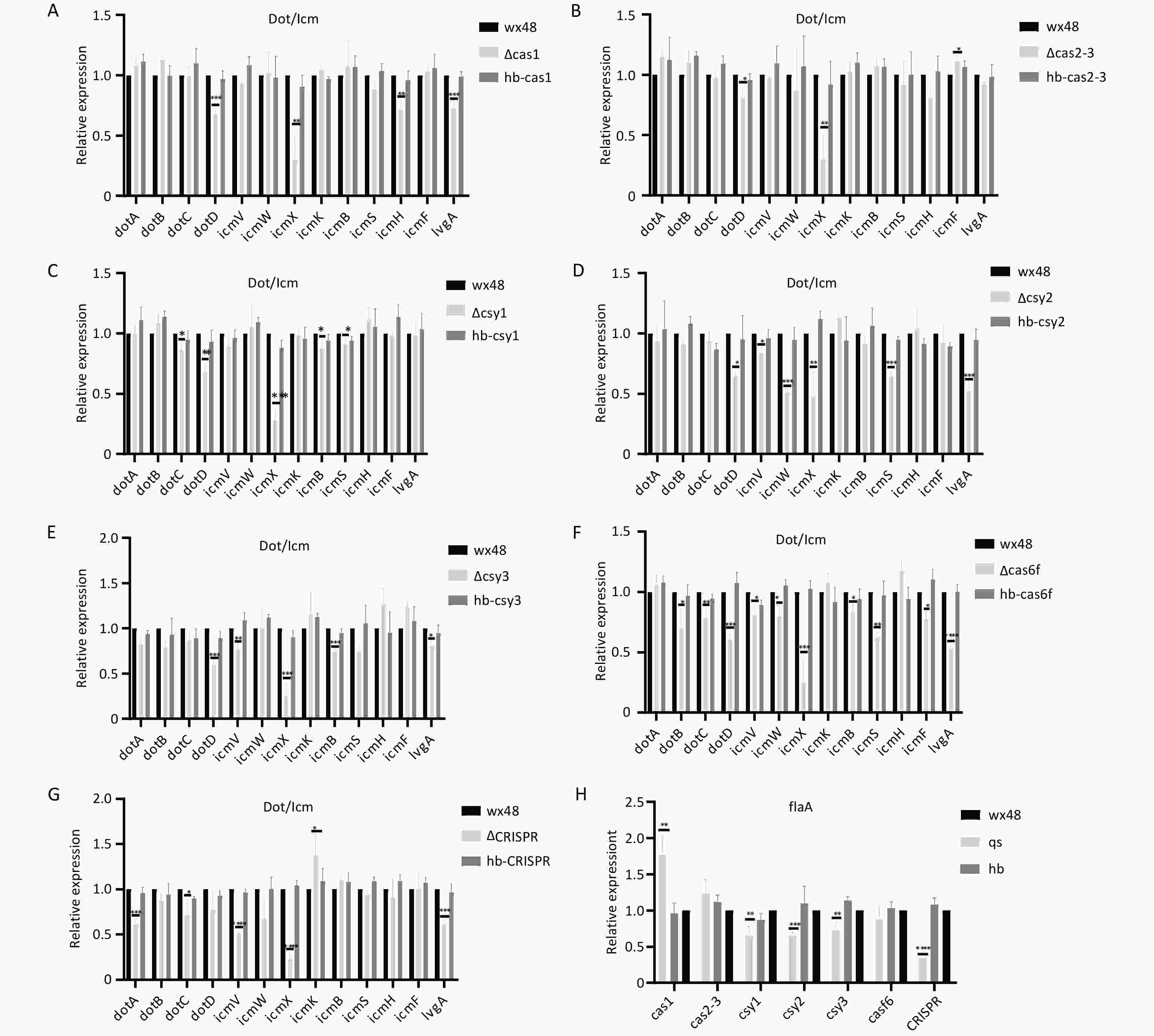
Figure 9. Expression levels of structural genes related to the Dot/Icm type IV secretion system in mutant strains. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Expression of structural genes related to the Dot/Icm type IV secretion system in (A) WT 48, ∆Cas1, and hb-Cas1; (B) WT 48, ∆Cas2-3, and hb-Cas2-3; (C) WT 48, ∆Csy1, and hb-Csy1; (D) WT 48, ∆Csy2, and hb-Csy2; (E) WT 48, ∆Csy3, and hb-Csy3; (F) WT48, ∆Cas6f, and hb-Cas6f; (G) E WT48, ∆CRISPR, and hb-CRISPR. (H) Expression of the flaA flagellin gene.
Flagellin may enhance the ability of Legionella to adhere to and invade host cells, increasing intracellular growth. Therefore, we examined the flagellin expression levels in each mutant strain. The expression of flagellin was moderately downregulated in ∆Csy1, ∆Csy2, ∆Csy3, and ∆CRISPR; upregulated in ∆Cas1; and unchanged in ∆Cas3-Cas2 and ∆Cas6f compared with that In the WT strain (Figure 9H). This finding suggests that the reduced intracellular proliferative capacity of the ∆Cas6f strain is unrelated to flagellin.
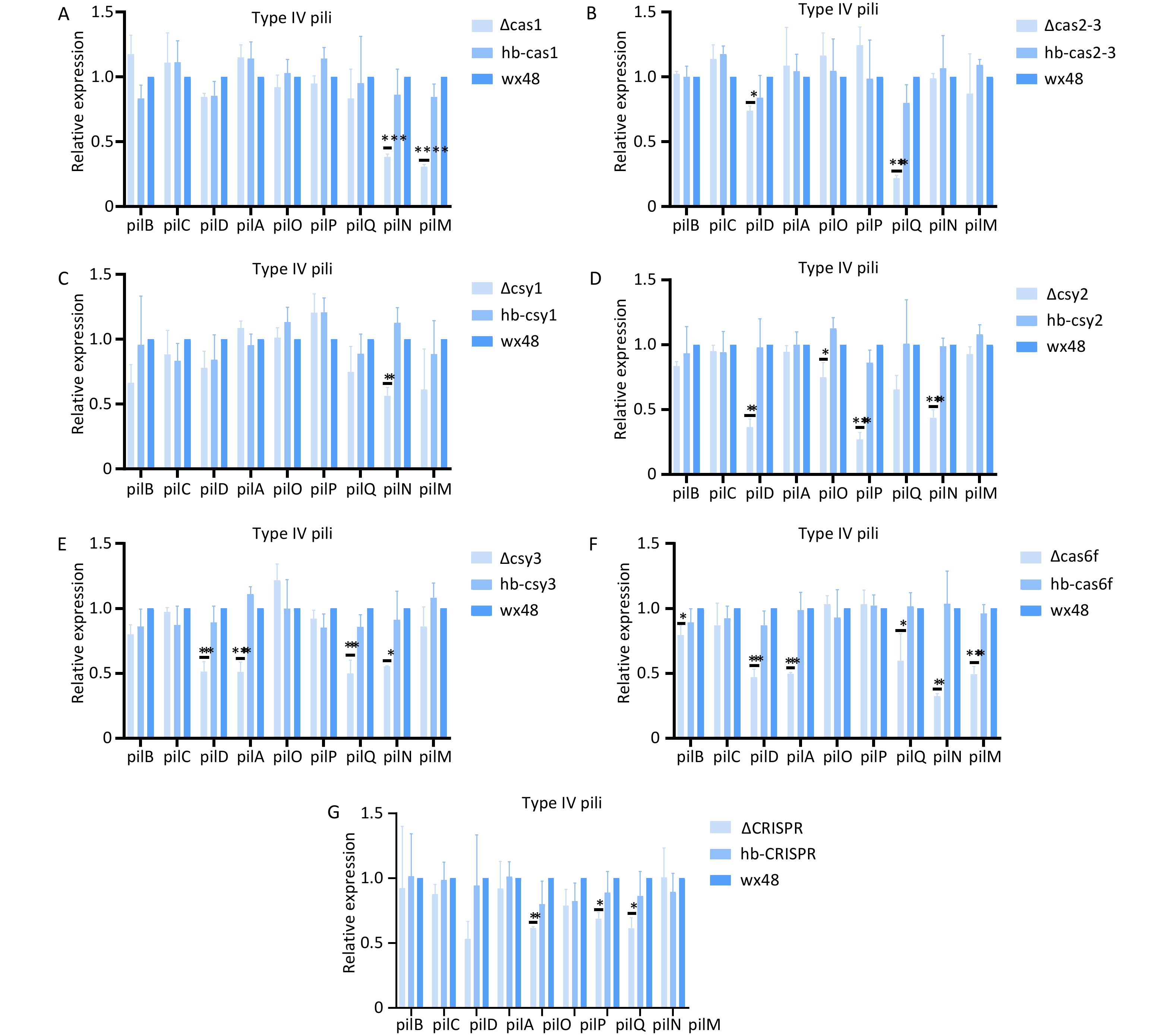
Deleting Cas6f led weakened the biofilm formation ability compared with that of the WT; therefore, we analyzed the expression levels of the structural genes in the type IV pili system. The expression levels of type IV pili-associated genes (pilB, pilD, pilA, pilQ, pilN, and pilM) were substantially downregulated in the Cas6f mutant compared with those in the WT (Figure 10F). Complementation with an intact Cas6f gene restored their expression levels to WT levels. These findings suggest that Cas6f deletion contributes to altering biofilm formation through modulating pilus-related gene networks. In addition, although the ∆Cas1, ∆Cas2-3, ∆Csy1, ∆Csy2, ∆Csy3, and ∆CRISPR deletions did not affect biofilm formation, the expression levels of the individual structural genes of T4P were slightly lower than those in the WT (Figure 10A–E,G), which may have been due to the mutual regulation of biofilm formation among the Cas genes in the CRISPR-Cas system.
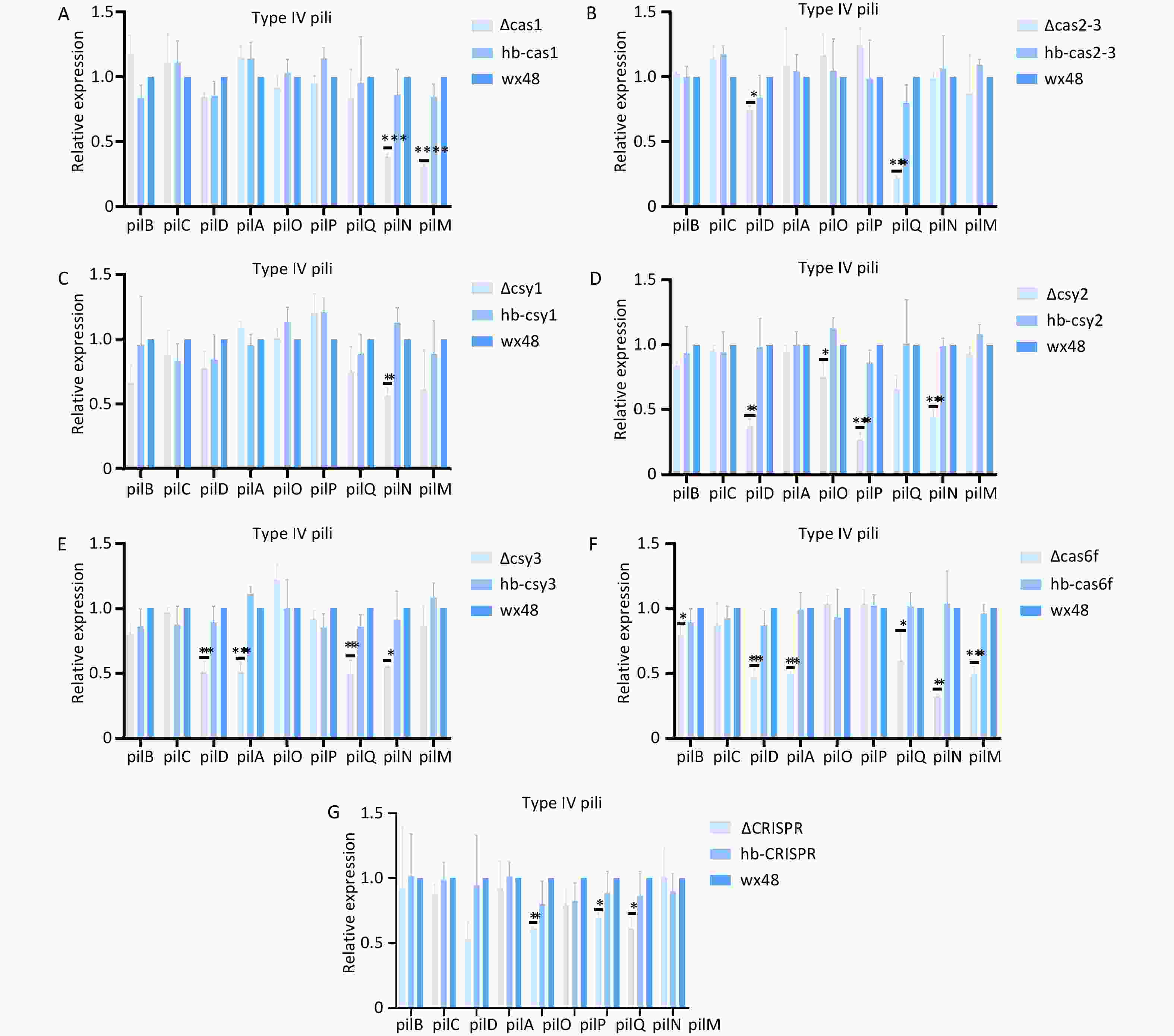
Figure 10. Expression levels of structural genes related to the type IV pili system in mutant strains. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Expression of structural genes related to the type IV pili system in (A) WT 48, ∆Cas1, and hb-Cas1; (B) WT 48, ∆Cas2-3, and hb-Cas2-3; (C) WT 48, ∆Csy1, and hb-Csy1; (D) WT 48, ∆Csy2, and hb-Csy2; (E) WT 48, ∆Csy3, and hb-Csy3; (F) WT48, ∆Cas6f, and hb-Cas6f; (G) WT48, ∆CRISPR, and hb-CRISPR.
-
The genomes of many prokaryotes, such as L. pneumophila, contain CRISPR arrays and Cas genes that enable their resistance to foreign genetic factors. Some bacteria use Cas proteins in their CRISPR/Cas systems to target their own genes, increasing their virulence during host cell invasion[36,37]. The pathogenesis of Legionella has been extensively studied, and the bacterial factors that promote intracellular infection and virulence have been identified[38,39]. However, relatively few studies have focused on the nonclassical functions of CRISPR-Cas in Legionella. Cas2 in L. pneumophila has nuclease activity and is essential for intracellularly infecting amoeba cells[32,33]. In addition, Cas2 is associated with thermotolerance[40]. The type II-B CRISPR-Cas system plays a crucial role in regulating the drug resistance and intracellular proliferation of L. pneumophila[31]. We evaluated the effects of the CRISPR-Cas system on the in vitro growth, drug resistance, biofilm formation, and intracellular proliferation of L. pneumophila by deleting its type I-F CRISPR-Cas genes. The type I-F CRISPR-Cas system of L. pneumophila regulates the expression of structural genes of the T4P system and Dot/Icm T4SSs, affecting the biofilm formation and intracellular proliferation of L. pneumophila. Therefore, the CRISPR-Cas system may be involved in regulating L. pneumophila virulence.
The gene distribution of the type I-F CRISPR-Cas system in Legionella was reported[34]. We analyzed the gene distribution of the type I-F CRISPR-Cas system using CRISPRCasFinder, and the results were similar to those of previous studies[34]. We also analyzed the relative expression levels of CRISPR-Cas-related genes using qRT-PCR. Deleting each Cas gene in the type I-F CRISPR-Cas system affects the expression levels of upstream and downstream Cas genes. This finding indicates that deleting one gene in the CRISPR-Cas system affects the expression levels of other genes in the system.
The CRISPR-Cas system, in addition to the defense function of exogenous nucleic acids, plays a role in regulating endogenous gene expression and host cell physiology. Biofilm production and colonization may be important for the environmental spread, survival, and pathogenesis of L. pneumophila[11,41,42]. Moreover, the type I-C CRISPR-Cas system in Streptococcus mutant strains may be associated with biofilm formation, and Cas3 deletion may hinder biofilm formation[43]. The biofilm formation ability of a WT Salmonella strain with type I-E CRISPR-Cas3 was a stronger than that of the ∆Cas3 mutant. Thus, Cas3 may be involved in regulating bacterial biofilm formation[44]. In this study, the biofilm formation ability of the ΔCas6f L. pneumophila mutant with a type I-F CRISPR-Cas system was weaker than that of the WT, whereas the biofilm formation ability of the complemented strain with the complete Cas6f gene was stronger than that of ΔCas6f. These findings collectively demonstrate that the CRISPR-associated gene Cas6f is involved in modulating biofilm formation in L. pneumophila.
The T4P system influences the biofilm formation and surface movement of Clostridioides difficile[45,46]. The type IV pili (T4P) system mediates key processes, such as bacterial twitching motility, DNA uptake, and biofilm formation. PilA is a core structural protein that is directly involved in assembling and functionally regulating the pilus.[45,47]. Therefore, we analyzed the effects of regulating Cas genes using the CRISPR-Cas system on the gene expression levels of the T4P system. Cas6f deletion led to a reduction in the expression levels of most genes in the T4P system. Thus, Cas6f may be indirectly involved in regulating of biofilm formation in L. pneumophila. Although deleting other genes impacted the expression levels of the individual structural genes of the T4P system, the biofilm formation ability did not notably differ compared with that of the WT strain, indicating that these genes are not essential for biofilm formation. Alternatively, these genes may maintain phenotypic stability through other unknown mechanisms that were not observed in our experiments.
The pathogenesis of L. pneumophila relies on the Dot/Icm type IV secretion system, which likely reflects its extensive engagement with the cellular processes of the host during the evolution of bacterial virulence[38,48]. Dot/Icm T4SS is crucial for Legionella-containing vacuole biogenesis and intracellular replication as well as a key virulence factor in the intracellular survival and pathogenicity of Legionella. A single L. pneumophila cell can transfer more than 300 proteins to the host cells via Dot/Icm T4SS effectors, which manipulate host cell functions to benefit the bacterium. This process is essential for the growth and intracellular transport of human macrophages and amoebas[49-52]. In this study, the ∆Cas1, ∆Cas2-3, ∆Csy1, ∆Csy2, ∆Csy3, and ∆CRISPR strains showed an ability to proliferate in macrophages that was similar to that of the WT strain; however, the proliferation ability of ∆Cas6f was much weaker. Moreover, L. pneumophila containing the complete type I-F CRISPR-Cas system exhibited a higher capacity to proliferate in macrophages than those lacking Cas6f. This suggests that Cas6f is involved in regulating intracellular proliferation. Cas6f forms a tight complex with cleaved crRNA products in Pseudomonas aeruginosa, which plays a functional role within the CRISPR pathway beyond pre-crRNA cleavage[53]. The Cas6f (also known as Csy4) in the type I-F CRISPR-Cas system is an endonuclease that binds with high affinity to repetitive stem-loop structures in pre-crRNAs by cutting these structures at the base to produce mature crRNAs[21,54,55,56]. Our qRT-PCR results demonstrated that deleting Cas6f decreased the expression levels of most genes related to intracellular proliferation in T4BSS of L. pneumophila compared with those in the WT. Although deletion of other genes affected the expression levels of individual structural genes in the T4BSS secretion system, the intracellular proliferation did not change. Therefore, we speculate that Cas6f is the main factor causing changes in intracellular proliferation via affecting the expression levels of Dot/Icm type IV secretion system structural genes, whereas the other genes do not affect intracellular proliferation.
The type I-F CRISPR-Cas system regulates the flaA expression levels. Notably, although the expression level of flaA in the ΔCas6f mutant did not differ from that of the WT, the intracellular proliferation capacity of the ΔCas6f mutant was considerably lower, suggesting that this phenotype is independent of the flagellin pathway. Flagellin enhances the adhesion to and invasion of host cells by L. pneumophila; however, this protein is not necessary for the intracellular proliferation of the bacteria[57]. In summary, the CRISPR-Cas system may affect the expression of the type IV secretion and the type IV pili (T4P) systems, causing changes in the intracellular proliferation and biofilm formation of L. pneumophila, with Cas6f playing an important role in this process.
-
The virulence mechanisms of Legionella have been extensively studied. The classical and nonclassical functions of the CRISPR-Cas system have been examined. We observed that the type I-F CRISPR-Cas system influenced the biofilm formation and intracellular proliferation of L. pneumophila and regulated the expression levels of the structural genes of the Dot/Icm T4SS and T4P systems, altering the pathogenicity of L. pneumophila. This finding suggests that the CRISPR-Cas system plays an important role in the virulence of L. pneumophila. This study expands our understanding of the biology of the CRISPR-Cas system and provides new targets for further understanding the pathogenic mechanisms of Legionella.
-
No additional data available.
Phenotypic Function of Legionella pneumophila Type I-F CRISPR-Cas
doi: 10.3967/bes2025.107
- Received Date: 2025-12-20
- Accepted Date: 2025-06-25
-
Key words:
- Legionella pneumophila /
- CRISPR-Cas /
- Biofilm /
- Intracellular proliferation
Abstract:
There is no conflict of interest in this article.
This study is exempt from ethical review.
| Citation: | Ting Mo, Hongyu Ren, Xianxian Zhang, Yunwei Lu, Zhongqiu Teng, Xue Zhang, Lupeng Dai, Ling Hou, Na Zhao, Jia He, Tian Qin. Phenotypic Function of Legionella pneumophila Type I-F CRISPR-Cas[J]. Biomedical and Environmental Sciences. doi: 10.3967/bes2025.107 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: