-
Antimicrobial resistance (AMR) has become a critical global public health challenge in the 21st century. Since the initial isolation of a blaNDM-1-carrying and carbapenem-resistant Klebsiella pneumoniae from an Indian hospital in 2009[1], the escalating prevalence of New Delhi metallo-β-lactamase (NDM)-encoding genes (blaNDM) has transformed carbapenem resistance into a worldwide phenomenon, transcending national and regional boundaries[2]. Up to 90 distinct NDM variants have been reported globally according to the NCBI GenBank Pathogens database. Plasmid-mediated horizontal gene transfer (HGT), which occurs both within and across bacterial species, has significantly accelerated the global dissemination of blaNDM-related genes and the associated resistance[3]. Carbapenem-resistant pathogens were responsible for 50,000–100,000 deaths globally in 2019[4]. Although NDM-1 has been relatively well characterized[5], the epidemiological profiles of other NDM variants require continued surveillance and in-depth investigation. The novel NDM-9 variant (GenBank accession no. KC999080) was first identified in 2013 from a clinically significant isolate of Klebsiella pneumoniae ST107 strain PPH1303 with a high level of resistance to carbapenems recovered from the urine culture of a pediatric patient in Beijing, China, who had acute lymphocytic leukemia and had undergone allogeneic stem cell transplantation[6].
To characterize the genomic epidemiology of foodborne carbapenem-resistant Salmonella harboring blaNDM genes across China and to elucidate the genetic environments and potential transmission mechanisms of these genes, we investigated the prevalence of blaNDM genes using PCR[7] among 2,877 foodborne Salmonella strains isolated from diverse food sources. This work was carried out across 31 provinces of the Chinese mainland in 2022, and included a positive strain, 22S130, which was isolated from retail chicken thigh sample in Hebei Province. It was sequenced using the QNome-3841hex (Qitan Tech, Chengdu, China) and Illumina Novaseq PE150 platform (Novogene, Beijing, China). Assembly of the Qitan genome was performed using flye v2.9, followed by error correction with minimap2 v2.29-r1283 and racon v1.5.0. Consed v29.0 was employed to generate the single, complete and closed chromosome/plasmid. Additionally, Serovar was predicted with SeqSero2. MLST v2.09, PlasmidFinder v2.1, and ResFinder v4.7.2 available at Center for Genomic Epidemiology (CGE) were utilized for multilocus sequence typing, plasmid replicon, and AMR genotypes identification, respectively. Antimicrobial susceptibility testing (AST) was determined by micro-broth dilution using Biofosun® Gram-negative panel (Fosun Diagnostics, Changsha, China). Furthermore, Clinical and Laboratory Standards Institute guidelines (CLSI M100-S34 and M31-A3) and European Committee on Antimicrobial Susceptibility Testing (EUCAST, v2025) were consulted for the resistance interpretation. To further analyze the genetic environment and perform plasmid structural comparative studies, Easyfig v2.2.2 and BLAST Ring Image Generator (BRIG, v0.95) were used, respectively.
In this study, 22S130, the positive strain isolated from the retail chicken thigh sample in Hebei Province, was identified as S. enterica serovar Indiana of the MLST type ST17. Among the selected 86 blaNDM gene positive Salmonella isolates from NCBI GenBank database, 20.93% (18/86) were identified as serovar Indiana ST17, followed by Typhimurium (18.60%, 16/86, 12 ST34, and 4 ST36), monophasic Typhimurium ST34 (15.12%, 13/86), Senftenberg ST14 (12.79%, 11/86), Kentucky ST198 (9.30%, 8/86), and other 13 serovars (Table 1). These data revealed that foodborne Salmonella carrying the blaNDM gene exhibited considerable serovar diversity, with S. Indiana ST17 constituting a significant proportion. While S. Indiana ST17 dominated the serovar diversity, the co-circulation of 16 additional serovars signifies a broad reservoir for blaNDM transmission, highlighting emergent threats from the Typhimurium (including its monophasic variant) and Senftenberg lineages.
Serovar Number Percentage MLST types Indiana 18 20.93% ST17 Typhimurium 16 18.60% ST34 (n = 12), ST36 (n = 4) I 4,[5],12:i:- 13 15.12% ST34 Senftenberg 11 12.79% ST14 Kentucky 8 9.30% ST198 Enteritidis 4 4.65% ST11 Mbandaka 3 3.49% ST413 Havana 2 2.33% ST578 Typhi 2 2.33% ST1 Othersa 9 10.47% - Note. aOthers included the following serovars of Salmonella: Corvallis, Geraldton, Goldcoast or Brikama, I -:e,h:1,5, I 4:-:-, London, Molade or Wippra Rissen, Stanley. Table 1. serovars of blaNDM-carrying Salmonella isolates retrived from NCBI
We observed that 22S130 exhibited severe multidrug resistance (MDR) against 18 antimicrobial agents belonging to 11 distinct classes. Excluding susceptibility to tigecycline and intermediate resistance to colistin, the strain demonstrated resistance to all the remaining 16 tested antimicrobial agents across 10 classes, including carbapenems (imipenem), the ultimate drug for clinically treating severe infections caused by MDR Enterobacteriaceae. Following the international expert consensus criteria for drug resistance classification[8], Salmonella isolate 22S130 qualified as an extensively drug resistant (XDR) isolate. Genomic analysis revealed multiple diverse resistance determinants of this isolate (Table 2). Seven β-lactamase genes, including blaCTX-M-55, three variants of blaTEM, blaOXA-1, and blaNDM-9, which collectively confer resistance to various β-lactams. With the exception of the chromosomally-encoded blaCTX-M-55, the remaining six β-lactamase genes were localized within plasmid p22S130. The chromosomally located aac(6')-Iaa gene, along with the plasmid-borne aac(6')-Ib-cr, aadA2, aph(4)-Ia, rmtB, and aac(3)-IV genes, collectively mediate aminoglycoside resistance. The mph(A), tet(A), fosA3, and ARR-3 genes located on the plasmid conferred resistance to azithromycin, tetracycline, fosfomycin, and rifampicin, respectively. Folate pathway inhibitor resistance involved genes such as sul1, sul2, and dfrA12 while resistance to phenicol was conferred by catB3 and floR. In addition to these antimicrobial resistance genes (ARGs), we identified chromosomal mutations contributing to quinolone resistance in gyrA (p.S83F, p.D87N) and parC (p.T57S, p.S80R). These mutations significantly enhanced the potential for quinolone resistance when combined with the plasmid-borne aac(6')-Ib-cr gene. Moreover, we observed the biocide tolerance gene qacE on plasmid p22S130.The convergence of XDR phenotype, carbapenemase (blaNDM-9), and enhanced fluoroquinolone resistance in S. Indiana ST17, highlights its role as a priority AMR vector.
Antimicrobial class Antimicrobial Agent (abbreviation) MIC (mg/L) R/I/Sa Resistance genes or point mutation Chromosome Plasmid β-Lactam combination agents Ampicillin/sulbactam (SAM) >64/32 R blaCTX-M-55 blaTEM-1B
blaTEM-214
blaTEM-206
blaTEM-141
blaOXA-1
blaNDM-9Penicillins Ampicillin (AMP) >64 R Cephalosporins Cefotaxime(CTX) >16 R Ceftazidime(CAZ) >64 R Cefazolin() >16 R Cefoxitin(FOX) >64 R Carbapenems Imipenem (IMP) 4 R Macrolides Azithromycin (AZM) 64 R mph(A) Aminoglycosides Gentamicin (GEN) >32 R aac(6')-Iaa, aac(6')-Ib-cr, aadA2, aph(4)-Ia,
rmtB, aac(3)-IVAmikacin (AK) >128 R Tetracyclines Tetracycline (TET) >32 R tet(A) Tigecycline (TGC)b ≤ 0.25 S (Fluoro)Quinolones Nalidixic acid (NAL) >64 R aac(6')-Ib-crf Ciprofloxacin (CIP) >4 R Folate pathway inhibitors Trimethoprim/sulfamethoxazole (SXT) >4/76 R sul1, sul2, dfrA12 Phenicols Chloramphenicol (CHL) >64 R catB3, floR Florfenicol (FFC) c,d >32 R Polymyxins Polymyxin E (Colistin, CT) d 1 I Fosfomycins Fosfomycin (FOS)e - - fosA3 Rifampicine - - ARR-3 Note. aR, resistant; I, intermediate; S, susceptible, R/I/S according to the CLSI guidelines M100-S34, 2024; bR/I/S according to EUCAST clinical breakpoints, 2025; cR/I/S according to the CLSI guidelines M31-A3, 2008; dUsed as a feed additive in animal production; eNo antimicrobial resistance phenotype data; faac(6')-Ib-cr also mediate resistance to fluoroquinolones such as ciprofloxacin, but MIC of ciprofloxacin does not always increase above ECOFF. Table 2. Antimicrobial susceptibility of Salmonella Indiana 22S130 to a panel of antimicrobial agents and aquired antimicrobial resistance-encoding genes identified in the bacterical genome with online retrieval in Resfinder database
The genetic environment related to blaNDM-9 in p22S130 is shown in Figure 1. Through the comparative analysis involving our blaNDM-9 fragment and ten homologous sequences from Enterobacteriaceae (including Salmonella, Escherichia coli, and Klebsiella pneumoniae; 9 plasmid-derived and 1 chromosomal), we identified a novel and highly conserved composite transposon structure harboring blaNDM-9, which was "IS26-blaNDM-9-bleMBL-trpF-dsbD-cutA-ISCR1.” This conservative structure was manifested in: (i) all the three bacterial species examined, (ii) diverse plasmid incompatibility types, and (iii) geographical origins and sources of isolation. This structural conservation suggests that the identified genetic environment may be prevalent across diverse Enterobacteriaceae, potentially functioning as a stable transmission vehicle for disseminating blaNDM-9. Two key features flanked the identified blaNDM-9 composite transposon in most of these 11 sequences: (1) a mercury resistance operon (merEDACPTR) appearing upstream and (2) an MDR transposon (ISCR1-sul1-aadA2-dfrA12-intI1-IS26) appearing downstream, implying potential co-selection under mercury exposure, a phenomenon documented in wastewater systems[9]. Notably, several sequences, including that of 22S130, lacked the upstream mer cluster, the region of which was replaced by diverse insertion sequences (ISs) (e.g., IS103, IS150, IS15, and IS26), suggesting potential complex recombination events mediated by these elements. This may reflect niche-specific adaptations, wherein antibiotic pressure supersedes mercury selection. While the functional implications of these flanking structures on the stability and dissemination of the blaNDM-9 transposon remain unclear and require further experimental validation to elucidate their mechanistic roles, they may facilitate co-selection under heavy metal or antibiotic pressure.
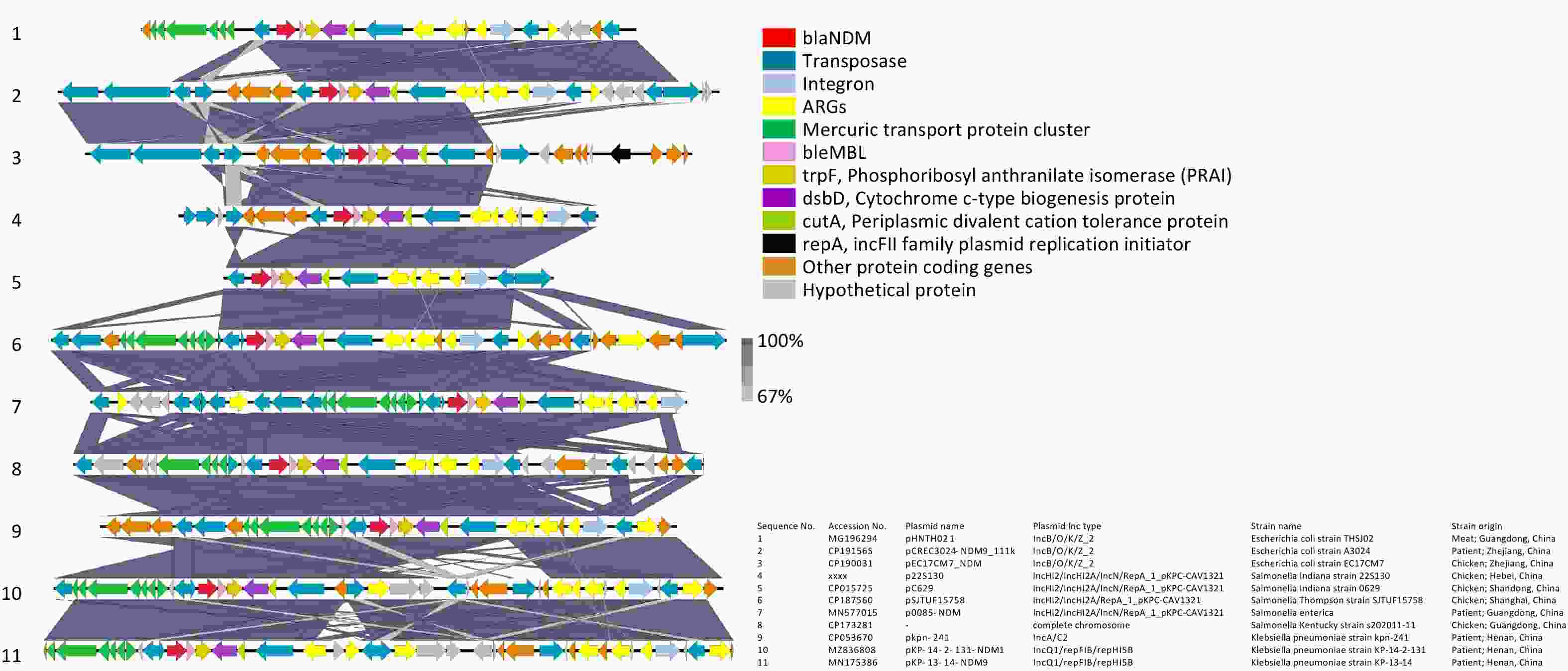
Figure 1. Genetic environments related to blaNDM-9 gene in bacterial plasmids. The figure was generated by Easyfig (v2.2.2). Confirmed and putative open reading frames (ORFs) are indicated by block arrows and their orientations with different colours, and arrow size is proportional to the predicted ORF length. blaNDM-9 gene is indicated by a red arrow, while genes encoding mobile elements (Insertion sequence, IS) are indicated by dark blue arrows. Regions of homology between the plasmids ranging from 67% to 100% are indicated by the graded shaded regions between sequences.
A comparison between the genetic environment flanking the blaNDM-9 gene in Salmonella 22S130 (this study) and that in K. pneumoniae PPH1303 (initial blaNDM-9 discovery)[6] revealed that the blaNDM-9 structure in both the strains contained an identical minimal mobile element (including blaNDM-9, bleMBL, and trpF) and shared conserved downstream dsbD and cutA genes. However, the genetic context adjacent to blaNDM-9 in our strain lacked the GroES and GroEL heat shock chaperonin-encoding genes and carried distinct ISs on the right flank. This suggests that the right-terminal genetic environment of blaNDM-9 in these two strains likely diverged because of an additional ISs insertion event.
Notably, the extracted fragments carrying blaNDM-9 exhibited exceptionally dense and diverse MGEs, including transposons (Tn3-like and IS26-flanked), integrons (intI1), and ISs (ISCR1 and IS26) among others, suggesting that the current configuration of these sequences may have undergone complex MGE-mediated homologous recombination events during their evolutionary history.
Concurrently, we also observed that all ten blaNDM-9-carrying plasmids in this study exhibited hybrid replicon types, with distinct differences in hybrid configurations existing across bacterial species: the three Escherichia coli-derived plasmids were of the IncB/O/K/Z_2 types, whereas the four Salmonella-derived plasmids exhibited the IncHI2/IncHI2A/IncN/RepA_1_pKPC-CAV1321 profiles, and the two Klebsiella pneumoniae-derived plasmids belonged to the IncQ1/repFIB/repHI5B types. Hybrid plasmid replicons have been reported to mediate various critical resistance mechanisms. Examples include IncFIB(K)/IncHI1B/IncX3 hybrid plasmids mediating NDM-IMP carbapenemase expression[10]; IncFIA/FIB/FIC(FII) hybrid plasmids facilitating the co-transfer of blaNDM-5 and fosA3[11]; and IncC-IncX3 hybrid plasmids co-carrying blaNDM-4, tet(X), and tmexCD3-toprJ3 that confer resistance to both carbapenems and tigecycline[12]. We speculate that the plasmid hybridization in this study may be associated with the diverse MGEs mentioned earlier, and with multiple homologous recombination events that catalyzed the fusion of distinct plasmid backbones. This hybrid configuration likely confers integration of varied replication initiation systems to maintain plasmid stability, expand the host range to facilitate cross-species transmission, enhance dissemination of host bacterial clones, and further facilitate accelerated global dissemination of the blaNDM-9 gene and carbapenem resistance. Three blaNDM-9-carrying Salmonella plasmids sharing high similarity with p22S130 were selected for comparative plasmid structural analysis between pSJTUF15758 (from S. Thompson, CP187560), p0085-NDM (from Salmonella spp., MN577015.1), and pC629 (from S. Indiana, CP015725, previously isolated from a whole chicken sample in Qingdao, China in our laboratory). These plasmids demonstrated nearly identical backbone structures (Supplementary Figure S1), with the blaNDM-9 gene consistently located in the distal region and exhibiting high sequence conservation in the blaNDM-9 genetic environment structure (Figure 1). Compared with p22S130, all three plasmids lacked a 130-165 kb segment encoding serine/threonine-protein kinase toxin HipA, DNA-binding protein H-NS, chromosome partition protein Smc, NA-cytosine methyltransferase, Tn3 family transposase TnAs1, tetracycline resistance protein TetC, and multiple putative proteins. This structural variation suggests p22S130 may have acquired enhanced tetracycline resistance through transposon activity, consistent with our strain's minimum inhibitory concentration (MIC) > 32 μg/mL.
Additionally, three Salmonella strains with a chromosomally integrated region identical to the ~17-kb blaNDM-9-containing region of p22S130 were identified: S. Indiana YZ21MCS4 (CP089313; chicken, Yangzhou, 2021), S. Indiana QT6365 (CP103966; diarrheal patient, Wenzhou, 2018), and S. Kentucky s202011_11 (CP173281; chicken, Guangzhou, 2020). Given that reports of the blaNDM-9 gene in Salmonella have thus far been predominantly confined to the serovar Indiana ST17 lineage (whether plasmid-borne or chromosomally integrated), we hypothesized that this lineage possesses competitive advantages over other Salmonella lineages, likely attributable to its abundance of MGEs flanking blaNDM-9 and its enhanced feasibility for forming hybrid plasmids. These genomic features not only facilitate inter-strain gene transfer but also enable vertical transmission of blaNDM via chromosomal integration events.
In this study, we conducted a genomic epidemiological analysis of an XDR Salmonella Indiana ST17 strain, which was isolated from retail chicken carrying the blaNDM-9 gene. In addition to harboring multiple resistance genes and exhibiting carbapenem resistance, this strain possessed a novel and highly conserved composite transposon structure harboring blaNDM-9. The unique genetic environment of this structure, combined with the plasmid's hybrid replicon types, likely collectively mediated the rapid blaNDM-9 dissemination. Our results demonstrate that Salmonella Indiana ST17 is emerging as a primary epidemic clone and reservoir for diverse blaNDM variants, and exhibits significant potential for disseminating carbapenem resistance via HGT, a process in which MGEs critically accelerate transmission dynamics. Given that ARGs have been categorized as emerging contaminants affecting public health and environmental safety, with the blaNDM gene being the pivotal and widespread one across multiple stages of the food chain, implementing AMR surveillance is imperative. The One Health framework, recognized as a vital tool for addressing the interconnections among humans, animals, and the environment[13], provides an essential strategy for elucidating carbapenem resistance mechanisms, tracking transmission pathways, and conducting comprehensive surveillance.
Genomic Epidemiology of Foodborne blaNDM-9 Gene-carrying Extensively Drug-resistant (XDR) Salmonella enterica Serovar Indiana ST17
doi: 10.3967/bes2025.147
- Received Date: 2025-07-17
- Accepted Date: 2025-09-19
The authors declare no competing interests in relation to this work.
| Citation: | Yujie Hu, Peiyuan Huang, Maosong Tian, Lei Zheng, Jun He, Bingbing Li, Jianyun Zhao, Séamus Fanning, Li Bai, Yinping Dong. Genomic Epidemiology of Foodborne blaNDM-9 Gene-carrying Extensively Drug-resistant (XDR) Salmonella enterica Serovar Indiana ST17[J]. Biomedical and Environmental Sciences. doi: 10.3967/bes2025.147 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: