-
The influenza virus, particularly the type A virus, is of great concern because it has a broad host range and causes substantial human morbidity and mortality. Influenza A virus could be classified into different groups or clusters based on the conserved structure of hemagglutinin (HA), including Group1 [H1a cluster (H1, H2, H5, and H6), H1b cluster (H11, H13, and H16), and H9 cluster (H8, H9, and H12)] and Group2 [H3 cluster (H3, H4, and H14) and H7 cluster (H7, H10, and H15)][1]. The viruses H1N1 and H3N2 circulate in humans, while the avian influenza viruses H9N2, H5N1, H6N1, H7N9, H5N6, and H10N8 occasionally infect humans and cause diseases[2]. The strain-specific antibodies (Abs) that directed toward the variable head region of HA predominated during viral infection or vaccination and exhibit neutralizing activities; by contrast, they are limited against newly-emerging viruses. Moreover, delayed production of hemagglutination inhibition (HI) Abs associated with the functional impairment of B and T cells and prolonged viral shielding have been observed in H5N1 and H7N9 infection[3,4]. Therefore, cross-reactive Abs is critical for mounting protective immunity against novel virus infection. A series of broad-neutralizing Abs (BnAbs) targeting the conserved epitopes of HA were recently isolated and found to show cross-protection to heterologous influenza viruses. Several types of epitope regions recognized by BnAbs, including the pocket of the receptor binding domain[5,6], the lateral patch of HA head[7], the hydrophobic patch in the HA stem[8,9], and the vestigial esterase (VE) domain[10], have been identified. Furthermore, the BnAbs primarily use specific variable heavy-chain (VH) or heavy-diversity (D) genes, such as VH1-69, VH3-30, VH1-18, VH3-23, VH6-1, VH1-2, VH4-59, VH3-7, D3-3, and D3-9 or their unique motifs, for HA binding[11-13]. VH1-69 is one of the genes most widely used by HA stem-targeted BnAbs, including CR6261, F10, CR9114, mAb1.12, and 27F3[8,9,14-16]. The unique motif in most HA stem-reactive VH1-69 Abs, IFY, which consists of the hydrophobic residues isoleucine (I) and phenylalanine (F) at positions 53 and 54 in the HCDR2 (complementarity determining regions, CDR) loop and the properly positioned tyrosine (Y) at 100 or 100a in HCDR3, binds to a hydrophobic groove adjacent to the fusion domain of HA2[17,18]. B cells with germline VH1-69-encoded B cell receptor (BCR) could be activated upon binding with HA[19,20]. These special features imply the existence of a specific selection and stimulation of B cells with VH1-69-encoded BCR in response to influenza virus. In another case, receptor-binding domain (RBD)-reactive BnAbs are usually encoded by the VH1-2, VH4-59, and VH1-69 genes. These Abs contact with the sialic acid moiety of RBD by the dipeptides VD(E) or PD at sites 106 and 107 in HCDR3[5,7]. The D3-9 gene segment is currently identified as a ‘SOS responder’ to influenza virus, similar to the VH1-69 gene[13], and uses the specific motif ‘LXYFXWL’ which binds with the HA stem.
The fully human Abs, 65C6, 100F4, FLA3.14, and FLD20.19 isolated from EBV-transformed B cells of recovered H5N1 patients were previously reported to neutralize different clades of H5 viruses[21-23]. However, the cross-subtype Abs response to the novel avian virus remains poor. To date, only one VH1-69 Ab with cross-neutralizing activity to H5N1 and seasonal H1N1 obtained by using the library constructed from the bone marrow of 5 Turkish H5N1 survivors has been reported[24]. Here, we expand peripheral blood CD19+B cells obtained from two human cases infected with clade 2.3.4 H5N1 virus in 2009 and isolated two BnAbs, i.e., 2-12D5, and 3-32G7.1. The genetic basis, biological functions, and epitopes of these BnAbs are then assessed and modeled.
-
The research protocol was approved by the Institutional Review Board of National Institute for Viral Disease Control and Prevention, China CDC, and written informed consent was obtained from the patients. Blood specimens from human cases infected with clade 2.3.4 H5N1 virus were collected on days 12 and 21 after disease onset in 2009, and peripheral blood mononuclear cells (PBMCs) were stored in liquid nitrogen. B cell culture, screening of Abs, and B cell cloning were performed as previously reported[25,26]. Briefly, CD19+ B cells were isolated from the recovered PBMCs using magnetic beads (STEMCELL, Vancouver, Canada) and cultured in 96-well plates at a density of 20 cells per well in the presence of 2 × 104 irradiated CD40L expressing 3T3 cells per well, 20 ng/mL hIL-2, 1 μg/mL CpG2006, and 10 ng/mL hIL-21 for 10 d[27]. H5-positive B cells were screened by an ELISA test with the supernatants of B cells binding to recombinant H5 (rH5) of A/Hubei/1/2011 (clade 2.3.2, HB-H5N1). The recombinant HAs used for ELISA were produced by using a baculovirus expression system and purified by Ni Sepharose (GE Healthcare Europe). Light-chain typing was carried by modified rH5-ELISA with HRP-anti-κ or anti-λ Abs (Southern Biotech, AL, USA). H5-positive B cells were further screened via the rH7 (A/Zhejiang/1/2013) binding test and neutralization test (NT) with H5N1 pseudotype particles (pps). H5N1 pps were generated as described previously[28], and the corresponding HA and NA of A/Guangxi/1/2008 (H5N1, clade 2.3.4) were used. Lysates of the cells in candidate wells were prepared, and the variable genes of heavy (H) and light (L) chains were amplified by RT-PCR and inserted into the IgG-Ab vector with the constant genes of Igγ1, κ1, or λ1. At least 10 variable gene clones for H/L (κ or λ) were analyzed from each candidate well. The Ab gene pairing was screened by transient transfection of human kidney 293T cells with each variable gene of H/L from the same candidate well for 48 h and assayed by rH5-ELISA binding. Abs were produced by transient transfection of human kidney 293T cells and subsequent purification by protein G columns from the culture supernatants.
-
Madin-Darby canine kidney (MDCK) cells were infected by influenza viruses at a multiplicity of infection (MOI) of 1–2. After 14 h, the cells were detached by 0.25% trypsin-EDTA and incubated with the tested Abs at 2.5 μg/mL for 1 h. The cells were washed and then stained with fluorescein-conjugated anti-human IgG serum. Stained cells were analyzed by using a FACS Aria II system with FACS Diva software (BD, San Diego, CA, USA). Infectivity was confirmed by anti-nucleoprotein (NP) staining. The viruses used in this work, including wild strains and reassortant viruses bearing HA and NA from the corresponding virus and the internal genes of A/Puerto Rico/8/1934, are described in Table 1. All viruses were propagated in embryonated chicken eggs, and virus titers were determined in terms of mean 50% tissue culture infective dose (TCID50) per milliliter on MDCK cells by NP ELISA.
Table 1. The binding and neutralizing activity of 2-12D5 and 3-37G7.1 to influenza viruses
Virus name Abbreviation HA Group/Cluster HA-binding IC50 (μg/mL) 2-12D5 3-32G7.1 2-12D5 3-32G7.1 A/Puerto Rico/8/34 (H1N1) PR8-H1N1 1/H1a + + 20 > 80 A/Guangdong/51/2008 (H1N1) GD-H1N1 1/H1a + + 20 40 A/Shanghai/1/2009 (H1N1pdm09) SH-H1N1 1/H1a + + 12.5 2.2 RG A/Hubei/1/2011 (H5N1, clade 2.3.2) RG HB-H5 1/H1a + + 20 10 RG A/Beijing/1/2003 (H5N1, clade 7) RG BJ-H5 1/H1a + + 5 2.5 RG A/Guangdong/1/2014 (H5N6, clade 2.3.4) GD-H5N6 1/H1a + + 20 2.5 A/waste water/GX/2/2010 (H6N2) GX-H6N2 1/H1a + + 2.5 > 80 A/Hong Kong/33982/2009 (H9N2) HK-H9N2 1/H9 + − 20 > 80 A/Env/JX-PYH/376/2014 (H11N3) JX-H11N3 1/H1b + − > 80 > 80 A/Env/JX-PYH/1102/2014 (H12N5) JX-H12N5 1/H9 + − > 80 > 80 A/Env/QHH/166/2012 (H13N8) QH-H13N8 1/H1b − − > 80 > 80 A/Hong Kong/01/1968 (H3N2) HK-H3N2 2/H3 − − > 160 > 160 A/Env/Guangdong/15188/2015 (H4N2) HN-H4N2 2/H3 − − > 160 > 160 RG A/Anhui/1/2013 (H7N9) AH-H7N9 2/H7 − − > 160 > 160 RG A/Guangdong/17SF003/2016 (H7N9) GD-H7N9 2/H7 − − > 160 > 160 RG A/Jiangxi/346/2013 (H10N8) JX-H10N8 2/H7 − − > 160 > 160 Note. ‘+’ denotes binding. ‘−’ denotes no binding. RG denotes a recombinant virus bearing the HA and NA from the corresponding virus and the internal genes of A/puerto Rico/8/1934. -
CR6261 was transiently expressed according to the VH/L sequence (GenBank HI919029.1/GenBank HI919031.1) and purified using protein G. ELISA plates were coated with the rH5 of HB-H5N1. Two fold serial dilution of 2-12D5, 3-32G7.1, and a control containing an unrelated Ab ranging from 5 μg/mL to 80 μg/mL, were mixed with biotinylated CR6261 at the EC50, respectively, and then added to the plates. After co-incubation for 1 h, binding of the biotinylated Abs was detected using a 1:200 dilution of HRP-streptavidin (R&D, MN, USA). The percentage of biotinylated CR6261 bound to rH5 of HB-H5N1 was calculated from the OD450 of biotinylated CR6261 with the test Abs divided by the OD450 of biotinylated CR6261 without Abs.
-
MDCK cells growing in 96-well plates were infected with influenza viruses (Table 1) for 1 h at 37 °C. The virus inocula were washed off, and the cells were overlaid with 2.4% Avicell supplemented with mAbs and 1 μg/mL TPCK-treated trypsin. Approximately 30–50 plaque-forming units (PFUs) of viruses were used, and the cells were processed by staining with anti-NP after 22 h of incubation. The number of plaques was counted by AID ELISPOT reader. The data were analyzed by using Prism software (GraphPad), and the concentration at which a mAb inhibited 50% of the plaques (IC50) was calculated using a nonlinear regression.
-
Cytotoxicity was determined as described previously[28]. Briefly, lung epithelial A549 cells were infected with the A/Shanghai/1/2009 (2009 pdmH1N1, SH-H1N1) or RG A/Guangdong/1/2014 (H5N6, GD-H5N6) viruses at an MOI of 5 for 14 h, detached, and then stained with different concentrations of the tested Abs for 1 h. Thereafter, the A549 cells were washed and incubated with PBMCs, which were isolated from healthy adult donors and pre-blocked with 10 μg/mL Ab against the HA-binding receptor NKp46 (9E2, Biolegend, CA, USA), at a 10:1 ratio in the presence of CD107a Abs (H4A3, BD Biosciences, San Diego, CA, USA) and monesin (Sigma-Aldrich, Saint Louis, MO, USA) in 0.5% low IgG FBS culture medium for 6 h. After incubation, the surface makers, CD3 of T cells, and CD56 of NK cells were stained. Cytotoxicity was analyzed by flow cytometry and calculated as the percentage of CD3-CD56+ NK cells positive for CD107a.
-
Serially diluted RG A/Beijing/1/2003 (H5N1, clade 7) viruses were pre-incubated with 20 μg/mL 2-12D5 or 10 μg/mL 3-32G7.1 for 1 h at 37 °C. The incubated mixtures were then added on MDCK cell monolayer. After 1 h of absorption, the cells were washed with PBS and cultured in DMEM containing 2 μg/mL TPCK-trypsin and 20 μg/mL 2-12D5 or 10 μg/mL 3-32G7.1. Cytopathic effect (CPE) in MDCK cells was monitored by daily observation and confirmed by hemagglutination assay. CPE-positive supernatants with the lowest virus titer were used for subsequent infection. Virus clones were purified from the supernatants from passages 10 and 15 by the plaque formation test. The clones were selected, and the HA gene was sequenced. The mutants were tested for neutralization and binding assay.
-
The 3D structures of the variable regions of 2-12D5 and 3-32G7.1 were predicted using Ab homology modeling software PIGS. In addition, comparison studies were performed using the known 81 HA-Ab complex structures in Protein Data Bank to obtain their binding modes with the HA structures of H1, H5, H6, and H9 (PDB codes: 5GJS, 5JW4, 5T08, and 1JSD). Results showed that the binding regions can be classified in two categories: the sialic acid binding cavity at the head of HA and the stem surface of HA. We assumed that these regions are the most likely binding regions. Thus, 3-32G7.1 and 2-12D5 were docked at these two regions using the well-established protein-protein docking tool ZDOCK. 2-12D5 and 3-32G7.1 were finally predicted to interact with the stem surface of HA by comparing the docking scores and structure complementarity. The corresponding 3D figures were drawn with PyMOL Molecular Graphics system (Delano Scientific, Palo Alto, CA, USA).
-
We expanded the peripheral blood CD19+ B cells of two patients infected with clade 2.3.4 H5N1 virus. Patient 1 was a 21-year-old female, and her PBMCs were sampled on days 12 after disease onset. Patient 2 was a 30-year-old male, and his PBMCs were sampled on days 21. Both patients recovered from infection. The H5N1 viruses of clade 2.3.4 were isolated from the two patients. The corresponding B cell cultures were initially screened by rH5, rH7-binding test, and NT with H5N1 pps. The results showed that B cells with pps NT activity or cross-reactivity are present in peripheral blood with very low proportions of 0.33% and 0.24%, respectively. The genes of 2-12D5 were selected from the B cells of Patient 1 with > 80% pps NT activity, while those of 3-32G7.1 were obtained from the B cells of Patient 2 that bound to H5 and H7.
The antigenic-binding profiles of 2-12D5 and 3-32G7.1, together with their in vitro neutralizing and ADCC activities, were characterized. 2-12D5 bound to cell surface-expressed HAs across different clusters of Group 1 HA, including H1, H5, H6, H9, H11, and H12, while 3-32G7.1 bound with H1, H5, and H6 (Table 1). We first performed HI tests of the two Abs against H5N1 and H7N9 and observed no activity (data not shown). We then carried out binding competitive ELISA between the mAbs with a biotin-labeled VH1-69 stem-directed mAb, CR6261. The binding of labelled CR6261 to H5 after pre-incubation with saturating concentrations of 2-12D5 and 3-32G7.1 decreased to 9.8% ± 1.9% and 21.1% ± 0.7%, respectively; by comparison, the binding of biotin-CR6261 with the control containing an unrelated Ab was 103.9% ± 0.3%. The reduced binding of CR6261 to rH5 implies that epitopes recognized by the two Abs overlap with the stem epitope of CR6261.
We measured the IC50 neutralization titers (i.e., the Ab concentration resulting in 50% reduction in infectivity) of the two Abs by using live viruses and tested their neutralizing effects by using plaque reduction tests. MDCK cells were inoculated with virus without TPCK-trypsin. After 1 h of absorption, unabsorbed viruses were washed off, and Avicell with the tested Abs and TPCK-trypsin were added to the MDCK cells. As shown in Table 1, 2-12D5 inhibited the formation of plaques by the H1, H5, H6, and H9 viruses, and 3-32G7.1 inhibited those by the seasonal H1, pdmH1 and H5 viruses.
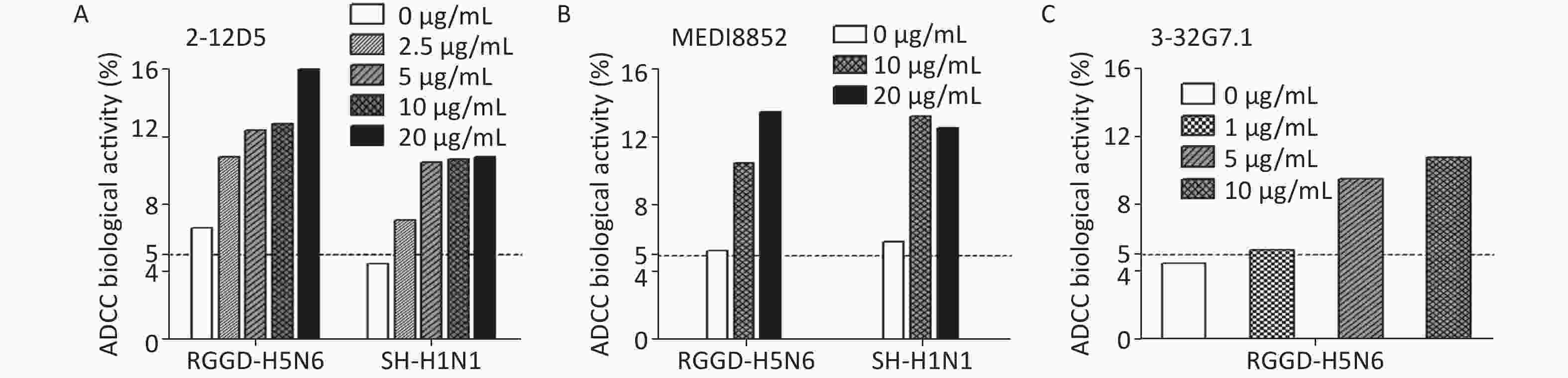
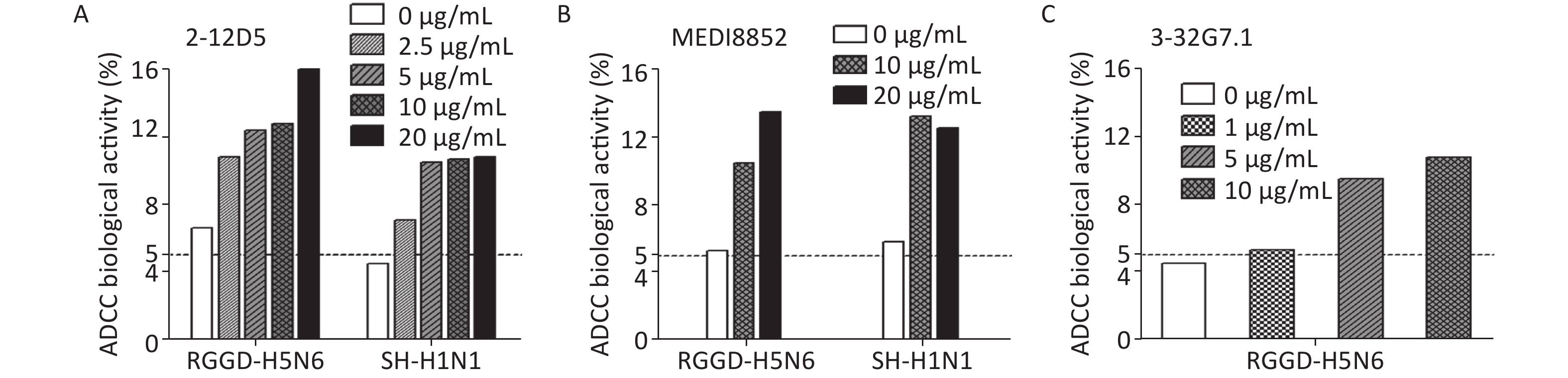
Besides targeting virus, the BnAbs interact with immune cells via ADCC or complement-dependent cytotoxicity (CDC) to cleave virus-infected cells in vivo. ADCC effects could be detected in healthy adults even without detectable neutralizing Abs against novel influenza viruses[29]. Because HA stem-targeting Abs are potent inducers of ADCC activity, we measured the abilities of the two Abs to activate the cytotoxicity of natural killer (NK) cells by CD107a staining. The percentage of activated CD107a+ NK cells remarkably increased upon co-culture with 2009 pdmH1N1- or H5N6-infected target cells pre-bound with 2-12D5 or 3-32G7.1 relative to that with no Ab (Figure 1). Interestingly, the effects of 2-12D5 were comparable with those of MEDI8852, which is currently known as the anti-flu BnAb with the greatest neutralizing and ADCC activities.
-
Previously, it was found that BnAbs triggered by H5 infection employ VH1-69 and VH4-46 germline genes, while the H5N1 vaccine induces VH1-18- and VH6-1-encoded BnAbs[11,24]. Here, we found that 2-12D5 utilizes VH1-69, which is a commonly used gene for neutralizing antibodies against many other viruses, and 3-32G7.1 utilizes VH1-2, which is a gene rarely used by anti-flu BnAbs (Tables 2–4). Two Abs carried a relatively low level of somatic hypermutations (SHMs) in the V genes (frequency is 5.90% and 2.78% for VH; 0.34% and 0.35% for VL), and 15 and 18 amino acids in HCDR3, respectively (Table 2). The VH1-69 gene detected in this work used VH1-69*01, which belongs to the F54-encoding alleles of the VH1-69 germline, including VH1-69 *01, *03, *05,*06, *07, *12, and *13[18]. Interestingly, the typical signature, IFY, an unique feature of VH1-69 BnAbs binding with the HA stem, was also found. Some signature mutations, including S30R in HCDR1 and P52aA in HCDR2, resulting in the high affinity described in a previous comprehensive study[19,20] were also found in 2-12D5 (Table 4).
Table 2. The gene characteristics of 2-12D5 and 3-32G7.1
Ab clone V gene Somatic mutation rate in V gene (% nucleotide) D gene J gene Somatic mutation rate in J gene (% nucleotide) Amino acid No. of CDR 2-12D5 VH1-69*01 5.90 DH2-15*01 JH4*02 8.33 8.8.15 Vλ4-69*01 0.34 − Jλ3*02 0.00 7.7.10 3-32G7.1 VH1-2*02 2.78 DH3-9*01 JH5*01 23.53 8.8.18 Vκ3-20*01 0.35 − Jκ4*01 7.89 7.3.9 Note. The VH, D, and JH sequences were compared by using the IMGT database. ‘−’ denotes the gene without the segment. Table 3. The VDJ and VJ gene and somatic hypermutations of VH1-69-, VH1-2-, and D3-9-encoded Abs against influenza virus
mAb VH DH JH SHMs in VH (aa) VL JL SHMs in VL (aa) Access #/reference CR6261 1-69*01 2-2*01 6*04 15 Vκ1-51*01 2*01/03*01 7 GenBank: HI919029.1/
GenBank: HI919031.1F10 1-69*01 NA NA 13 Vκ10-54*01 3*02 5 PDB: 3FKU_T MAb1.12 1-69*06 NA 6 21 − − 14 [16] CR9114 1-69*06 3-10*01 6*02 17 Vκ1-44*01 7-*01 11 GenBank:JX213639.1/
GenBank: JX213640.127F3 1-69*06 NA NA 14 − − − [15] 2-12D5 1-69*01 2-15*01 4*02 12 Vλ4-69*01 3*02 1 The study CH65 1-2 1-1 6 12 Vκ3-21 2 − [6] 3-32G7.1 1-2 3-9*01 5*-01 3 Vκ3-20 4*01 2 The study FI6v3 3-30*03F/
3-30*18/
3-30-5*013-9*01 4*02 8 Vκ4-1*01 J1*01 32 GenBank: JN234435.1/
GenBank: JN234444.1S9-3-37 1-18 3-9 − − Vκ2-24 − − [13] 31.b.09 1-18 3-9 − − − − − [13] 31.d.01 3-30 3-9 − − − − − [13] 54.e.01 7-4-1 3-9 − − − − − [13] 56.ND.12 3-23 3-9 − − − − − [13] 56.h.01 3-23 3-9 − − − − − [13] 02-1A05 1-2 3-9 4 − Vκ3-20 − − [31] 04-1B12 1-2 3-9 4 − Vλ2-14 − − [31] 27-1C08 1-2 3-9 6 − Vλ2-14 − − [31] Note. ‘−’ denotes not available. Table 4. The molecular signature of VH1-69- and VH1-2-encoded Abs
Abs/gemline gene CDR1-IMGT FR2-IMGT CDR2-IMGT FR3-IMGT CDR3-IMGT 26------30--33 34----------40----------------------50 5152a----------57 58-60------------------70------------------80--82abc---------------90-92 93------100----------102 VH1-69*01 GGTFSSYA ISWVRQAPGQGLEWMGG IIPIFGTA NYAQKFQGRVTITADESTSTAYMELSSLRSEDTAVYYC AR F10 EV....F. ..............L.. .S.M...P ...............Q..R....D.R............ ..SP---SYICSGGTCVFDH CR6261 ..P.R... ...........P..... .......T K..P...........DFAG.V.............M... .KHM---GYQVRET---MDV 2-12D5 .V..R.F. F................ .TA...VV ..........................G.T......... .KEAG---YYS--GRSHFDD 27C3 ....GN.. .N............... .S....PP K...........S..I.S.....D....S.D....... ..KLEPPYYFYSY----MDV CR9114 ...SNN.. ............D.... .S....ST A...........S..IFSN......N..T.......F. ..--HGNYYYYSG----MDV VH1-69*06 ........ ................. ........ ...............K...................... .. VH1-2*02 GYTFTGYY MHWVRQAPGQGLEWMGW INPNSGGT NYAQKFQGWVTMTRDTSISTAYMELSRLRSDDTAVYYC AR 3-32G7.1 GYTFTGYY IHWVRQAPGQGLEWMGW INPNSGGT NYAQKFQARVTMTKDTSISTAYMELSRLRSDDTAVYYC ARGDHVLRYFDWQSSEPS CH65 GYTFTDYH INWVRQAPGQGLEWMGW IHPNSGDT NYAQKFQGWVTMTRDTAISTAYMEVNGLKSDDTAVYYC ARGGLEPRSVDYYYYGMDV VRC01 GYEFIDCT LNWIRLAPGKRPEWMGW LKPRGGAV NYARPLQGRVTMTRDVYSDTAFLELRSLTVDDTAVYFC TRGKNCDYNWDFEH FI6 GFTFSTA MHWVRQAPGRGLEWVAV ISYDGNYK YYADSVKGRFSISRDNSNNTLHLEMNTLRTEDTALYYC AKDSQLRSLLYEWLSQGYFDYW S9-3-37 − − − − LGYFDWL 31.b.09 − − − − HILTGF 31.d.01 − − − − DSLFQLRYFDWLLGDAFNI 54.e.01 − − − − DPLQLILRYFDWLFKPLDY 56.ND.12 − − − − GAHLGRIELRYFEWLRKDYYYYGMDV 56.h.01 − − − − GAHLGRIELRYFEWLRKDYYYYGMDV 02-1A05 − − − − CARDSGMRYFDWLSGYFDFW 04-1B12 − − − − CARVGKLQYFDWPAHYFDYW 27-1C08 − − − − CARDKGLRYFDWLSGGMDVW Note. The amino acid sequences of VH1-69-encoded Abs, CR6261, F10, CR9114, and 27F3 (green background), VH1-2-encoded CH65 and VRC01 (white background), D3-9-encoded S9-3-37, 31.b.09, 31.d.01, 54.e.01, 56.ND.12, 56.h.01, 02-1A05, 04-1B12, and 27-1C08 (grey background) as well as 2-12D5 and 3-32G7.1 in the study, which were highlighted with red color were aligned. Residue positions are according to Kabat numbering. Dots indicate identical residues. The CDRs defined by IMGT were marked. The sites previously reported to mediate the binding to the HA glycoprotein of influenza virus are highlighted by red color and the amino acids in the corresponding residues in this study are highlighted in green color. '−' denotes not available. The sequence of D gene in CDR3 of Abs, 3-32G7.1, FI6, S9-3-37, 31.b.09, 31.d.01, 54.e.01, 56.ND.12, 56.h.01, 02-1A05, 04-1B12, and 27-1C08, was labeled with underline. The F/L at the residue 100e interacts with the HA stem, which are highlighted by blue color. 3-32G7.1 carried significantly fewer SHMs compared with 2-12D5 (Tables 3 and 4) and showed a VH1-2+D3-9 lineage. Attention related to the VH1-2 gene has focused on the CD4-binding mAb VRC01, which presents an unusual breadth of neutralizing activity against HIV. This Ab binds to CD4 via its HCDR2 and acquires a large number of somatic mutations from several years of infection[30]. VH1-2- or VH1-2+D3-9-encoded BnAbs recognize the conserved epitopes via different contacting interfaces; for example, the CH65 encoded by the VH1-2 gene incorporates its CDRH3 with HA RBD[6], and 27-1C08 uses the specific motif ‘LXYFXWL’ in the D segment to interact with the HA stem[31]. Specifically, the amino acid ‘F’ in the motif bind to the HA stem. The Ab 3-32G7.1 detected in this work targeted the HA stem, and the motif ‘VLRYFDW’ and the unique ‘F’ were found in its D segment. No somatic mutations were found in its HCDR1 and HCDR2 (Table 4).
-
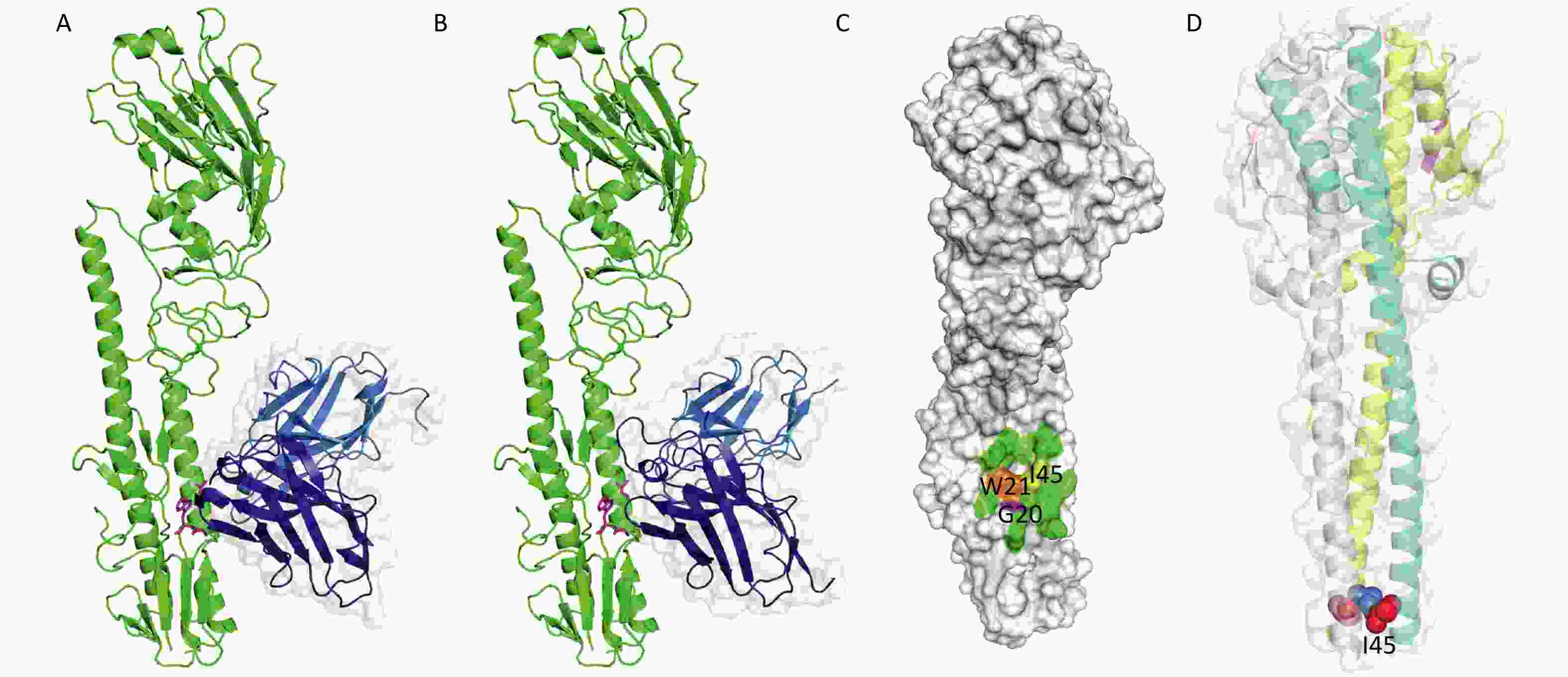
We modeled the 3D structures and binding-modes of 2-12D5 and 3-32G7.1 using PIGS[32] and ZDOCK[33] by comparing the homology of HA regions and CDR fragments from previously reported X-ray crystal structures (PDB codes: 5GJS, 5GJT, 5JW3, and 5JW4) (Figure 2A and 2B). Although their CDRs are obviously different, both BnAbs were predicted to target the HA stem region, which is a common epitope observed in many HA-Abs complexes. This region is formed by two parallel alpha helices and the surrounding loops with a hydrophobic hollow area in the center (Figure 2C). This area consists of a group of conserved residues (I45, W21, and G20 at HA2) in various HA subtypes, which allow the Abs to neutralize various influenza viruses. A critical residue next to the hollow area is a glycosylation site (N38, T38, or S38) in H3 and H7, compared with H38 in H1, H5, H6, and H9. The presence of the glycan hinders their binding to Group 2 HA stem. Thus, most of the BnAbs that neutralize H1, H5, H6, and H9 do not work in H3 or H7.
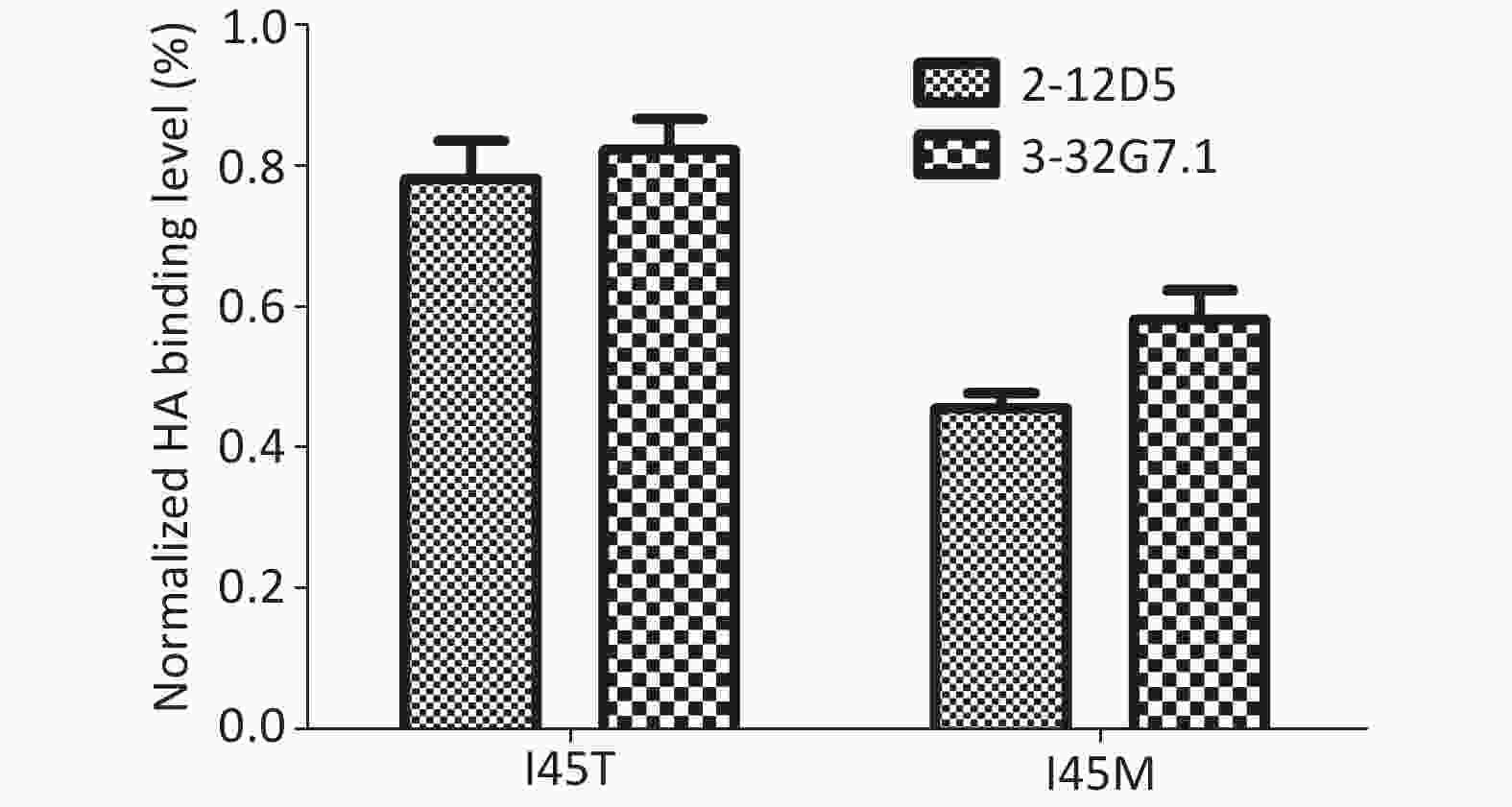
The escape variants of RG-A/Beijing/1/2003 (H5N1, clade 7) were generated to confirm their epitope localization. Variants carrying I45M and I45T in HA2 were detected in the presence of 20 μg/mL 2-12D5 and 10 μg/mL 3-32G7.1, respectively. The mutants were resistant to Ab neutralization (IC50 > 160 μg/mL, Table 5) and showed reduced binding capacities to the two Abs (45.7% ± 2.1%, 58.3% ± 4.2%; 78.1% ± 5.5%, and 82.3% ± 4.4% of the bindings of WT virus for 2-12D5 and 3-32G7.1, respectively, Figure 3). Interestingly, I45T escape mutants against 3-32G7.1 were observed at earlier passages and showed a frequency higher than that of 2-12D5 (Table 5).
Table 5. IC50 neutralization titers against RG A/Beijing/1/2003 (H5N1, Clade 7) and its mutants
Escape mutation Selected by the Abs HA domain Mutation occurance among clones (P10) Mutation occurance among clones (P15) IC50 neutralization titers (μg/mL) 2-12D5 3-32G7.1 I45 no − 5/5 5/5 5 2.5 I45M 2-12D5 Helix A of HA2 2/32 5/5 > 160 > 160 I45T 3-32G7.1 Helix A of HA2 31/32 5/5 > 160 > 160 Note. ‘−’denotes not available. The HA stem is vital for HA conformational changes or membrane fusion during viral infection. A variety of escape mutations detected in HA stem-reactive BnAbs, including HA2 H111L of the H5N1 virus for CR6261; V466I or R526G in HA of the 2009 pdmH1N1 virus for CR9114; S123G, N460S, and N203V in HA, and E329K in NA of seasonal H1N1 for F10; T318K in HA of H1N1 virus, and HA2 V52E of H2N2 virus for C179, suggest that crucial residues for infection may vary among different subtypes of viruses[8,34,35]. An HA1 N21S/Y detected in mutants escaping from AbS9-3-37 with a D3-9-encoded HCDR3 suggests that the residue is associated with HA fusion activity but not a loss of a direct Ag–Ab interaction could be involved[36]. Further studies on the escape mechanism of HA stem-BnAbs are needed.
The known crystal structure of HA (PDB code: 5JW4) demonstrates that I45 in HA2 locates in the alpha helix in the HA stem, which is in a core region of the antigen-Ab binding interface (Figure 2C). After viral absorption, fusion occurs between viral and host cell membranes via HA refolding of its secondary and tertiary structures. The crystal structure of HA at the fusion state (PDB code: 1HTM) shows that I45 is embedded in the hydrophobic core of the triple helix (Figure 2D). Our findings that the substitution at Site 45 in HA2 of H5N1 for the two HA stem-reactive BnAbs did support the critical roles of I45.
-
In our study, we assayed the early-phase heterosubtypic Ab response in H5N1 survivors, which is rarely documented. We discovered cross-neutralizing anti-HA Ab genes from peripheral blood B cell repertoires even in the presence of very low proportions (e.g., 0.3%) of total B cells. Specifically, cross-reactive B cells were present in our samples at a much earlier time than that reported in previous studies on H5N1 survivors, which had a sampling time point of 1–4 months post-infection (poi.)[21-24]. This finding implies that the genes could be present as early as 12 d after H5N1 infection, consistent with the rapid responses of memory B cells upon re-exposure to the antigen.
Interestingly, although the BnAbs were obtained from B cells at certain time point from two H5N1 cases, they appeared to share a number of consensus genetic properties with other BnAbs[5,13,17,19]. We discovered that the two Abs belong to different lineages but utilize the most common pathogen-responding genes, namely, VH1-69 or D3-9. Anti-influenza BnAbs are known to use different binding modes or motifs to bind to the HA stem and acquire cross-reactivity within or between groups[11,13,17,18,20,37]. Similar to most other VH1-69 BnAbs, the 2-12D5 observed in our study is of the VH1-69-D2-15-JH4 lineage from an acute case on days 12 post-infection and possesses the unique HA stem-targeted IFY motif. Moreover, this Ab bears low levels of SHMs in VH and two known high-affinity-associated SHMs, namely, S30R in HCDR1 and P52aA in HCDR2, and exhibits very broad neutralizing activity. Conversely, 3-32G7.1, which was obtained from another case on days 21 post-infection, was of the VH1-2-D3-9-JH5 lineage, had no SHMs in HCDR1 and HCDR2, but carried an LRYFDW region encoded by the D3-9 segment, which is very similar to the recently identified HA stem-binding motif-LXYFXW, particularly, with the F at 100e[13]. Only 0.01%–0.1% of the LXYFXW sequence is found in the B cell repertoires of healthy donors, and it is likely specific to Group1 HA triggering and binding[13]. To date, several VH genes rearranged with D3-9, including VH1-18, VH3-30, and VH1-2 have been observed in anti-flu BnAbs[11,13,31]. 3-32G7.1 in this study demonstrated genetic similarity to the VH1-2+D3-9 BnAb, 27-1C08, thus supporting the hypothesis that D3-9 is a specific Ab lineage triggered by the influenza virus; in addition, the D3-9-encoded HCDR3 region appears to be specific for the binding of the HA stem. The germline nature of 3-32G7.1 suggests that this Ig class could be elicited relatively quickly because of the low number of SHMs required for affinity maturation as compared with those of VH1-69 BnAbs. Importantly, along with the accelerating data for genetic basis for BnAbs, the B cell receptor-targeting immunogens are proposed for influenza vaccine strategy. The low SHM rate detected in this work implies that VH1-69 and D3-9 are ideal candidate germlines that could be easily activated by vaccination or infection.
The epitopes of the Abs in the current study are notably located at the stem domain of Group1 HA, similar to most BnAbs. Modeling and selection of escaped variants against the Abs revealed that 2-12D5 and 3-32G7.1 target the hydrophobic groove at the N terminus of HA2. A fixed mutation occurs in residue 45, which is highly conserved across subtypes. Previous data reveal that HA stem BnAbs neutralize viral infection by inhibiting membrane fusion or preventing HA cleavage, NA inhibition activity, or ADCC in vivo[10,38-40]. Residue 45 of HA2 is located in the stem region surrounding the fusion peptide, and mutations at nearby residues 47, 51, 54, 58, 59, 63, and 70 of H1, H3, H5, and H7 could significantly affect the fusion pH[41,42]. We found that 2-12D5 and 3-32G7.1 contact with this region and afford a certain degree of cross-protection by neutralizing the virus itself and clearing virus-infected cells in vitro. Although the efficacy of BnAbs in patients is unclear, we show evidence that cross-reactive B cells could be triggered by novel H5N1 viruses, even at a very early time, consistent with patient survival. More importantly, the breadth of antigen binding and function of the Abs from H5N1 patients in our study is much broader than that described in previous reports[21,23,24]. Our findings support the currently proposed vaccination design that the conserved antigenic site in the HA stem domain is a protective epitope. The encouraging finding that the immunogenicity of the HA stem-subunit in vivo could be enhanced by its increasing concentration in draining lymph nodes supports the hypothesis that it is a potential epitope for human vaccination[43]. We observed the escape variants as those have been identified for most HA-stem BnAbs. Further study of HA stem epitopes as vaccine candidates and analyses of their cost-effectiveness is needed.
The data on the conserved antigenic site in the HA stem domain and the specific gene uses of VH1-69 or VH1-2+D3-9 could provide potential strategies for vaccine development.
-
The authors declare no conflicts of interest.
-
We acknowledge MedImmune LLC (Gaithersburg, MD 20878, USA) to provide MEDI8852. We thank Prof. Patrick C Wilson, University of Chicago, for providing IgG-Ab vector with the constant genes of Igγ1, κ1, or λ1. We thank Dr. QIN Kun, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, for his kind helps in experiment design, technical supporting, and manuscript revising.
doi: 10.3967/bes2020.014
Cross-neutralizing Anti-hemagglutinin Antibodies Isolated from Patients Infected with Avian Influenza A (H5N1) Virus
-
Abstract:
Objective To recover broad-neutralizing monoclonal antibodies (BnAbs) from avian influenza A (H5N1) virus infection cases and investigate their genetic and functional features. Methods We screened the Abs repertoires of expanded B cells circulating in the peripheral blood of H5N1 patients. The genetic basis, biological functions, and epitopes of the obtained BnAbs were assessed and modeled. Results Two BnAbs, 2-12D5, and 3-37G7.1, were respectively obtained from two human H5N1 cases on days 12 and 21 after disease onset. Both Abs demonstrated cross-neutralizing and Ab-dependent cellular cytotoxicity (ADCC) activity. Albeit derived from distinct Ab lineages, i.e., VH1-69-D2-15-JH4 (2-12D5) and VH1-2-D3-9-JH5 (3-32G7.1), the BnAbs were directed toward CR6261-like epitopes in the HA stem, and HA2 I45 in the hydrophobic pocket was the critical residue for their binding. Signature motifs for binding with the HA stem, namely, IFY in VH1-69-encoded Abs and LXYFXW in D3-9-encoded Abs, were also observed in 2-12D5 and 3-32G7.1, respectively. Conclusions Cross-reactive B cells of different germline origins could be activated and re-circulated by avian influenza virus. The HA stem epitopes targeted by the BnAbs, and the two Ab-encoding genes usage implied the VH1-69 and D3-9 are the ideal candidates triggered by influenza virus for vaccine development. -
Key words:
- VH1-69 /
- D3-9 /
- Avian influenza A (H5N1) virus /
- Cross-neutralizing /
- Antibody
-
Figure 1. Antibody-dependent cellular cytotoxicity (ADCC) mediated by 2-12D5 and 3-32G7.1.
(A) 2-12D5, (B) MEDI8852, and (C) 3-32G7.1 induced ADCC activities against 2009 pdmH1N1 (A/Shanghai/1/2016)- or H5N6 (A/Guangdong/1/2014)-infected lung epithelial A549 cells. NK cells were gated, and activated CD3−CD56+NK cells expressing CD107a were detected by FACS. One representative data point was obtained from two independent experiments.
Figure 2. Binding modes of 2-12D5 and 3-32G7.1.
The binding modes of (A) 2-12D5 and (B) 3-32G7.1 with the H5 structure (PDB code: 5JW4). Heavy chains are highlighted in deep blue, and light chains are highlighted in aqua marine. I45, W21, and G20 are indicated by magenta sticks. (C) The interface of pre-fusion HA with Abs is located at the stem region of HA. I45, W21, and G20 are indicated in yellow, orange, and magenta, respectively. (D) The triple helix formed during membrane fusion and three I45 (highlighted in colored spheres at the bottom) in HA2 (PDB code: 1HTM) maintained its structure via hydrophobic force.
Figure 3. Binding of 2-12D5 and 3-32G7.1 with the mutants of RG A/Beijing/1/2003 (H5N1).
The mutants of RG A/Beijing/1/2003 (H5N1) were cloned and used to infect MDCK cells. The surface HAs of virus-infected MDCK cells were stained with 2.5 μg/mL 2-12D5 and 3-32G7.1. Binding of the tested Abs was measured by staining with polyclonal anti-human IgG Fc serum and FACS detection. NP stain was used for sequential normalization. The relative HA-positive percentage is expressed as the normalized binding with mutants to that of wild virus, and the data are expressed as the means of three replicates with the standard error.
Table 1. The binding and neutralizing activity of 2-12D5 and 3-37G7.1 to influenza viruses
Virus name Abbreviation HA Group/Cluster HA-binding IC50 (μg/mL) 2-12D5 3-32G7.1 2-12D5 3-32G7.1 A/Puerto Rico/8/34 (H1N1) PR8-H1N1 1/H1a + + 20 > 80 A/Guangdong/51/2008 (H1N1) GD-H1N1 1/H1a + + 20 40 A/Shanghai/1/2009 (H1N1pdm09) SH-H1N1 1/H1a + + 12.5 2.2 RG A/Hubei/1/2011 (H5N1, clade 2.3.2) RG HB-H5 1/H1a + + 20 10 RG A/Beijing/1/2003 (H5N1, clade 7) RG BJ-H5 1/H1a + + 5 2.5 RG A/Guangdong/1/2014 (H5N6, clade 2.3.4) GD-H5N6 1/H1a + + 20 2.5 A/waste water/GX/2/2010 (H6N2) GX-H6N2 1/H1a + + 2.5 > 80 A/Hong Kong/33982/2009 (H9N2) HK-H9N2 1/H9 + − 20 > 80 A/Env/JX-PYH/376/2014 (H11N3) JX-H11N3 1/H1b + − > 80 > 80 A/Env/JX-PYH/1102/2014 (H12N5) JX-H12N5 1/H9 + − > 80 > 80 A/Env/QHH/166/2012 (H13N8) QH-H13N8 1/H1b − − > 80 > 80 A/Hong Kong/01/1968 (H3N2) HK-H3N2 2/H3 − − > 160 > 160 A/Env/Guangdong/15188/2015 (H4N2) HN-H4N2 2/H3 − − > 160 > 160 RG A/Anhui/1/2013 (H7N9) AH-H7N9 2/H7 − − > 160 > 160 RG A/Guangdong/17SF003/2016 (H7N9) GD-H7N9 2/H7 − − > 160 > 160 RG A/Jiangxi/346/2013 (H10N8) JX-H10N8 2/H7 − − > 160 > 160 Note. ‘+’ denotes binding. ‘−’ denotes no binding. RG denotes a recombinant virus bearing the HA and NA from the corresponding virus and the internal genes of A/puerto Rico/8/1934. Table 2. The gene characteristics of 2-12D5 and 3-32G7.1
Ab clone V gene Somatic mutation rate in V gene (% nucleotide) D gene J gene Somatic mutation rate in J gene (% nucleotide) Amino acid No. of CDR 2-12D5 VH1-69*01 5.90 DH2-15*01 JH4*02 8.33 8.8.15 Vλ4-69*01 0.34 − Jλ3*02 0.00 7.7.10 3-32G7.1 VH1-2*02 2.78 DH3-9*01 JH5*01 23.53 8.8.18 Vκ3-20*01 0.35 − Jκ4*01 7.89 7.3.9 Note. The VH, D, and JH sequences were compared by using the IMGT database. ‘−’ denotes the gene without the segment. Table 3. The VDJ and VJ gene and somatic hypermutations of VH1-69-, VH1-2-, and D3-9-encoded Abs against influenza virus
mAb VH DH JH SHMs in VH (aa) VL JL SHMs in VL (aa) Access #/reference CR6261 1-69*01 2-2*01 6*04 15 Vκ1-51*01 2*01/03*01 7 GenBank: HI919029.1/
GenBank: HI919031.1F10 1-69*01 NA NA 13 Vκ10-54*01 3*02 5 PDB: 3FKU_T MAb1.12 1-69*06 NA 6 21 − − 14 [16] CR9114 1-69*06 3-10*01 6*02 17 Vκ1-44*01 7-*01 11 GenBank:JX213639.1/
GenBank: JX213640.127F3 1-69*06 NA NA 14 − − − [15] 2-12D5 1-69*01 2-15*01 4*02 12 Vλ4-69*01 3*02 1 The study CH65 1-2 1-1 6 12 Vκ3-21 2 − [6] 3-32G7.1 1-2 3-9*01 5*-01 3 Vκ3-20 4*01 2 The study FI6v3 3-30*03F/
3-30*18/
3-30-5*013-9*01 4*02 8 Vκ4-1*01 J1*01 32 GenBank: JN234435.1/
GenBank: JN234444.1S9-3-37 1-18 3-9 − − Vκ2-24 − − [13] 31.b.09 1-18 3-9 − − − − − [13] 31.d.01 3-30 3-9 − − − − − [13] 54.e.01 7-4-1 3-9 − − − − − [13] 56.ND.12 3-23 3-9 − − − − − [13] 56.h.01 3-23 3-9 − − − − − [13] 02-1A05 1-2 3-9 4 − Vκ3-20 − − [31] 04-1B12 1-2 3-9 4 − Vλ2-14 − − [31] 27-1C08 1-2 3-9 6 − Vλ2-14 − − [31] Note. ‘−’ denotes not available. Table 4. The molecular signature of VH1-69- and VH1-2-encoded Abs
Abs/gemline gene CDR1-IMGT FR2-IMGT CDR2-IMGT FR3-IMGT CDR3-IMGT 26------30--33 34----------40----------------------50 5152a----------57 58-60------------------70------------------80--82abc---------------90-92 93------100----------102 VH1-69*01 GGTFSSYA ISWVRQAPGQGLEWMGG IIPIFGTA NYAQKFQGRVTITADESTSTAYMELSSLRSEDTAVYYC AR F10 EV....F. ..............L.. .S.M...P ...............Q..R....D.R............ ..SP---SYICSGGTCVFDH CR6261 ..P.R... ...........P..... .......T K..P...........DFAG.V.............M... .KHM---GYQVRET---MDV 2-12D5 .V..R.F. F................ .TA...VV ..........................G.T......... .KEAG---YYS--GRSHFDD 27C3 ....GN.. .N............... .S....PP K...........S..I.S.....D....S.D....... ..KLEPPYYFYSY----MDV CR9114 ...SNN.. ............D.... .S....ST A...........S..IFSN......N..T.......F. ..--HGNYYYYSG----MDV VH1-69*06 ........ ................. ........ ...............K...................... .. VH1-2*02 GYTFTGYY MHWVRQAPGQGLEWMGW INPNSGGT NYAQKFQGWVTMTRDTSISTAYMELSRLRSDDTAVYYC AR 3-32G7.1 GYTFTGYY IHWVRQAPGQGLEWMGW INPNSGGT NYAQKFQARVTMTKDTSISTAYMELSRLRSDDTAVYYC ARGDHVLRYFDWQSSEPS CH65 GYTFTDYH INWVRQAPGQGLEWMGW IHPNSGDT NYAQKFQGWVTMTRDTAISTAYMEVNGLKSDDTAVYYC ARGGLEPRSVDYYYYGMDV VRC01 GYEFIDCT LNWIRLAPGKRPEWMGW LKPRGGAV NYARPLQGRVTMTRDVYSDTAFLELRSLTVDDTAVYFC TRGKNCDYNWDFEH FI6 GFTFSTA MHWVRQAPGRGLEWVAV ISYDGNYK YYADSVKGRFSISRDNSNNTLHLEMNTLRTEDTALYYC AKDSQLRSLLYEWLSQGYFDYW S9-3-37 − − − − LGYFDWL 31.b.09 − − − − HILTGF 31.d.01 − − − − DSLFQLRYFDWLLGDAFNI 54.e.01 − − − − DPLQLILRYFDWLFKPLDY 56.ND.12 − − − − GAHLGRIELRYFEWLRKDYYYYGMDV 56.h.01 − − − − GAHLGRIELRYFEWLRKDYYYYGMDV 02-1A05 − − − − CARDSGMRYFDWLSGYFDFW 04-1B12 − − − − CARVGKLQYFDWPAHYFDYW 27-1C08 − − − − CARDKGLRYFDWLSGGMDVW Note. The amino acid sequences of VH1-69-encoded Abs, CR6261, F10, CR9114, and 27F3 (green background), VH1-2-encoded CH65 and VRC01 (white background), D3-9-encoded S9-3-37, 31.b.09, 31.d.01, 54.e.01, 56.ND.12, 56.h.01, 02-1A05, 04-1B12, and 27-1C08 (grey background) as well as 2-12D5 and 3-32G7.1 in the study, which were highlighted with red color were aligned. Residue positions are according to Kabat numbering. Dots indicate identical residues. The CDRs defined by IMGT were marked. The sites previously reported to mediate the binding to the HA glycoprotein of influenza virus are highlighted by red color and the amino acids in the corresponding residues in this study are highlighted in green color. '−' denotes not available. The sequence of D gene in CDR3 of Abs, 3-32G7.1, FI6, S9-3-37, 31.b.09, 31.d.01, 54.e.01, 56.ND.12, 56.h.01, 02-1A05, 04-1B12, and 27-1C08, was labeled with underline. The F/L at the residue 100e interacts with the HA stem, which are highlighted by blue color. Table 5. IC50 neutralization titers against RG A/Beijing/1/2003 (H5N1, Clade 7) and its mutants
Escape mutation Selected by the Abs HA domain Mutation occurance among clones (P10) Mutation occurance among clones (P15) IC50 neutralization titers (μg/mL) 2-12D5 3-32G7.1 I45 no − 5/5 5/5 5 2.5 I45M 2-12D5 Helix A of HA2 2/32 5/5 > 160 > 160 I45T 3-32G7.1 Helix A of HA2 31/32 5/5 > 160 > 160 Note. ‘−’denotes not available. -
[1] Nobusawa E, Aoyama T, Kato H, et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology, 1991; 182, 475−85. doi: 10.1016/0042-6822(91)90588-3 [2] Bui CM, Chughtai AA, Adam DC, et al. An overview of the epidemiology and emergence of influenza A infection in humans over time. Arch Public Health, 2017; 75, 15. doi: 10.1186/s13690-017-0182-z [3] Wang X, Fang S, Lu X, et al. Seroprevalence to avian influenza A(H7N9) virus among poultry workers and the general population in southern China: a longitudinal study. Clin Infect Dis, 2014; 59, e76−83. doi: 10.1093/cid/ciu399 [4] Gu J, Xie Z, Gao Z, et al. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet, 2007; 370, 1137−45. doi: 10.1016/S0140-6736(07)61515-3 [5] Schmidt AG, Therkelsen MD, Stewart S, et al. Viral receptor-binding site antibodies with diverse germline origins. Cell, 2015; 161, 1026−34. doi: 10.1016/j.cell.2015.04.028 [6] Whittle JR, Zhang R, Khurana S, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA, 2011; 108, 14216−21. doi: 10.1073/pnas.1111497108 [7] Raymond DD, Bajic G, Ferdman J, et al. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci USA, 2018; 115, 168−73. doi: 10.1073/pnas.1715471115 [8] Throsby M, Van Den Brink E, Jongeneelen M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One, 2008; 3, e3942. doi: 10.1371/journal.pone.0003942 [9] Sui J, Hwang WC, Perez S, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nal Struct Mol Biol, 2009; 16, 265−73. [10] Bangaru S, Zhang H, Gilchuk IM, et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat Commun, 2018; 9, 2669. doi: 10.1038/s41467-018-04704-9 [11] Joyce MG, Wheatley AK, Thomas PV, et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell, 2016; 166, 609−23. doi: 10.1016/j.cell.2016.06.043 [12] Kallewaard NL, Corti D, Collins PJ, et al. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell, 2016; 166, 596−608. doi: 10.1016/j.cell.2016.05.073 [13] Wu NC, Yamayoshi S, Ito M, et al. Recurring and adaptable binding motifs in broadly neutralizing antibodies to influenza virus are encoded on the D3-9 segment of the Ig gene. Cell Host Microbe, 2018; 24, 569−78 e4. doi: 10.1016/j.chom.2018.09.010 [14] Dreyfus C, Laursen NS, Kwaks T, et al. Highly conserved protective epitopes on influenza B viruses. Science, 2012; 337, 1343−8. doi: 10.1126/science.1222908 [15] Lang S, Xie J, Zhu X, et al. Antibody 27F3 broadly targets influenza A group 1 and 2 hemagglutinins through a further variation in VH1-69 antibody orientation on the HA stem. Cell reports, 2017; 20, 2935−43. doi: 10.1016/j.celrep.2017.08.084 [16] Wyrzucki A, Bianchi M, Kohler I, et al. Heterosubtypic Antibodies to Influenza A Virus Have Limited Activity against Cell-Bound Virus but Are Not Impaired by Strain-Specific Serum Antibodies. Journal of Virology, 2015; 89, 3136−44. [17] Avnir Y, Tallarico AS, Zhu Q, et al. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS Pathog, 2014; 10, e1004103. doi: 10.1371/journal.ppat.1004103 [18] Avnir Y, Watson CT, Glanville J, et al. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci Rep, 2016; 6, 20842. doi: 10.1038/srep20842 [19] Pappas L, Foglierini M, Piccoli L, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature, 2014; 516, 418−22. doi: 10.1038/nature13764 [20] Lingwood D, Mctamney PM, Yassine HM, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature, 2012; 489, 566−70. doi: 10.1038/nature11371 [21] Hu H, Voss J, Zhang G, et al. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J Virol , 2012; 86, 2978−89. doi: 10.1128/JVI.06665-11 [22] Qian M, Hu H, Zuo T, et al. Unraveling of a neutralization mechanism by two human antibodies against conserved epitopes in the globular head of H5 hemagglutinin. J Virol, 2013; 87, 3571−7. doi: 10.1128/JVI.01292-12 [23] Simmons CP, Bernasconi NL, Suguitan AL, et al. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med, 2007; 4, e178. doi: 10.1371/journal.pmed.0040178 [24] Kashyap AK, Steel J, Oner AF, et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A, 2008; 105, 5986−91. doi: 10.1073/pnas.0801367105 [25] Tiller T. Single B cell antibody technologies. New biotechnology, 2011; 28, 453−7. doi: 10.1016/j.nbt.2011.03.014 [26] Tiller T, Meffre E, Yurasov S, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods, 2008; 329, 112−24. doi: 10.1016/j.jim.2007.09.017 [27] Tiller T, Schuster I, Deppe D, et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. MAbs, 2013; 5, 445−70. doi: 10.4161/mabs.24218 [28] Du N, Zhou J, Lin X, et al. Differential activation of NK cells by influenza A pseudotype H5N1 and 1918 and 2009 pandemic H1N1 viruses. J Virol, 2010; 84, 7822−31. doi: 10.1128/JVI.00069-10 [29] Jegaskanda S, Vandenberg K, Laurie KL, et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity in intravenous immunoglobulin as a potential therapeutic against emerging influenza viruses. J Infect Dis, 2014; 210, 1811−22. doi: 10.1093/infdis/jiu334 [30] Scheid JF, Mouquet H, Ueberheide B, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science, 2011; 333, 1633−7. doi: 10.1126/science.1207227 [31] Andrews SF, Joyce MG, Chambers MJ, et al. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol, 2017; 2. [32] Marcatili P, Olimpieri PP, Chailyan A, et al. Antibody modeling using the prediction of immunoglobulin structure (PIGS) web server [corrected]. Nat Protoc, 2014; 9, 2771−83. doi: 10.1038/nprot.2014.189 [33] Pierce BG, Wiehe K, Hwang H, et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics, 2014; 30, 1771−3. doi: 10.1093/bioinformatics/btu097 [34] Anderson CS, Ortega S, Chaves FA, et al. Natural and directed antigenic drift of the H1 influenza virus hemagglutinin stalk domain. Sci Rep, 2017; 7, 14614. doi: 10.1038/s41598-017-14931-7 [35] Prachanronarong KL, Canale AS, Liu P, et al. Mutations in influenza A virus neuraminidase and hemagglutinin confer resistance against a broadly neutralizing hemagglutinin stem antibody. J Virol, 2019; 93. [36] Yamayoshi S, Yasuhara A, Ito M, et al. Differences in the ease with which mutant viruses escape from human monoclonal antibodies against the HA stem of influenza A virus. J Clin Virol, 2018; 108, 105−11. doi: 10.1016/j.jcv.2018.09.016 [37] Ekiert DC, Bhabha G, Elsliger MA, et al. Antibody recognition of a highly conserved influenza virus epitope. Science, 2009; 324, 246−51. doi: 10.1126/science.1171491 [38] Chen YQ, Lan LY, Huang M, et al. Hemagglutinin stalk-reactive antibodies interfere with influenza virus neuraminidase activity by steric hindrance. J Virol, 2019; 93. [39] Dilillo DJ, Palese P, Wilson PC, et al. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest, 2016; 126, 605−10. doi: 10.1172/JCI84428 [40] Kosik I, Angeletti D, Gibbs JS, et al. Neuraminidase inhibition contributes to influenza A virus neutralization by anti-hemagglutinin stem antibodies. J Exp Med, 2019; 216, 304−16. [41] Herfst S, Schrauwen EJ, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science, 2012; 336, 1534−41. doi: 10.1126/science.1213362 [42] Mair CM, Ludwig K, Herrmann A, et al. Receptor binding and pH stability - how influenza A virus hemagglutinin affects host-specific virus infection. Biochim Biophys Acta, 2014; 1838, 1153−68. doi: 10.1016/j.bbamem.2013.10.004 [43] Angeletti D, Kosik I, Santos JJS, et al. Outflanking immunodominance to target subdominant broadly neutralizing epitopes. Proc Natl Acad Sci USA, 2019; 116, 13474−9. doi: 10.1073/pnas.1816300116 -




 下载:
下载:



 Quick Links
Quick Links