-
Human methylenetetrahydrofolate reductase (MTHFR), an essential enzyme in folate metabolism, catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. Several polymorphisms in the MTHFR gene have been found to be associated with reduced enzyme activity and related to folic acid intake in relevant populations [1]. The most clinically significant polymorphism is C677T (also known as rs1801133), which involves a C to T transition at nucleotide position 677 in exon 4 of the MTHFR gene [2]. Individuals can have one of three genotypes at this locus: wild-type homozygous CC, heterozygous CT, and mutant homozygous TT. The enzymatic activity of MTHFR is decreased to 65% in individuals with the heterozygous genotype and to 30% in those with the mutant homozygous genotype [2]. Therefore, the genotypic identification of MTHFR polymorphisms in the clinical setting is helpful for informing personalized nutrition supplementation or treatment decisions.
Traditional methods for detecting single nucleotide polymorphisms (SNP) include Sanger sequencing and commercial kits [3]. With the rapid advancement of technologies for studying molecular biology, a range of new genotyping methods have been developed, such as the GoldMag nanoparticle-based lateral flow assay, fluorescence-based polymerase chain reaction (PCR) melting curve method, and fluorescence-based PCR-capillary electrophoresis [4-6]. However, existing detection methods typically have some shortcomings. For example, DNA sequencing, the gold-standard assay, is costly and time-consuming, and TaqMan fluorescence probe-based PCR requires a long detection time and high sample quality. In this study, we developed a new, quick, and convenient one-step assay to detect the genotype of the polymorphism at locus position 677 of the MTHFR gene.
Recently, clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) proteins have shown significant potential for rapid and sensitive nucleic acid detection [7]. Cas12 is an endonuclease that can accurately bind to and cleave a double-stranded DNA target under the guidance of a single-stranded guide RNA (sgRNA) [8]. In many assays, the CRISPR/Cas12 system is used to identify SNP mutations, such as those in severe acute respiratory syndrome coronavirus 2 and drug-resistant Salmonella enterica [9,10]. CRISPR/Cas12b is the third most efficient genome editing system after CRISPR/Cas9 and CRISPR/Cas12a. Compared with these other two CRISPR/Cas systems, the CRISPR/Cas12b-based method does not tolerate any mismatch between the sgRNA and target DNA sequence and therefore has higher specificity and accuracy when used to detect DNA polymorphisms [11]. The genotyping process based on CRISPR/Cas12b is initiated by the specific recognition of the target DNA by the sgRNA. The trans-cleavage activity of Cas12b efficiently cleaves nonspecific single-stranded DNAs (ssDNAs), including those modified with fluorophore and quencher (FQ) groups. By cleaving the FQ-labeled ssDNA, the target gene can be detectable as a fluorescence signal. Although Cas12b has been used in some studies for SNP detection [10], it has hardly been used in the diagnostic field. In this study, we aimed to establish a CRISPR/Cas12b-based system for genotyping the polymorphism at locus position 677 of the MTHFR gene.
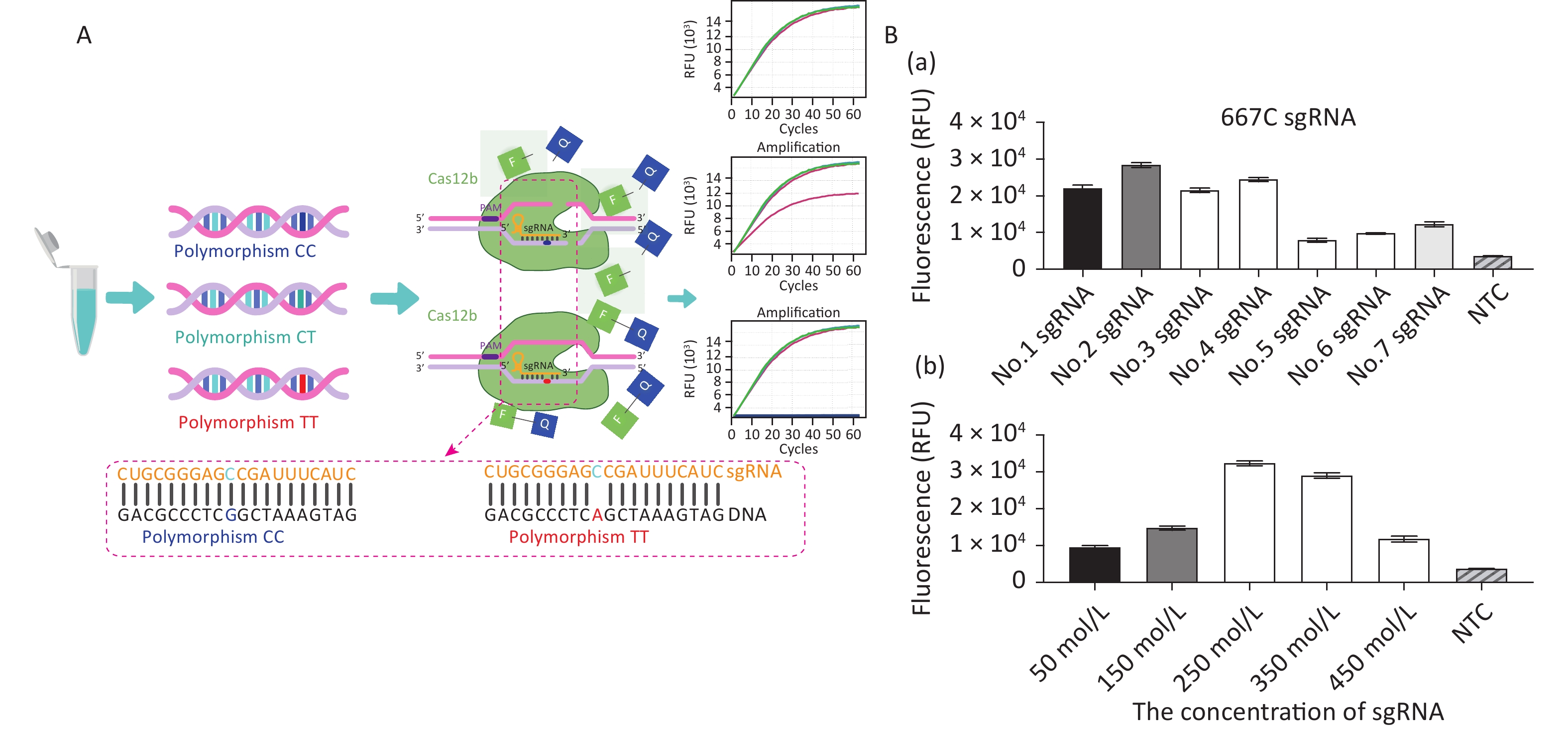
The principles of CRISPR/Cas12b-mediated MTFHR C677T genotyping are illustrated in Figure 1A. The assay system consists of the Cas12b protein from Alicyclobacillus acidiphilus (AapCas12b) (BestEnzymes Biotech Co., Ltd., Lianyungang, China), sgRNA targeting the C677T SNP, and an FQ reporter probe, all of which can be premixed in a tube. To differentiate 677C from 677T, a 677C-specific sgRNA sequence was designed (Supplementary Table S1, available in www.besjournal.com). A DNA sample is mixed with the AapCas12b protein, FQ reporter probe, and sgRNA, after which the fluorescence signal is recorded. If the polymorphism is present, the specific Cas12b–sgRNA will bind to the target DNA to form a ternary complex, which in turn activates the trans-cleavage activity of Cas12b and subsequent cleavage of the FQ reporter probe to produce a fluorescent signal for detection.
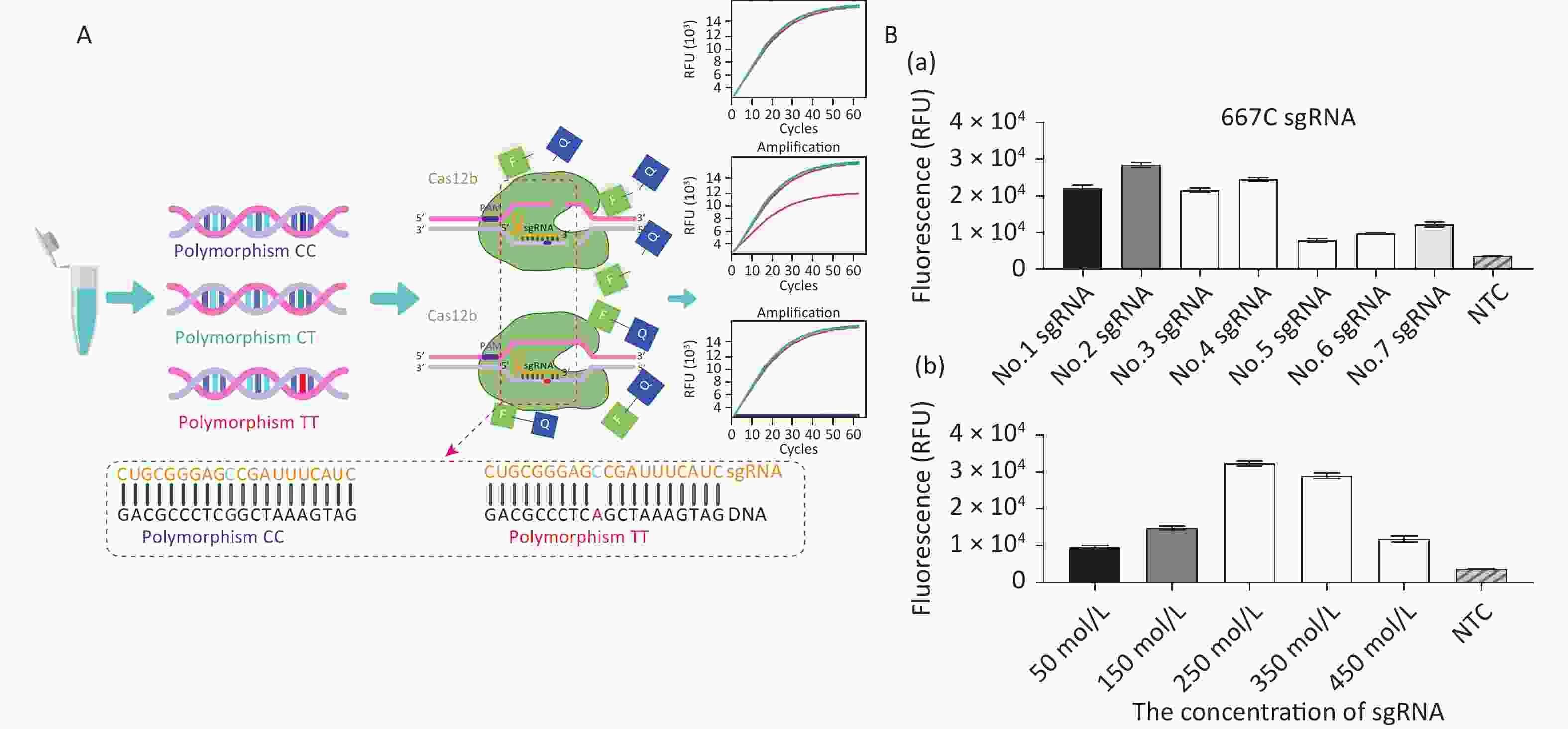
Figure 1. Establishment of the CRISPR/Cas12b-mediated assay for genotyping the MTHFR C677T single nucleotide polymorphism locus. (A) Schematic diagram of the CRISPR/Cas12b-based detection process. The left panel shows the three polymorphic alleles: CC, CT, and TT. The lower panel shows the partial 677C sgRNA sequence that targets the MTHFR gene sequence. Base pairs are represented as vertical lines. In the presence of target DNA containing the polymorphism 677CC, the 677C sgRNA binds to the target sequence to form a ternary complex, which in turn activates the trans-cleavage activity of Cas12b and subsequently cleaves the FQ reporter to produce the fluorescence signal for detection. The 677C sgRNA does not bind with DNA containing the polymorphic T allele; therefore, the trans-cleavage activity of Cas12b is not activated and no fluorescence signal is produced. (B) Optimization of the sgRNA conditions, including the screening of seven candidate sgRNAs and their optimal concentration. The goal of optimization was to screen the reaction system with the highest CRISPR/Cas12b efficiency. For this detection with different sgRNAs, the 20 μL reaction mixture contained AapCas12b protein (final concentration 600 nmol/L), 1× Cas reaction buffer, RNase inhibitor (final concentration 1 U/μL), and 104 copies of standard MTHFR-677C-DNA. The bar graphs represent the different fluorescence values at the end point of 30 min of continuous signal acquisition at 48 °C. Error bars represent the results from three independent experiments.
For double-stranded DNA cleavage, the CRISPR/Cas12b endonuclease requires a protospacer-adjacent motif (PAM) sequence-TTN-which should be located in an appropriate position as its distance from the start site of the target sequence can influence the recognition efficiency [12]. We analyzed the genomic DNA (gDNA) sequence of MTFHR and screened for the PAM sequence upstream of the specific SNP site. Seven sgRNAs were designed and prepared to select the optimal one for highest cutting efficiency. Following the recommendations of the AapCas12b product-accompanying manual (BestEnzymes Biotech Co.), the 20 μL reaction mixture contained 1× Cas reaction buffer, 250 nmol/L AapCas12b protein, 250 nmol/L sgRNA, 20 U RNase inhibitor, and 104 copies of target DNA. During the method establishment and optimization period, we used the MTHFR-677C-DNA standard as the target DNA. This standard is a previously constructed plasmid containing the MTHFR C677T polymorphism fragment [13]. The reaction mixtures were then quickly transferred to the CFX Connect Real-Time System (Bio-Rad, USA) and incubated at 48 °C. Fluorescence signals were detected every 30 s for 30 min. The results showed that sgRNA No. 2, which was specific for 677CC, had the highest fluorescence signal and the best detection efficiency (Figure 1B-a). The detection system was further optimized by performing the assay with a range of sgRNA concentrations (50–450 nmol/L), whereupon 250 nmol/L sgRNA was finalized as the concentration for optimal reaction detection (Figure 1B-b).
To prove the principle of the CRISPR/Cas12b-mediated assay and its sensitivity in distinguishing polymorphisms, 104 copies of standard plasmids containing genotype CC or TT, as well as a 1:1 molar ratio mix of the two, were tested using the assay. As evident from the fluorescence signal curve in Figure 2A, the system could clearly distinguish between the three genotype samples. There was no increasing fluorescence signal detected in the no-template control (NTC) group; a weak signal increase with a relative fluorescence unit (RFU) value of less than 4.5 × 103 for genotype TT; and a significantly increasing fluorescence signal with an RFU value of up to 3.2 × 104 for genotype CC. For genotype CT, the maximum signal value (1.7 × 104) fell in between those of genotypes CC and TT. These results suggest that the Cas12b-mediated assay can distinguish MTHFR polymorphisms at locus position 677.
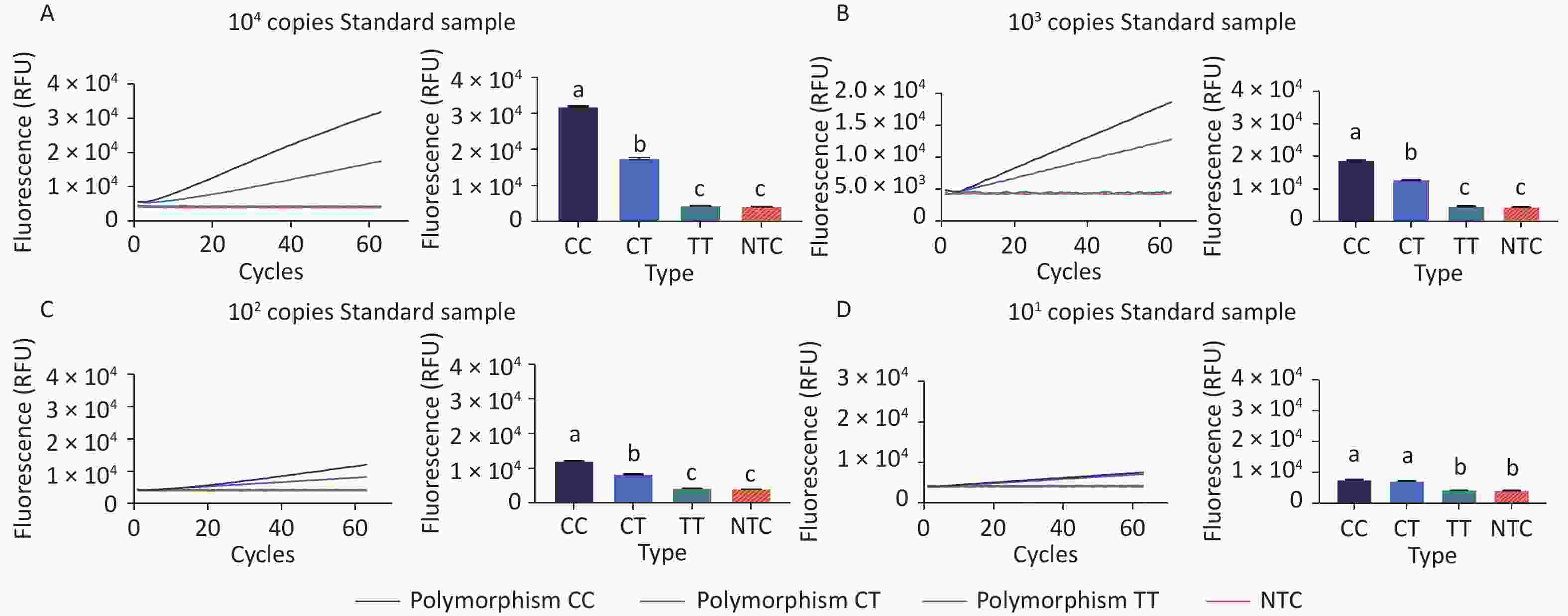
Figure 2. Proof of the principle behind the CRISPR/Cas12b-based assay and detection sensitivity test. Fluorescence curves for the three genotypes detected using this assay are shown. The fluorescence curves were from samples with 104 (A), 103 (B), 102 (C), and 101 copies (D), respectively. The bar graph represents the different fluorescence values at the end point of the 30 min continuous signal acquisition period. These experiments were independently repeated three times. Different letters indicate significant differences among the treatments. Multiple comparisons of the means were performed using Tukey’s test at a significance level of 0.05.
The sensitivity of this assay was determined using 10-fold serial dilutions of the fragments at 103, 102, and 10 copies. Standard plasmids containing different copies of the CC, TT, and CT genotypes were tested using the assay. As shown in Figure 2D, the samples with 10 copies per reaction still had fluorescence levels that were distinguishable from that of the NTC group at the signal detection time point of 30 min. However, the fluorescence signals from genotype CC could not be distinguished from those of genotype CT at the time point of 30 min. As shown in Figure 2C, distinct genotyping of all three polymorphisms was achieved for samples with concentrations as low as 102 copies per reaction.
To analyze the accuracy of our new CRISPR/Cas12b-mediated genotyping assay, we compared its results with data from a fluorescence probe-based PCR assay, which is one of the most commonly used methods in clinical practice. The PCR method was performed according to the instructions of the Human MTHFR Gene Polymorphism Detection Kit (YZY Medical Technology Co., Ltd., Wuhan, China), which is widely used for MTHFR C677T polymorphism genotyping in clinical practice. Eight human gDNA samples from the Second People’s Hospital of Lianyungang and standard plasmids (representing 3 polymorphism genotypes) containing 104 copies of MTHFR were simultaneously analyzed and evaluated by blind analysis. Our new assay showed a high level of concordance with the reference PCR method, with an overall agreement of 100% (Figure 3 and Table 1), indicating its suitability for use on clinical specimens.
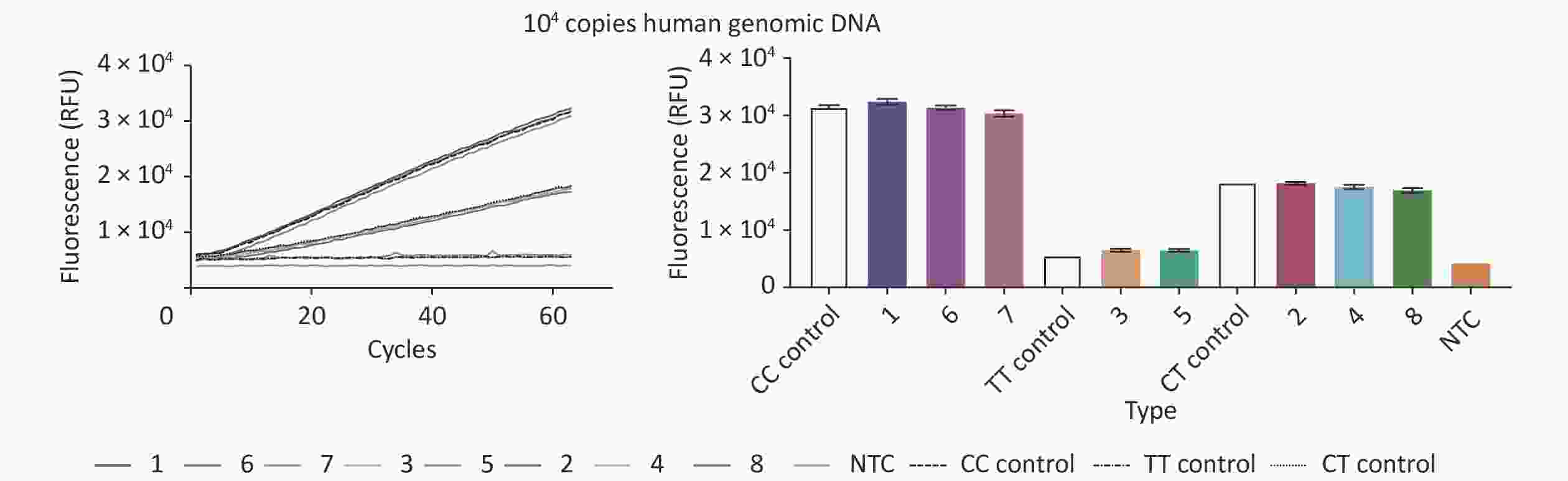
Figure 3. Validation of the CRISPR/Cas12b-mediated genotyping assay using eight human genomic DNA samples. The CRISPR/Cas12b-based assay was used to detect the MTHFR C677T polymorphism in eight human genomic DNA samples. The fluorescence curves of the eight samples and standard plasmids (representing 3 polymorphism genotypes) containing 104 copies of MTHFR are shown. The bar graph represents the different fluorescence values at the end point of the 30 min continuous signal acquisition period. These experiments were independently repeated three times.
Table 1. Analysis of MTHFR C677T polymorphisms in human genomic DNA samples
Sample No. Genotyping Results Fluorescence Probe PCR 667 CRISPR/Cas12b 1 CC CC 2 CT CT 3 TT TT 4 CT CT 5 TT TT 6 CC CC 7 CC CC 8 CT CT In this study, we have established an accurate, rapid, and convenient method to detect the genotype of the polymorphism at locus position 677 of the MTHFR gene. Our assay, which takes approximate 30 min to complete, requires only a single step of adding 1 μL of human gDNA sample (concentration > 10 ng/μL) to a premixed system containing the AapCas12b protein, C677T SNP site-targeting sgRNA, and FQ reporter probe and waiting for the emission of the fluorescence signal. When applied to nucleic acid detection, the CRISPR/Cas system is typically combined with an amplification procedure to increase the detection limit. The sensitivity of our new assay was sufficient to genotype the MTHFR C677T polymorphism in human gDNA samples. The concentration of human gDNA extracted by commercial kits is generally greater than 10 ng/µL, which is equivalent to approximately 3 × 103 copies/µL by calculation; that is, 1 µL of human gDNA contains 3 × 103 copies of the MTHFR gene, which is much higher than the sensitivity of the method (102 copies per reaction). Consequently, our assay does not require an amplification step before Cas12b-mediated genotyping. As mentioned above, Cas12b cannot tolerate any mismatch between the sgRNA and target DNA sequences, resulting in higher specificity and accuracy when used for DNA polymorphism detection [11]. Compared with traditional methods, our assay does not require complicated hands-on experiments, complex instruments, or long time courses. Considering that different types of quantitative PCR instruments or microplate readers may lead to differences in the acquisition of fluorescence signals, we can overcome this problem with the help of standard test materials; that is, plasmids containing CC, TT, or CT polymorphism genotypes. These plasmids were labeled with the concentration and copy number information. Using the same number of copies of human gDNA or standard plasmids in the reaction system, the fluorescence signals or curves of the clinical samples can be compared with those of the standards to ascertain the genotype. Therefore, the method we established has a wide range of usage scenarios.
In summary, we have established a simple and accurate one-step assay based on the CRISPR/Cas12b system for MTHFR C677T genotyping in 30 min. Our assay provides an alternative tool for a more accurate and reliable diagnosis of folate-associated metabolic disorders that warrant the development of personalized medicine. In future, we will explore the effectiveness of the assay in clinical trials.
-
Table S1. Sequences of all 7 sgRNAs
sgRNA Sequence (5’–3’) MTHFR-sgRNA1 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC GCGGGAGCCGAUUUCAUCAU MTHFR-sgRNA2 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC CUGCGGGAGCCGAUUUCAUC MTHFR-sgRNA3 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC GUCUGCGGGAGCCGAUUUCA MTHFR-sgRNA4 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC GGAGCCGAUUUCAUCAUCAC MTHFR-sgRNA5 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC UCUGCGGGAGCCGAUUUCAU MTHFR-sgRNA6 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC GUGUCUGCGGGAGCCGAUUU MTHFR-sgRNA7 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC AGCCGAUUUCAUCAUCACGC
doi: 10.3967/bes2023.070
A Rapid and Accurate CRISPR/Cas12b-Mediated Genotyping Assay for the Methylenetetrahydrofolate Reductase Gene Polymorphism C677T
-
&These authors contributed equally to this work.
注释: -
Figure 1. Establishment of the CRISPR/Cas12b-mediated assay for genotyping the MTHFR C677T single nucleotide polymorphism locus. (A) Schematic diagram of the CRISPR/Cas12b-based detection process. The left panel shows the three polymorphic alleles: CC, CT, and TT. The lower panel shows the partial 677C sgRNA sequence that targets the MTHFR gene sequence. Base pairs are represented as vertical lines. In the presence of target DNA containing the polymorphism 677CC, the 677C sgRNA binds to the target sequence to form a ternary complex, which in turn activates the trans-cleavage activity of Cas12b and subsequently cleaves the FQ reporter to produce the fluorescence signal for detection. The 677C sgRNA does not bind with DNA containing the polymorphic T allele; therefore, the trans-cleavage activity of Cas12b is not activated and no fluorescence signal is produced. (B) Optimization of the sgRNA conditions, including the screening of seven candidate sgRNAs and their optimal concentration. The goal of optimization was to screen the reaction system with the highest CRISPR/Cas12b efficiency. For this detection with different sgRNAs, the 20 μL reaction mixture contained AapCas12b protein (final concentration 600 nmol/L), 1× Cas reaction buffer, RNase inhibitor (final concentration 1 U/μL), and 104 copies of standard MTHFR-677C-DNA. The bar graphs represent the different fluorescence values at the end point of 30 min of continuous signal acquisition at 48 °C. Error bars represent the results from three independent experiments.
Figure 2. Proof of the principle behind the CRISPR/Cas12b-based assay and detection sensitivity test. Fluorescence curves for the three genotypes detected using this assay are shown. The fluorescence curves were from samples with 104 (A), 103 (B), 102 (C), and 101 copies (D), respectively. The bar graph represents the different fluorescence values at the end point of the 30 min continuous signal acquisition period. These experiments were independently repeated three times. Different letters indicate significant differences among the treatments. Multiple comparisons of the means were performed using Tukey’s test at a significance level of 0.05.
Figure 3. Validation of the CRISPR/Cas12b-mediated genotyping assay using eight human genomic DNA samples. The CRISPR/Cas12b-based assay was used to detect the MTHFR C677T polymorphism in eight human genomic DNA samples. The fluorescence curves of the eight samples and standard plasmids (representing 3 polymorphism genotypes) containing 104 copies of MTHFR are shown. The bar graph represents the different fluorescence values at the end point of the 30 min continuous signal acquisition period. These experiments were independently repeated three times.
Table 1. Analysis of MTHFR C677T polymorphisms in human genomic DNA samples
Sample No. Genotyping Results Fluorescence Probe PCR 667 CRISPR/Cas12b 1 CC CC 2 CT CT 3 TT TT 4 CT CT 5 TT TT 6 CC CC 7 CC CC 8 CT CT S1. Sequences of all 7 sgRNAs
sgRNA Sequence (5’–3’) MTHFR-sgRNA1 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC GCGGGAGCCGAUUUCAUCAU MTHFR-sgRNA2 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC CUGCGGGAGCCGAUUUCAUC MTHFR-sgRNA3 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC GUCUGCGGGAGCCGAUUUCA MTHFR-sgRNA4 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC GGAGCCGAUUUCAUCAUCAC MTHFR-sgRNA5 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC UCUGCGGGAGCCGAUUUCAU MTHFR-sgRNA6 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC GUGUCUGCGGGAGCCGAUUU MTHFR-sgRNA7 GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC AGCCGAUUUCAUCAUCACGC -
[1] Khan IA, Shaik NA, Kamineni V, et al. Evaluation of gestational diabetes mellitus risk in south indian women based on MTHFR (C677T) and FVL (G1691A) mutations. Front Pediatr, 2015; 3, 34. [2] Liew SC, Gupta ED. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: epidemiology, metabolism and the associated diseases. Eur J Med Genet, 2015; 58, 1−10. doi: 10.1016/j.ejmg.2014.10.004 [3] Kim S, Misra A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng, 2007; 9, 289−320. doi: 10.1146/annurev.bioeng.9.060906.152037 [4] Chen KY, Xu JX, Wang MM, et al. Single probe PCR melting curve analysis MTHFR C677T SNP sites. Anal Biochem, 2021; 619, 114102. doi: 10.1016/j.ab.2021.114102 [5] Cheng HL, Chiou SS, Liao YM, et al. Genotyping of two single nucleotide polymorphisms in 5, 10-methylenetetrahydrofolate reductase by multiplex polymerase chain reaction and capillary electrophoresis. J Chromatogr A, 2011; 1218, 2114−20. doi: 10.1016/j.chroma.2010.08.080 [6] Hui WL, Zhang SN, Zhang C, et al. A novel lateral flow assay based on GoldMag nanoparticles and its clinical applications for genotyping of MTHFR C677T polymorphisms. Nanoscale, 2016; 8, 3579−87. doi: 10.1039/C5NR07547E [7] Kellner MJ, Koob JG, Gootenberg JS, et al. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc, 2019; 14, 2986−3012. doi: 10.1038/s41596-019-0210-2 [8] Yang YH, Wang DD, Lü P, et al. Research progress on nucleic acid detection and genome editing of CRISPR/Cas12 system. Mol Biol Rep, 2023; 50, 3723−38. doi: 10.1007/s11033-023-08240-8 [9] Fasching CL, Servellita V, Mckay B, et al. COVID-19 variant detection with a high-fidelity CRISPR-Cas12 enzyme. J Clin Microbiol, 2022; 60, e0026122. doi: 10.1128/jcm.00261-22 [10] Yang H, Yang S, Xia XH, et al. Sensitive detection of a single-nucleotide polymorphism in foodborne pathogens using CRISPR/Cas12a-signaling ARMS-PCR. J Agric Food Chem, 2022; 70, 8451−7. doi: 10.1021/acs.jafc.2c03304 [11] Li LX, Li SY, Wu N, et al. HOLMESv2: A CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth Biol, 2019; 8, 2228−37. doi: 10.1021/acssynbio.9b00209 [12] Jain I, Minakhin L, Mekler V, et al. Defining the seed sequence of the Cas12b CRISPR-Cas effector complex. RNA Biol, 2019; 16, 413−22. doi: 10.1080/15476286.2018.1495492 [13] Si XX, Gu QH, Zhao CJ, et al. Highly sensitive genotyping of MTHFR C677T polymorphisms using a novel RPA-LDR-qPCR assay. Acta Biochim Biophys Sin (Shanghai), 2022; 54, 1753−6. doi: 10.3724/abbs.2022151 -
 22416+Supplementary Materials.pdf
22416+Supplementary Materials.pdf

-




 下载:
下载:





 Quick Links
Quick Links