-
Biliary atresia (BA) is one of the most common and enigmatic forms of neonatal cholestasis and is characterized by inflammation and obstruction of extrahepatic bile ducts resulting in biliary cirrhosis, which occurs in 1 in 8,000 to 1 in 18,000 live births[1]. Revealing of the mechanisms underlying BA is crucial for early diagnosis, intervention, and better prognosis. The etiology of BA remains unclear, and currently, noncoding and epigenetic factors, rather than classic genetic alterations, are assumed to be responsible for genetic predisposition to BA.
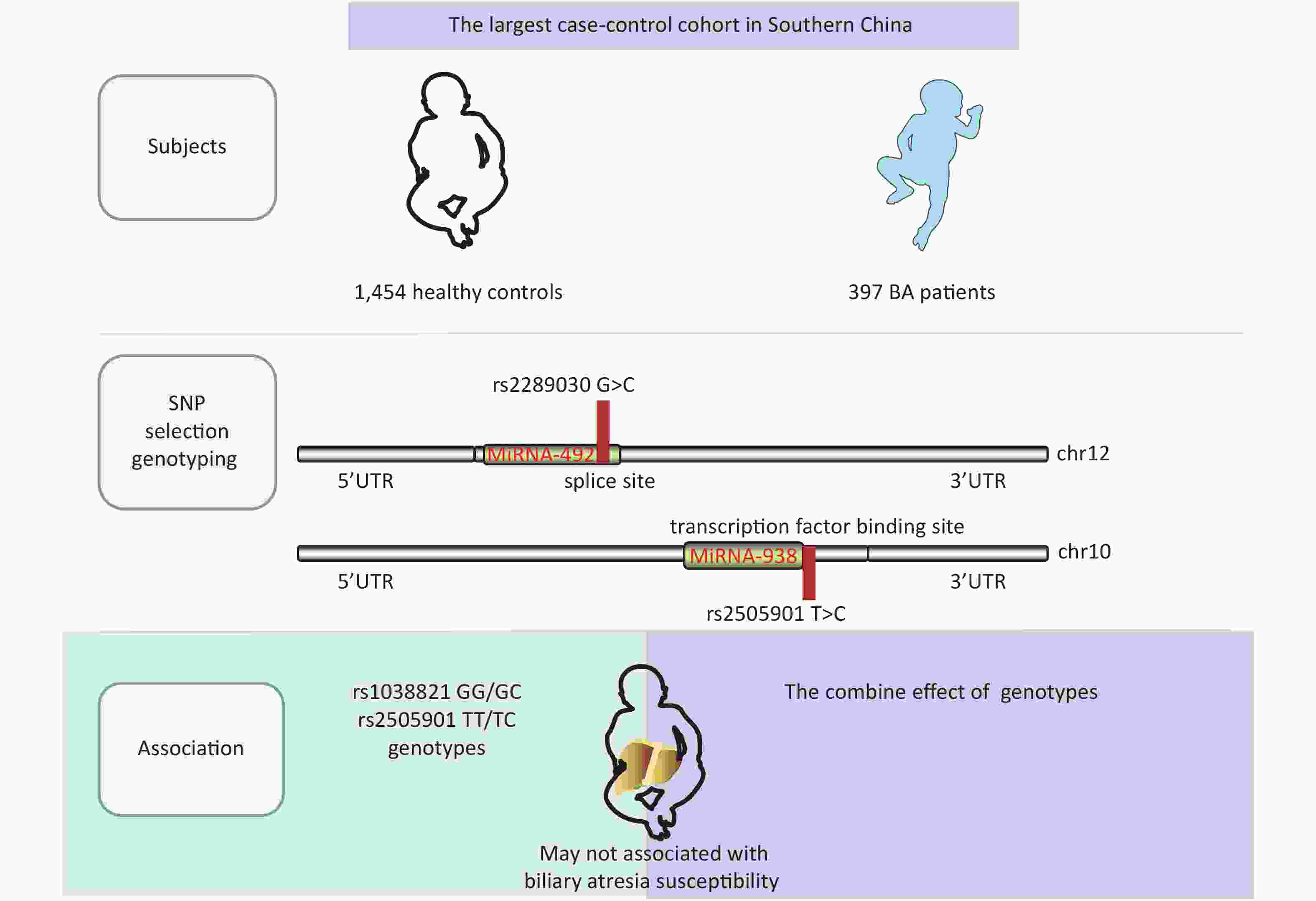
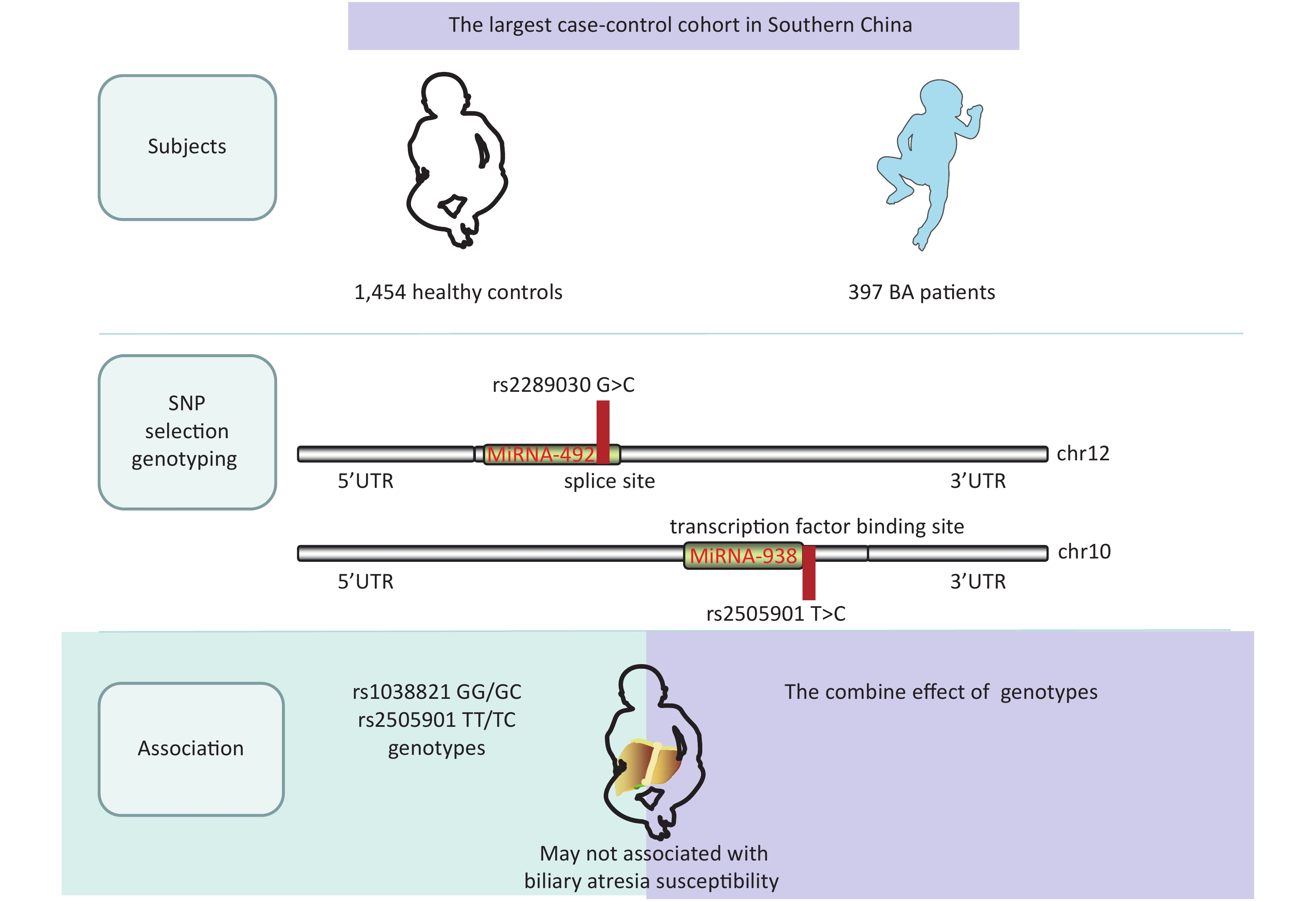
A single nucleotide polymorphism (SNP) is a variation of a single nucleotide in an organism’s genome. MicroRNA (miRNA) plays an important role in various clinical diseases by suppressing or degrading target mRNAs. We investigated miRNA-938 and miRNA-492 in this study, which have been reported to be regulators involved in various gastrointestinal cancers[2]. Some BA cases develop cholangiocarcinoma and hepatocellular carcinoma after a Kasai operation[3], and previous studies suggested that the miRNA-492 G>C rs2289030 polymorphism may affect susceptibility to various liver cancers including hepatocellular carcinoma[4]. Previous studies predominantly investigated genetic polymorphisms in miRNA-499 and miRNA-222, all of which were significantly correlated not only with cancer but also with BA[5, 6]. Considering the significant roles of miRNAs in cancer and BA, it is of great importance to further explore potential associations of miRNA-938 T>C (rs2505901) and miRNA-492 rs2289030 G>C polymorphisms in BA.
We conducted a case-control study with 500 patients and 1,463 controls (Table 1) and with 486 patients and 1,457 controls (Table 2). Patients were enrolled at the Guangzhou Women and Children’s Medical Center from March 2001 to June 2019. BA cases were confirmed using intraoperative cholangiograms, and children with bile duct disconnection were diagnosed with BA. Healthy children who had received a routine physical examination in the same hospital and matched the cases regarding age and sex were randomly selected as controls. The exclusion criteria of controls were as follows: (1) non-southern Chinese; (2) physiological jaundice; and (3) other types of liver diseases or autoimmune disease or children who had undergone liver transplantation. The basic clinical information of this cohort is shown in Supplementary Table S1, available in www.besjournal.com. At the same time, a fully healthy control was randomly selected from the same center to match the patients regarding age, sex, and ethnicity. Informed consent was obtained from the participants or their guardians in accordance with the relevant laws and regulations. This study was approved by the Institutional Review Board of Guangzhou Women and Children’s Medical Center (ethics approval number: #34500). All genotypes and alleles of miRNA SNPs were distributed equally between controls and BA groups (Supplementary Figure S1, available in www.besjournal.com).
Table 1. Association between miRNA-492 rs2289030 G>C polymorphism and biliary atresia susceptibility
Genotype Cases (N = 500) Controls (N = 1,463) Pa Crude OR (95% CI) P Adjusted OR (95% CI)b Pb rs2289030 G>C (HWE = 0.059) GG 270 (54.00) 828 (56.60) 1.00 1.00 GC 199 (39.80) 527 (36.02) 1.16 (0.94−1.43) 0.178 1.14 (0.90−1.45) 0.289 CC 31 (6.20) 108 (7.38) 0.88 (0.58−1.34) 0.554 0.77 (0.48−1.22) 0.261 Additive 0.663 1.04 (0.88−1.22) 0.661 0.99 (0.82−1.19) 0.901 Dominant 230 (46.00) 635 (43.40) 0.313 1.11 (0.91−1.36) 0.313 1.07 (0.85−1.35) 0.565 Recessive 469 (93.80) 1355 (92.62) 0.374 0.83 (0.55−1.25) 0.374 0.73 (0.46−1.14) 0.166 Note. OR, odds ratio; CI, confidence interval, HWE, Hardy-Weinberg equilibrium. a χ2 test for genotype distributions between biliary atresia patients and controls. b Adjusted for age and gender. Table 2. Association between miRNA-938 rs2505901 T>C polymorphism and biliary atresia susceptibility
Genotype Cases (N = 486) Controls (N = 1,457) Pa Crude OR (95% CI) P Adjusted OR (95% CI)b Pb rs2505901 T>C (HWE = 0.714) TT 278 (57.20) 850 (58.34) 1.00 1.00 TC 187 (38.48) 529 (36.31) 1.08 (0.87−1.34) 0.478 1.08 (0.85−1.38) 0.533 CC 21 (4.32) 78 (5.35) 0.82 (0.50−1.36) 0.446 0.88 (0.50−1.54) 0.660 Additive 0.973 1.00 (0.84−1.19) 0.973 1.02 (0.84−1.24) 0.858 Dominant 208 (42.80) 607 (41.66) 0.660 1.05 (0.85−1.29) 0.659 1.06 (0.84−1.34) 0.649 Recessive 465 (95.68) 1379 (94.65) 0.370 0.80 (0.49−1.31) 0.371 0.86 (0.49−1.48) 0.579 Note. OR, odds ratio; CI, confidence interval, HWE, Hardy-Weinberg equilibrium. a χ2 test for genotype distributions between biliary atresia patients and controls. b Adjusted for age and gender. Table S1. The detailed clinical information of the subjects in this study
Variables Cases (n = 506) Controls (n = 1,473) Subjects Age range (Months), n (%) 2.088 ± 1.934 18.61 ± 19.75 ≤ 3 470 (92.89) 521 (35.37) > 3 36 (7.11) 952 (64.63) Gender, n (%) Females 214 (42.29) 506 (34.35) Males 292 (57.71) 967 (65.65) Clinical manifestation, n (%) Cystic biliary atresia (CBA) 44 (8.70) non-CBA 462 (91.30) We used a TIANamp Blood DNA Kit (TianGen Biotech, Beijing, China) to isolate genomic DNA from 2 mL venous blood samples, according to the manufacturer’s instructions. A real-time quantitative PCR instrument (Applied Biosystems, Foster City, CA, USA) was used to amplify PCR products which were subsequently genotyped regarding the chosen SNPs using a TaqMan approach. In addition, a blind method was used for genotyping, 10% duplication was selected at random for a different genotyping test, and the results of the repeated tests were fully consistent with the original results. A t-test was employed to test differences in age. Frequency distributions of sex and genotype between patients and controls were tested using a chi-square test. A goodness-of-fit test was used to estimate the Hardy–Weinberg equilibrium (HWE) in controls. After adjusting for age and sex, we performed an unconditional multiple logistic regression model to assess associations of miRNA-492 rs2289030 G>C and miRNA-938 rs2505901 T>C polymorphism with BA susceptibility according to odds ratios (ORs) and 95% confidence intervals (CIs). A stratified analysis was conducted based on age, sex, and clinical stage. Data were analyzed using two-tailed tests in SAS statistical software. Statistical significance is reported at P < 0.05.
Table 1 summarizes genotypic and allele distribution of miRNA-492 rs2289030 polymorphisms among BA cases and controls. The genotypic and allele distribution of this polymorphism in the controls indicated a HWE (P > 0.05). Frequencies of the rs2289030 G allele were considerably higher in BA cases (73.9%) than in controls (26.1%). We fitted a dominant model of GG + GC vs. CC, a recessive model of GG vs. CC + CG, and an additive model of CC vs. GG. Negative results were found in the dominant model (P = 0.565; OR = 0.313; 95% CI: 0.85–1.35), in the recessive model (P = 0.166; OR = 0.73; 95% CI: 0.46–1.14), and in the additive model (P = 0.901; OR = 0.99; 95% CI: 0.82–1.19).
The genotype distribution of miRNA-938 rs2505901 T>C polymorphism in BA patients and controls is shown in Table 2. The genotype distribution was in a HWE in controls (P = 0.714); however, no significant association of the miRNA-938 rs2505901 T>C polymorphism with BA susceptibility was observed (dominant model of TT + TC vs. CC: adjusted OR = 1.06; 95% CI: 0.84–1.34; P = 0.649; recessive model of TT vs. TC + CC: adjusted OR = 0.86; 95% CI: 0.49–1.48; P = 0.579; T vs. C: adjusted OR = 0.88: 95% CI: 0.50–1.54; P = 0.858). We used ORs and 95% CIs to quantify the relationship by fitting multivariate logistic regression models. Contrary to our prediction, neither the miRNA-492 nor the miRNA-938 allele was associated with the risk of BA.
After decades of genetics exploration, the genetic predisposition factors of BA remain complicated, and substantial gene associations were discovered in genome-wide association studies (GWAS). GWAS on Chinese and Thai population identified the 10 most strongly BA-associated SNPs and showed that rs17095355 on 10q24, which is downstream of the SNP XPNPEP1, was strongly associated with BA[7]. Previous studies predominantly focused on SNPs that are closely associated with inflammatory mediators such as XPNPEP1 or with proteins that are abundantly expressed in biliary epithelia due to the proposed immune pathogenic pathway and disrupted biliary morphogenic development. Therefore, the current study is the first association study to address the relationship between BA risk and a gastroenterological cancer miRNA gene polymorphism.
In this study, we investigated the effects of miRNA-492 G>C (rs2289030) and miRNA-938 T>C (rs2505901) polymorphisms on the risk of BA for the following reasons: (1) the polymorphism of miRNA-492 G>C (rs2289030) which is located in the third nucleotide position of from the 3’ untranslated region of miRNA-492, probably changes miRNA-492 targets. miRNA-938 T>C (rs2505901) is located on a seed sequence of mature miRNA, and both polymorphisms are located in mature miRNA and result in a change which may affect its function. (2) As only two reported SNPs were studied, we cannot exclude the possibility that other variants are associated with BA risk. A different explanation for this lacking correlation may be genetic heterogeneity. (3) Emerging evidence suggests that transforming growth factor-β1 (TGF-β1) expression levels in the liver are correlated with hepatic fibrogenesis in BA patients. A TGF-β1-induced epithelial mesenchymal transition (EMT) mechanism was confirmed in BA cases resulting in biliary dysmorphogenesis which frequently occurred in various cancers[8]. miRNA-938 is associated with the TGF-β signaling pathway[9], and miRNA-492 was proposed to promote EMT processes[10].
Our study has a few limitations. As a hospital-based retrospective study, hospital admission bias and information were inevitably biased regarding patient selection and information collection. Further, the association between a single microRNA polymorphism and BA susceptibility may be weak, and quantitative trait loci should be considered in view of the complex interactions between gene polymorphisms. Moreover, given the moderate effects of this locus, our sample size may have been to small to detect such an association. Therefore, further studies should be conducted with a larger sample of patients and should include genotype association analysis, stratified subgroup analysis, and strict quality monitoring by follow-up studies.
In conclusion, we demonstrated that miRNA-492 G>C (rs2289030) and miRNA-938 T>C (rs2505901) polymorphisms were not significantly associated with BA susceptibility, thus further research is required to investigate the contribution of microRNA gene variation to BA susceptibility.
Competing Interests The authors have no conflict of interest to declare in relation to this article.
Author Contributions All authors contributed significantly to this work. LS, RZL, YT, and MF performed the research study, and XF O collected the data; YT and LS analyzed the data; RZL and RZZ conceived the study; RZL and LS wrote the manuscript; MF prepared the Tables; HMX revised the manuscript and provided constructive comments. All authors reviewed the manuscript and have read and approved the final version.
Abbreviations SNP, single nucleotide polymorphism; HWE, Hardy-Weinberg equilibrium; OR, odds ratio; CI, confidence interval; TGF-β1, transforming growth factor β1; EMT, epithelial mesenchymal transition.
doi: 10.3967/bes2021.080
Association of miRNA-492 rs2289030 G>C and miRNA-938 rs2505901 T>C Gene Polymorphisms with Biliary Atresia Susceptibility
-
-
Table 1. Association between miRNA-492 rs2289030 G>C polymorphism and biliary atresia susceptibility
Genotype Cases (N = 500) Controls (N = 1,463) Pa Crude OR (95% CI) P Adjusted OR (95% CI)b Pb rs2289030 G>C (HWE = 0.059) GG 270 (54.00) 828 (56.60) 1.00 1.00 GC 199 (39.80) 527 (36.02) 1.16 (0.94−1.43) 0.178 1.14 (0.90−1.45) 0.289 CC 31 (6.20) 108 (7.38) 0.88 (0.58−1.34) 0.554 0.77 (0.48−1.22) 0.261 Additive 0.663 1.04 (0.88−1.22) 0.661 0.99 (0.82−1.19) 0.901 Dominant 230 (46.00) 635 (43.40) 0.313 1.11 (0.91−1.36) 0.313 1.07 (0.85−1.35) 0.565 Recessive 469 (93.80) 1355 (92.62) 0.374 0.83 (0.55−1.25) 0.374 0.73 (0.46−1.14) 0.166 Note. OR, odds ratio; CI, confidence interval, HWE, Hardy-Weinberg equilibrium. a χ2 test for genotype distributions between biliary atresia patients and controls. b Adjusted for age and gender. Table 2. Association between miRNA-938 rs2505901 T>C polymorphism and biliary atresia susceptibility
Genotype Cases (N = 486) Controls (N = 1,457) Pa Crude OR (95% CI) P Adjusted OR (95% CI)b Pb rs2505901 T>C (HWE = 0.714) TT 278 (57.20) 850 (58.34) 1.00 1.00 TC 187 (38.48) 529 (36.31) 1.08 (0.87−1.34) 0.478 1.08 (0.85−1.38) 0.533 CC 21 (4.32) 78 (5.35) 0.82 (0.50−1.36) 0.446 0.88 (0.50−1.54) 0.660 Additive 0.973 1.00 (0.84−1.19) 0.973 1.02 (0.84−1.24) 0.858 Dominant 208 (42.80) 607 (41.66) 0.660 1.05 (0.85−1.29) 0.659 1.06 (0.84−1.34) 0.649 Recessive 465 (95.68) 1379 (94.65) 0.370 0.80 (0.49−1.31) 0.371 0.86 (0.49−1.48) 0.579 Note. OR, odds ratio; CI, confidence interval, HWE, Hardy-Weinberg equilibrium. a χ2 test for genotype distributions between biliary atresia patients and controls. b Adjusted for age and gender. S1. The detailed clinical information of the subjects in this study
Variables Cases (n = 506) Controls (n = 1,473) Subjects Age range (Months), n (%) 2.088 ± 1.934 18.61 ± 19.75 ≤ 3 470 (92.89) 521 (35.37) > 3 36 (7.11) 952 (64.63) Gender, n (%) Females 214 (42.29) 506 (34.35) Males 292 (57.71) 967 (65.65) Clinical manifestation, n (%) Cystic biliary atresia (CBA) 44 (8.70) non-CBA 462 (91.30) -
[1] Chardot C. Biliary atresia. Orphanet J Rare Dis, 2006; 1, 28. doi: 10.1186/1750-1172-1-28 [2] Naccarati A, Pardini B, Stefano L, et al. Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis, 2012; 33, 1346−51. doi: 10.1093/carcin/bgs172 [3] Uno S, Kataoka TR, Okajima H, et al. Perihilar cholangiocarcinoma in an explanted liver after Kasai operation for biliary atresia: a case report and literature review. Pathol Int, 2020; 70, 888−92. doi: 10.1111/pin.13016 [4] Al-Qahtani AA, Al-Anazi MR, Nazir N, et al. Association of single nucleotide polymorphisms in microRNAs with susceptibility to hepatitis B virus infection and HBV-related liver complications: a study in a Saudi Arabian population. J Viral Hepat, 2017; 24, 1132−42. doi: 10.1111/jvh.12749 [5] Shan YH, Shen N, Han LZ, et al. MicroRNA-499 Rs3746444 polymorphism and biliary atresia. Dig Liver Dis, 2016; 48, 423−8. doi: 10.1016/j.dld.2015.11.014 [6] Shen WJ, Dong R, Chen G, et al. microRNA-222 modulates liver fibrosis in a murine model of biliary atresia. Biochem Biophys Res Commun, 2014; 446, 155−9. doi: 10.1016/j.bbrc.2014.02.065 [7] Garcia-Barceló MM, Yeung MY, Miao XP, et al. Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24. 2. Hum Mol Genet, 2010; 19, 2917−25. doi: 10.1093/hmg/ddq196 [8] Xiao YT, Zhou Y, Chen YW, et al. The expression of epithelial-mesenchymal transition-related proteins in biliary epithelial cells is associated with liver fibrosis in biliary atresia. Pediatr Res, 2015; 77, 310−5. doi: 10.1038/pr.2014.181 [9] Cho SH, Ahn EH, An HJ, et al. Association of miR-938G> A polymorphisms with Primary Ovarian Insufficiency (POI)-Related Gene Expression. Int J Mol Sci, 2017; 18, 1255. doi: 10.3390/ijms18061255 [10] Wang Z, Liu YH, Wang M, et al. Effects of miR-492 on migration, invasion, EMT and prognosis in ovarian cancer by targeting SOX7. J Buon, 2020; 25, 797−804. -




 下载:
下载:



 Quick Links
Quick Links