-
As early as 1962, the relationship between mitochondria and human disease was concerned[1]. In the meantime, a number of molecular epidemiological studies have been published on the associations between mtSNPs and neurological, psychiatric[2], or metabolic diseases especially cardiovascular diseases. In our previous study, we reported the associations between mitochondrial gene mutations and essential hypertension in Chinese Han population[3-5]. Norikoetareported significant associations between mtSNPS and myocardial infarction[6], atherosclerotic cerebral infarction[7], metabolic syndrome[8] and longevity among the Japanese[9-10] and significant associations between mitochondrial haplogroup T and coronary artery disease, as well as diabetic retinopathy in Middle European Caucasians were indicated[11]. However, the relationship between mitochondrial gene mutations and coronary heart disease in the Chinese Han population, especially in young people, has not been reported. In our study, C5263T single-nucleotide mutation of the mitochondrial ND2 gene was observed in the Chinese Han family of coronary heart disease attacked at young age. 5263 is a nucleotide position in the gene encoding the ND2 protein and ND2 protein encoded by the ND2 gene is a subunit of Complex Ⅰ. The C5263T mutation refered to the substitution of the cytosine at nucleotide 5263 by a thymidine. The resultant change of the encoded alanine to valine might alter the ND2 protein Complex Ⅰ to induce mitochondrial calcium overload and increase production of reactive oxygen species (ROS), thus causing myocardial apoptosis, sustained myocardial damage, and electrophysiological disorders[12]. Therefore, we hypothesized that C5263T mutation might be involved in the occurrence and development of premature CHD in young people.
-
The inclusion criteria[13-14] were as follows: typical clinical presentations of paroxysmal retrosternal pain, including a tightness, squeezing, or pressure sensation and a burning sensation that could radiate to the left arm, jaw, neck, back, shoulder, or left forearm ulnar side, that was intermittent or persistent and was sustained for > 10-20 min, with suspected acute coronary syndrome; angiographic results indicating that at least one coronary artery had a decrease in diameter of ≥ 50% as observed from more than 2 different angles; and a clinical diagnosis of myocardial infarction. Based on age, patients were divided into the case group (≤ 45 years) and the experimental group (age of on-set great than 45 years).
-
Patients who had recent surgery, trauma, acute infection, and other combined chronic diseases and malignant tumours were excluded. All included patients signed informed consent forms.
-
Collection of Clinical Data General information, including the patient's age, gender, and body mass index (BMI), was collected. Cubital vein samples of patients were collected to detect creatine kinase (CK), cardiac troponin T (cTnT), B-type natriuretic peptide (BNP), total cholesterol (TC), triglyceride (TG), fibrinogen (Fib), high-sensitivity C-reactive protein (hs-CRP), low density lipoprotein-cholesterol (LDL-C), glycated haemoglobin, high density lipoprotein-cholesterol (HDL-C), and D-dimer (DD). Biochemical tests were performed using an automatic biochemical analyser (Hitachi 7600DDP, Japan) in the Department of Biochemistry in the General Hospital of the Chinese People's Liberation Army. Coagulation tests were performed using an STA automatic coagulation analyser (STA-Evolution, France) in the Clinical Laboratory Department of the General Hospital of the Chinese People's Liberation Army.
Coronal Angiography and Evaluation Criteria Coronal angiography was performed using the Judkins method through the right femoral artery or the right radial artery route. Multi-position projection and multi-site radiography were performed. The imaging diagnosis was confirmed by 2 cardiovascular intervention specialists. According to the evaluation criteria, CHD was diagnosed when the left main coronary artery and 3 epicardial coronary arteries and their large branches had maximum stenosis diameters of ≥ 50%.
Mitochondrial Genome Sequencing Mitochondrial DNA in whole blood was extracted using a reagent kit (Promega Wizard, A1120, USA). Primers were designed to amplify all 24 DNA fragments in mitochondria. PCR products were purified using the QIAEXⅡ purification reagent kit (Qiagen) and processed using the BigDye Terminator Cycle sequencing reaction reagent kit. Sequencing and analysis were directly performed using the ABI3700DNA automatic sequencing instrument. Comparison and analysis of DNA sequencing results and corresponding protein sequences were performed using the SeqWeb program GAP (GCG). Comparison of homology was performed using BLAST from the National Center for Biotechnology Information (NCBI). All sequencing results were compared to the 2013 edition of the Cambridge Reference Sequence[15].
Pedigree Analysis According to the regulations of the Ethics Committee of the General Hospital of the Chinese People's Liberation Army, signed informed consent was obtained from all subjects. Family members and relatives of the 2 young CHD patients with the mitochondrial C5263T single nucleotide mutation were investigated in detail, and pedigree maps were plotted.
Establishment of Lymphocyte Cell Lines from the Subjects Venous blood samples from the 5 subjects (Ⅰ1, Ⅱ1, Ⅱ2, Ⅲ1, Ⅲ2) in the pedigrees of one proband harbouring C5263T mutation were collected from the median cubital vein using EDTA as the anti-coagulant. Mononuclear cells from the peripheral venous blood samples were infected with Epstein-Barr virus (EBV) in this study. B lymphocyte proliferation was induced through the CpG DNA sequence, and T lymphocyte proliferation was inhibited using cyclosporine A. Finally, corresponding immortalized B lymphocyte cell lines were established from the subjects.
Detection of Cellular Reactive Oxygen Species (ROS) Levels A ROS detection reagent kit (Beyotime) was used. The instruments used were a BD FACSVerse flow cytometer and a Stratostabletop high-speed refrigerated centrifuge. 2', 7'-Dichloro-dihydro-fluorescein diacetate (DCFH-DA) was diluted in serum-free RPMI 1640 culture medium. Then, the cells were washed to remove the DCFH-DA, resuspended, and placed in the flow cytometer for monitoring and recording.
Detection of the Mitochondrial Membrane Potential (MMP) An MMP detection reagent kit was used (Beyotime). The instruments used were a BD FACSVerse flow cytometer, Vortex-5 vortex mixer, and Stratostabletop high-speed refrigerated centrifuge. After stimulation, the cell suspension was mixed thoroughly. JC-1 working solution was added, mixed thoroughly and incubated. The cells were centrifuged, washed with JC-1 buffer, and resuspended.
Statistical Methods All statistical analyses were performed using the SPSS 22.0 (Statistical Product and Service Solutions 22.0) statistical software. Measurement data were expressed as and were examined using the t test, ANOVA or the Wilcoxon rank sum test. Count data were presented as the number of cases and the percentage and were examined using the χ2 test. P < 0.05 indicated a statistically significant difference.
-
The percentage of males was significantly higher among young CHD patients than older CHD patients (P = 0.0025). The differences in BMI (P = 0.130), CK (P = 0.180), cTnT (P = 0.453), BNP (P = 0.705), TC (P = 0.023), TG (P= 0.569), Fib (P = 0.135), hs-CRP (P = 0.257), LDL-C (P = 0.541), blood glucose (P = 0.757), HDL-C (P = 0.190), and DD (P = 0.267) between the case and experimental groups were not significant (Table 1). There are no significant differences among BMI, TC, TG, Fib, hs-CRP, LDL-C, blood glucose, HDL-C, and DD except CK, cTNT and BNP, which stand for myocardial damage.
Table 1. Comparison of the Blood Biochemical Indicators between the Groups
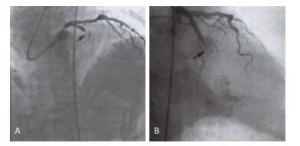
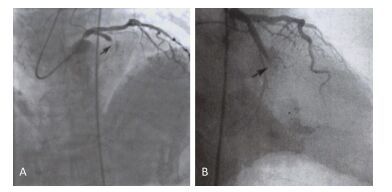
Items Case Group (Young CHD) Experimental Group (Older CHD) Control Group (Non-CHD) P1 P2 P3 115 100 100 Gender (male) 110 (96%) 73 (73%) 50 (50%) 0.0025* 0.001* 0.722 Age (y) 37 ± 8 69 ± 19 45 ± 25 0.005* 0.072 0.065 BMI (kg/m2) 27.260 ± 4.429 26.441 ± 4.003 25.123 ± 2.987 0.130 0.250 0.654 CK (U/L) 412.250 ± 932.448 116.263 ± 228.850 45.235 ± 37.336 0.180 0.001* 0.001* cTnT (ng/mL) 1.432 ± 4.801 1.518 ± 0.051 0.002 ± 0.005 0.453 0.001* 0.001* BNP (pg/mL) 430.061 ± 1100.702 657.453 ± 481.990 145.859 ± 170.33 0.705 0.051 0.062 TC (mmol/L) 4.451 ± 1.354 4.038 ± 0.943 4.208 ± 0.552 0.053 0.223 0.365 TG (mmol/L) 2.133 ± 1.762 1.978 ± 1.353 1.523 ± 1.044 0.569 0.078 0.061 Fib (g/L) 3.136 ± 0.837 3.108 ± 0.749 2.744 ± 0.965 0.135 0.123 0.115 hs-CRP (mg/dL) 0.543 ± 0.852 0.619 ± 1.629 0.212 ± 0.158 0.257 0.075 0.062 LDL-C (mg/dL) 2.774 ± 1.104 2.672 ± 0.761 1.511 ± 0.526 0.541 0.056 0.051 blood glucose (mmol/L) 6.466 ± 1.728 5.907 ± 1.104 4.859 ± 1.158 0.190 0.093 0.125 HDL-C (mmol/L) 0.966 ± 0.254 0.989 ± 0.271 1.025 ± 0.056 0.757 0.799 0.852 D-dimer (µg/mL) 0.355 ± 0.542 0.401 ± 0.683 0.258 ± 0.026 0.467 0.087 0.056 Note.BMI: body mass index, CK: creatine kinase, cTnT: cardiac troponin t, BNP: brain natriuretic peptide, TC: total cholesterol, TG: triglyceride, Fib: fibrinogen, hs-CRP: high-sensitivity C-reactive protein, LDL-C: low-density lipoprotein cholesterol, HDL-C: high-density lipoprotein cholesterol. Independent sample t test. ANOVA method, P < 0.05 represent significantly different. P1 stands for the comparison between Case group and Experimental group, P2 stands for the comparison between Case group and Control group, P3 stands for the comparison between Experimental group and Control group. *P < 0.05. Two young CHD patients with the mitochondrial C5263T single-nucleotide mutation underwent coronary angiography. Patient A carrying the mitochondrial C5263T single nucleotide mutation had 100% stenosis at the single nucleotide mutation had 100% stenosis at the proximal end of the circumflex branch (Figure 1B). Proximal end of the anterior descending branch (Figure 1A), and Patient B carrying the mitochondrial C5263T.
-
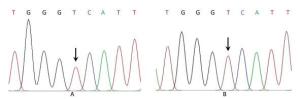
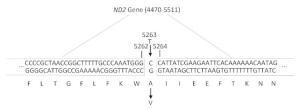
Mitochondrial gene full sequence scanning of all patients and comparison with the 2013 edition of the Revised Cambridge Reference Sequence resulted in the detection of a series of gene point mutations. All mitochondrial mutation results were compared with reported mitochondrial mutations [comparison source: MitoMap (http://www.mitomap.org)]. A total of 165 types of point mutations were discovered. Two young CHD patients had the C5263T single-nucleotide mutation in the ND2 gene (Figures 2 and 3). This mutation is highly evolutionarily conserved among higher animals such as humans and gorillas and was not detected in the experimental group. The mitochondrial gene full sequence scanning results of these 2 patients (Table 2) revealed that the HV2 gene had 2 mutation sites, the OHR gene had 1 mutation site, the HV3 gene had 1 mutation site, the RNR1 gene had 2 mutation sites, and the RNR2 gene had 2 mutation sites. Among the mutation sites, 24 were located in protein coding regions. However, with the exception of C5263T, the mutation sites exhibited poor conservation and have not been associated with CHD in the literature.
Table 2. Analysis of the Full Sequence Scanning of Patients Carrying the Mitochondrial C5263T Mutation
Gene Mutation Site Base Change Amino Acid Change Conservationa Reported Previouslybc Number 1 Number 2 HV2 A73G A-G Y 1 - G94A G-A Y 1 - OHR A263G A-G Y 1 1 HV3 T489C T-C Y 1 - RNR1 A750G A-G Y 1 1 A1438G A-G Y 1 1 RNR2 A2706G A-G Y 1 1 G3010A G-A Y 1 - T3109 deletion T-DEL Y 1 - ND1 G3316A G-A syn Y 1 - A3837T A-T syn Y - 1 C3970T C-T syn Y - 1 ND2 A4769G A-G syn Y 1 1 C4883T C-T syn Y 1 - C5178A C-A L-M L/S/T/T/S Y 1 - C5263T C-T A-V A/A/I/L/L Y 1 1 CO1 T5963C T-C syn Y 1 - T6392C T-C syn Y - 1 C7028T C-T syn Y 1 1 T7741C T-C syn Y - 1 G8020A G-A syn Y - 1 ATP6 C8575T C-T syn Y 1 A8701G A-G T-A T/S/L/L/Q Y 1 A8860G A-G T-A T/A/A/A/T Y 1 CO3 C9536T C-T syn Y 1 T9540C T-C syn Y 1 C9764T C-T syn Y 1 ND3 G10310A G-A syn Y 1 A10398G A-G T-S T/A/A/A/A Y 1 C10400T C-T syn Y 1 T10873C T-C syn Y 1 ND4 C11215T C-T syn Y 1 G11719A G-A syn Y 1 1 G11914A G-A syn Y 1 Note. aConservation of amino acid for polypeptides or nucleotide for rRNAs in humans (H), gorillas (O), mice (M), chickens (C), and zebrafish, (Z); bCambridge Reference Sequence; con-line mitochondrial gene database. -
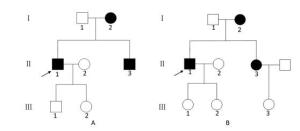
Among the probands in the 3-generation pedigree of Patient A carrying the C5263T point mutation (Figure 4A, made by Photoshop CS6), Ⅱ1 and Ⅱ3 both died of acute coronary syndrome. Among the probands in the 3-generation pedigree of Patient B carrying the C5263T point mutation (Figure 4B, made by Photoshop CS6), Ⅰ1 died of acute coronary syndrome, and Ⅱ3 did not participate in this study for personal reasons. Members in the third generation of these two family lineages did not yet have coronary artery disease due to their young ages. The penetrance rates of maternal members in the above family lineages were 100% and 75%, respectively. The maternal inheritance pattern suggests that the mitochondrial C5263T point mutation might participate in the occurrence and development of CHD among young people in these two 3-generation family lineages.
-
The detection results were listed in The ROS level was significantly higher in the cells from the mutation group than the non-mutation group (4750.82 ± 1045.55 vs. 3888.58 ± 487.60, P = 0.022) (Table 3). The ratio between the JC-1 monomers and multimers was expressed in the form of P2/P3, which was inversely proportional to the MMP. The MMP in the mutation group was significantly lower than the MMP in the non-mutation group (0.77 ± 0.57 vs. 0.35 ± 0.14, P = 0.045).
Table 3. ROS Levels and MMP in the Cells from the Mutation and non-mutation Group
Group Mutation Group Non-mutation t P ROS levels (U/well) 4750.82 ± 1045.55 3888.58 ± 487.60 2.301 0.022* Δψ 0.77 ± 0.57 0.35 ± 0.14 2.043 0.045* P 0.045* 0.386 Note. P value stands for the results of the comparison between mutation group and non-mutation group in ROS and MMP. *P < 0.05 represent significantly different. ROS: reactive oxygen species. Δψ: mitochondrial membrane potential. -
Environmental factors that influence CHD including CK (P = 0.180), cTnT (P = 0.453), TG (P = 0.569), Fib (P = 0.135), hs-CRP (P = 0.257), LDL-C (P = 0.541), and glycated haemoglobin (P = 0.190) as well as BMI did not differ significantly between the case group and the control group. Herein, we considered that genetic factors might account for a larger proportion of premature CHD patients in this study. The percentage of males was significantly higher among young Chinese Han CHD patients (n = 115) than older Chinese Han CHD patients (n = 100). Higher oestrogen levels in young women are thought to have a certain protective function in the cardiovascular system[16].
Full sequence scanning of patients in these 2 groups detected the mitochondrial ND2 gene C5263T mutation only in the case group. The ND2 protein is a subunit of Complex Ⅰ, and 5243 is a nucleotide site in the gene encoding the ND2 protein. The C5243T mutation occurs when the cytosine at the 5243 site is substituted by thymidine, which changes the encoded alanine to valine. This amino acid substitution might cause a change in the ND2 protein of Complex Ⅰ, thus inducing a series of changes in mitochondrial functions. Mitochondrial dysfunction has been implicated in the pathogenesis of several diseases including coronaryatherosclerotic characterised by free radical-induced damage[17-19]. In the study, this mutation might associate with increasing production of mitochondrial ROS and lower MMP levels, induce the opening of the mitochondrial permeability transition pore (mPTP)[20-22] cause myocardial apoptosis, sustained myocardial damage and electrophysiological disorders, and induce the occurrence of acute coronary syndromes.
A meta-analysis of studies from other countries showed that the mitochondrial respiratory chain Complex Ⅰ was the major site of ROS production[23-24]. Mitochondrial respiratory chain Complex Ⅰ damage caused by stress reactions, such as myocardial ischaemia, can cause premature reduction of oxygen by electrons and lead to massive production of ROS. The mechanism may involve Complex Ⅰ electron leakage caused by the destruction of the lipid bilayer in mitochondria by ROS, thus inducing a vicious cycle of massive production of ROS[24-25]. A large amount of ROS opens the mPTP channel to induce myocardial apoptosis. In addition, a large amount of ROS can reduce cellular bioenergy to cause arrhythmia and myocardial necrosis, apoptosis, and fibrosis; activate calcium-dependent potassium channels and couple the PKG-1α signal transduction in smooth muscle cells to function on vascular endothelial cells to participate in the process of atherosclerosis[26-27]; and alter voltage-dependent potassium channels and thiol redox signalling transduction to inhibit the formation of coronary collateral circulation[28-30]. Therefore, we speculated that the mitochondrial gene C5243T mutation promotes ROS production by changing the structure of Complex Ⅰ to participate in the occurrence and development of CHD in young people.
Today, young people are subject to long-term chronic fatigue and sympathetic nerve stimulation; therefore, human cells are persistently in a stressed state. Eukaryotic cells in a stress environment can activate the ROS system to induce mitochondria to produce ROS such as superoxide. The increase in ROS induced by chronic stress in young people further promotes the early onset of CHD. In pathological injury under stress status or pressure overload, a variety of signalling molecules and ionic mechanisms induce increases in calcium ion levels in cardiomyocytes[31]; the elevated calcium ions promote ROS production, and the excessive ROS further increases calcium ion and ROS levels through stimulation of mitochondrial calcium signalling and regulation of calcium regulatory proteins to finally form a positive feedback of calcium ion and ROS. In addition, to induce mitochondrial dysfunction through the activation of the mPTP pathway, calcium ions can also cause the activation and elevation of Ca2+/calmodulin-dependent protein kinases Ⅱ (CaMⅡ) through the Ca2+/CaM-dependent activation pathway. CaMⅡ can cause arrhythmia and myocardial apoptosis through induction of the elongation of the action potential duration and early after-depolarisation, thus causing sustained damage of cardiomyocytes and electrophysiological disorders[32-33]. Therefore, we speculated that the mitochondrial ND2 gene C5243T mutation had synergistic effects with calmodulin gene mutations of polymorphism to increase the incidence of CHD or even sudden death in young people.
In summary, the mitochondrial gene C5243T mutation might promote ROS production and calcium overload by changing the structure of Complex Ⅰ to participate in the occurrence and development of CHD in young people. These findings provide a theoretical basis for the early screening and discovery of susceptible populations and early prevention measures for CHD in young people.
Limitations Because of limited funding and time, the sample size of this study was relatively small, and we were going to expand the sample size in the future. At present, there were very few researches on coronary heart disease in young people, so it was meaningful to present the research results in the geneticist's perspective.
doi: 10.3967/bes2017.037
The Association between the C5263T Mutation in the Mitochondrial ND2 Gene and Coronary Heart Disease among Young Chinese Han People
-
Abstract:
Objective This study aimed to investigate the genetic background of mitochondrial genes in young patients with Coronary heart disease (CHD) to provide a foundation for the early prevention of young patients with CHD. Methods 115 cases of young (≤ 45 years) CHD Chinese Han patients (case group), 100 cases of older ( > 45 years) Chinese Han CHD patients (experimental group) hospitalized and 100 cases of healthy people through physical examination (control group) at the General Hospital of PLA between January 2014 and December 2015 were selected. General information, clinical assessment, pedigree analysis, and mitochondrial full sequence scanning were performed.The pedigrees of one patient harbouring the C5263T mutation were recruited. Mitochondrial functional analysis including cellular reactive oxygen species (ROS) levels and mitochondrial membrane potential (MMP) were performed on pedigrees with the C5263T mutation (mutation group) and without the mutation (non-mutation group). Results The differences in biochemical tests (P < 0.05) between the case group and experimental group were not significant. The C5263T single-nucleotide mutation of the mitochondrial ND2 gene was observed in 2 young CHD patients in the case group. The premature CHD of these 2 patients followed a pattern of maternal inheritance. The mutation group (Ⅰ1, Ⅱ2) had higher ROS levels (4750.82 ± 1045.55 vs. 3888.58 ± 487.60, P = 0.022) and lower MMP levels (P = 0.045) than the non-mutation group (Ⅱ1, Ⅲ1, Ⅲ2). Conclusion We speculated that the mitochondrial C5263T mutation might be associated with the occurrence CHD in Chinese Han young people. -
Key words:
- Mitochondrion /
- Coronary heart disease /
- Young
注释:1) AUTHOR CONTRIBUTIONS: -
Table 1. Comparison of the Blood Biochemical Indicators between the Groups
Items Case Group (Young CHD) Experimental Group (Older CHD) Control Group (Non-CHD) P1 P2 P3 115 100 100 Gender (male) 110 (96%) 73 (73%) 50 (50%) 0.0025* 0.001* 0.722 Age (y) 37 ± 8 69 ± 19 45 ± 25 0.005* 0.072 0.065 BMI (kg/m2) 27.260 ± 4.429 26.441 ± 4.003 25.123 ± 2.987 0.130 0.250 0.654 CK (U/L) 412.250 ± 932.448 116.263 ± 228.850 45.235 ± 37.336 0.180 0.001* 0.001* cTnT (ng/mL) 1.432 ± 4.801 1.518 ± 0.051 0.002 ± 0.005 0.453 0.001* 0.001* BNP (pg/mL) 430.061 ± 1100.702 657.453 ± 481.990 145.859 ± 170.33 0.705 0.051 0.062 TC (mmol/L) 4.451 ± 1.354 4.038 ± 0.943 4.208 ± 0.552 0.053 0.223 0.365 TG (mmol/L) 2.133 ± 1.762 1.978 ± 1.353 1.523 ± 1.044 0.569 0.078 0.061 Fib (g/L) 3.136 ± 0.837 3.108 ± 0.749 2.744 ± 0.965 0.135 0.123 0.115 hs-CRP (mg/dL) 0.543 ± 0.852 0.619 ± 1.629 0.212 ± 0.158 0.257 0.075 0.062 LDL-C (mg/dL) 2.774 ± 1.104 2.672 ± 0.761 1.511 ± 0.526 0.541 0.056 0.051 blood glucose (mmol/L) 6.466 ± 1.728 5.907 ± 1.104 4.859 ± 1.158 0.190 0.093 0.125 HDL-C (mmol/L) 0.966 ± 0.254 0.989 ± 0.271 1.025 ± 0.056 0.757 0.799 0.852 D-dimer (µg/mL) 0.355 ± 0.542 0.401 ± 0.683 0.258 ± 0.026 0.467 0.087 0.056 Note.BMI: body mass index, CK: creatine kinase, cTnT: cardiac troponin t, BNP: brain natriuretic peptide, TC: total cholesterol, TG: triglyceride, Fib: fibrinogen, hs-CRP: high-sensitivity C-reactive protein, LDL-C: low-density lipoprotein cholesterol, HDL-C: high-density lipoprotein cholesterol. Independent sample t test. ANOVA method, P < 0.05 represent significantly different. P1 stands for the comparison between Case group and Experimental group, P2 stands for the comparison between Case group and Control group, P3 stands for the comparison between Experimental group and Control group. *P < 0.05. Table 2. Analysis of the Full Sequence Scanning of Patients Carrying the Mitochondrial C5263T Mutation
Gene Mutation Site Base Change Amino Acid Change Conservationa Reported Previouslybc Number 1 Number 2 HV2 A73G A-G Y 1 - G94A G-A Y 1 - OHR A263G A-G Y 1 1 HV3 T489C T-C Y 1 - RNR1 A750G A-G Y 1 1 A1438G A-G Y 1 1 RNR2 A2706G A-G Y 1 1 G3010A G-A Y 1 - T3109 deletion T-DEL Y 1 - ND1 G3316A G-A syn Y 1 - A3837T A-T syn Y - 1 C3970T C-T syn Y - 1 ND2 A4769G A-G syn Y 1 1 C4883T C-T syn Y 1 - C5178A C-A L-M L/S/T/T/S Y 1 - C5263T C-T A-V A/A/I/L/L Y 1 1 CO1 T5963C T-C syn Y 1 - T6392C T-C syn Y - 1 C7028T C-T syn Y 1 1 T7741C T-C syn Y - 1 G8020A G-A syn Y - 1 ATP6 C8575T C-T syn Y 1 A8701G A-G T-A T/S/L/L/Q Y 1 A8860G A-G T-A T/A/A/A/T Y 1 CO3 C9536T C-T syn Y 1 T9540C T-C syn Y 1 C9764T C-T syn Y 1 ND3 G10310A G-A syn Y 1 A10398G A-G T-S T/A/A/A/A Y 1 C10400T C-T syn Y 1 T10873C T-C syn Y 1 ND4 C11215T C-T syn Y 1 G11719A G-A syn Y 1 1 G11914A G-A syn Y 1 Note. aConservation of amino acid for polypeptides or nucleotide for rRNAs in humans (H), gorillas (O), mice (M), chickens (C), and zebrafish, (Z); bCambridge Reference Sequence; con-line mitochondrial gene database. Table 3. ROS Levels and MMP in the Cells from the Mutation and non-mutation Group
Group Mutation Group Non-mutation t P ROS levels (U/well) 4750.82 ± 1045.55 3888.58 ± 487.60 2.301 0.022* Δψ 0.77 ± 0.57 0.35 ± 0.14 2.043 0.045* P 0.045* 0.386 Note. P value stands for the results of the comparison between mutation group and non-mutation group in ROS and MMP. *P < 0.05 represent significantly different. ROS: reactive oxygen species. Δψ: mitochondrial membrane potential. -
[1] LUFT R, IKKOS D, PALMIERI G, et al. A case of severe hypermetabolism of nonthyroid origin with a defect in the maintenance of mitochondrial respiratory control: a correlated clinical, biochemical, and morphological study. J Clin Invest, 1962; 41, 1776-804. doi: 10.1172/JCI104637 [2] Ueno H, Nishigaki Y, Kong QP, et al. Analysis of mitochondrial DNA variants in Japanese patients with schizophrenia. Mitochondrion, 2009; 9, 385-93. doi: 10.1016/j.mito.2009.06.003 [3] Zhu HY, Wang SW, Liu L, et al. A mitochondrial mutation A4401G is involved in the pathogenesis of left ventricular hypertrophy in Chinese hypertensives. Eur J Hum Genet, 2009; 17, 172-8. doi: 10.1038/ejhg.2008.151 [4] Zhu HY, Wang SW, Liu L, et al. Genetic variants in mitochondrial tRNA genes are associated with essential hypertension in a Chinese Han population. Clinica Chimica Acta, 2009; 410, 64-9. doi: 10.1016/j.cca.2009.09.023 [5] Zhu HY, Wang SW, Martin LJ, et al. The role of mitochondrial genome in essential hypertension in a Chinese Han population. Eur J Hum Genet, 2009; 17, 1501-6. doi: 10.1038/ejhg.2009.63 [6] Nishigaki Y, Yamada Y, Fuku N, et al. Mitochondrial haplogroup N9b is protective against myocardial infarction in Japanese males. Hum Genet, 2007; 120, 827-36. doi: 10.1007/s00439-006-0269-z [7] Nishigaki Y, Yamada Y, Fuku N, et al. Mitochondrial haplogroup A is a genetic risk factor for atherothrombotic cerebral infarction in Japanese females. Mitochondrion, 2007; 7, 72-9. doi: 10.1016/j.mito.2006.11.002 [8] Tanaka M, Fuku N, Nishigaki Y, et al. Women with mitochondrial haplogroup N9a are protected against metabolic syndrome. Diabetes, 2007; 56, 518-21. doi: 10.2337/db06-1105 [9] Alexe G, Fuku N, Bilal E, et al. Enrichment of longevity phenotype in mtDNA haplogroups D4b2b, D4a, and D5 in the Japanese population. Hum Genet, 2007; 121, 347-56. doi: 10.1007/s00439-007-0330-6 [10] Bilal E, Rabadan R, Alexe G, et al. Mitochondrial DNA haplogroup D4a is a marker for extreme longevity in Japan. PLoS One, 2008; 3, e2421. doi: 10.1371/journal.pone.0002421 [11] Kofler B, Mueller EE, Eder W, et al. Mitochondrial DNA haplogroup T is associated with coronary artery disease and diabetic retinopathy: a case control study. BMC Med Genet, 2009; 10, 35. http://www.biomedcentral.com/imedia/1544545222509253_manuscript.pdf [12] Muntean DM, Sturza A, Danila MD, et al. The Role of Mitochondrial Reactive Oxygen Species in Cardiovascular Injury and Protective Strategies. Oxid Med Cell Longev, 2016; 2016, 8254942. http://downloads.hindawi.com/journals/omcl/2016/8254942.pdf [13] O'Gara PT, Kushner FG, Ascheim DD, et al. 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol, 2013; 6, e78-e140. https://www.guideline.gov/summaries/summary/39429 [14] Jneid H, Anderson JL, Wright RS, et al. 2012 ACCF/AHA focused update of the guideline for the management of patients with unstable angina/non-ST-elevation myocardial infarction (updating the 2007 guideline and replacing the 2011 focused update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol, 2012; 60, 645-81. doi: 10.1016/j.jacc.2012.06.004 [15] Bandelt HJ, Kloss-Brandstatter A, Richards MB, et al. The case for the continuing use of the revised Cambridge Reference Sequence (rCRS) and the standardization of notation in human mitochondrial DNA studies. J Hum Genet, 2014; 59, 66-77. doi: 10.1038/jhg.2013.120 [16] Meyer MR, Barton M. Estrogens and Coronary Artery Disease: New Clinical Perspectives. Adv Pharmacol, 2016; 77, 307-60. doi: 10.1016/bs.apha.2016.05.003 [17] Monsalve M, Borniquel S, Valle I, et al. Mitochondrial dysfunction in human pathologies. Front Biosci, 2007; 12, 1131-53. doi: 10.2741/2132 [18] Tan AL, Forbes JM, Cooper ME. AGE, RAGE, and ROS in diabetic nephropathy. Semin Nephrol, 2007; 27, 130-43. doi: 10.1016/j.semnephrol.2007.01.006 [19] Schleicher E, Friess U. Oxidative stress, AGE, and atherosclerosis. Kidney Int Suppl, 2007; 106, S17-S26. http://www.kidney-international.org/article/S0085-2538(15)52547-7/pdf [20] Hurst S, Hoek J, Sheu SS. Mitochondrial Ca2+ and regulation of the permeability transition pore. J Bioenerg Biomembr, 2017; 49, 27-47. doi: 10.1007/s10863-016-9672-x [21] El-Hattab AW, Scaglia F. Mitochondrial Cardiomyopathies. Front Cardiovasc Med, 2016; 3, 25. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3783139/pdf/20130900s00007p385.pdf [22] Dorsch M, Behmenburg F, Raible M, et al. Morphine-Induced Preconditioning: Involvement of Protein Kinase A and Mitochondrial Permeability Transition Pore. PLoS One, 2016; 11, e151025. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4788451/ [23] Lee HL, Chen CL, Yeh ST, et al. Biphasic modulation of the mitochondrial electron transport chain in myocardial ischemia and reperfusion. Am J Physiol Heart Circ Physiol, 2012; 302, H1410-22. doi: 10.1152/ajpheart.00731.2011 [24] Paradies G, Petrosillo G, Pistolese M, et al. Decrease in mitochondrial complex Ⅰ activity in ischemic/reperfused rat heart: involvement of reactive oxygen species and cardiolipin. Circ Res, 2004; 94, 53-9. doi: 10.1161/01.RES.0000109416.56608.64 [25] Gadicherla AK, Stowe DF, Antholine WE, et al. Damage to mitochondrial complex Ⅰ during cardiac ischemia reperfusion injury is reduced indirectly by anti-anginal drug ranolazine. Biochim Biophys Acta, 2012; 1817, 419-29. doi: 10.1016/j.bbabio.2011.11.021 [26] Zhang DX, Borbouse L, Gebremedhin D, et al. H2O2-induced dilation in human coronary arterioles: role of protein kinase G dimerization and large-conductance Ca2+-activated K+ channel activation. Circ Res, 2012; 110, 471-80. doi: 10.1161/CIRCRESAHA.111.258871 [27] Liu Y, Zhao H, Li H, et al. Mitochondrial sources of H2O2 generation play a key role in flow-mediated dilation in human coronary resistance arteries. Circ Res, 2003; 93, 573-80. doi: 10.1161/01.RES.0000091261.19387.AE [28] Pung YF, Sam J, Stevanov K, et al. Mitochondrial oxidative stress corrupts coronary collateral growth by activating adenosine monophosphate activated kinase-alpha signaling. Arterioscler Thromb Vasc Biol, 2013; 33, 1911-9. doi: 10.1161/ATVBAHA.113.301591 [29] Zuo L, Pannell BK, Re AT, et al. Po2 cycling protects diaphragm function during reoxygenation via ROS, Akt, ERK, and mitochondrial channels. Am J Physiol Cell Physiol, 2015; 309, C759-66. http://paper.medlive.cn/literature/1950224 [30] Ohanyan V, Yin L, Bardakjian R, et al. Requisite Role of Kv1. 5 Channels in Coronary Metabolic Dilation. Circ Res, 2015; 11, 612-21. http://circres.ahajournals.org/content/early/2015/07/29/CIRCRESAHA.115.306642?related-urls=yeslCIRCRESAHA.115.306642v1 [31] Song J, Liu X, Zhai P, et al. A putative mitochondrial calcium uniporter in A. fumigatus contributes to mitochondrial Ca2+ homeostasis and stress responses. Fungal Genet Biol, 2016; 94, 15-22. doi: 10.1016/j.fgb.2016.07.001 [32] Akar FG, O'Rourke B. Mitochondria are sources of metabolic sink and arrhythmias. Pharmacol Ther, 2011; 131, 287-94. doi: 10.1016/j.pharmthera.2011.04.005 [33] Kim JB, Kim C, Choi E, et al. Particulate air pollution induces arrhythmia via oxidative stress and calcium calmodulin kinase Ⅱ activation. Toxicol Appl Pharmacol, 2012; 259, 66-73. doi: 10.1016/j.taap.2011.12.007 -




 下载:
下载:



 Quick Links
Quick Links