-
Periodontitis belongs to periodontal diseases, which is a chronic inflammatory disease mainly involving periodontal supporting tissue[1]. Chronic periodontitis is the most common type of periodontitis, accounting for approximately 95% of patients with periodontitis[2]. The disease progression of chronic periodontitis is relatively slow. In most cases, microorganisms and their products first accumulate in gingiva for a long time, causing gingival inflammation. Once gingivitis is not treated properly, periodontal tissues including the periodontal ligament, cementum and alveolar bone could be damaged, resulting in the occurrence of periodontitis[3]. Periodontitis occurs primarily in adults but the incidence rates among young people increases, seriously affecting people’s life quality[4,5]. It is noteworthy that periodontitis affects not only oral tissues, but also many systemic diseases. Besides, periodontitis has been considered as a strong risk factor for diabetes and cardiovascular disease[6].
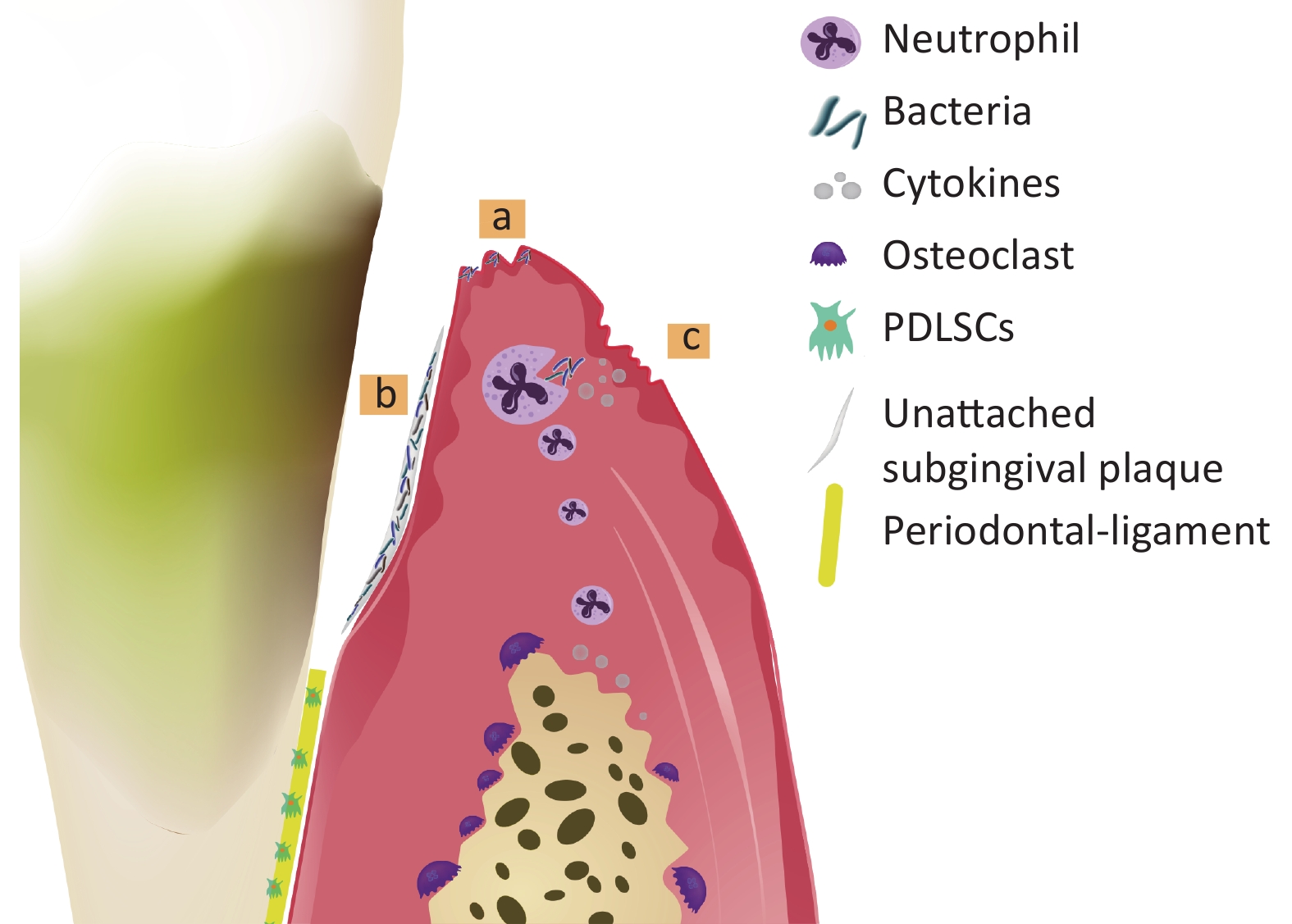
At present, periodontitis is acknowledged to be a multifactorial disease with complicated pathogenesis. Bacteria, host and environment are all involved in the occurrence and development of periodontitis[7]. Firstly, subgingival plaque formed by bacteria and their products is the initiation factor of periodontitis, which could potentially accelerate the destruction of alveolar bone[8,9]. Secondly, the host immune responses prevent microorganisms from entering periodontal tissue in the early stage of infection, but excessive cytokines, prostaglandins and matrix metalloproteinases production lead to periodontal supporting tissue damage and bone loss[10]. Periodontitis often occurs when the balance between microorganism and host defense is broken[11,12]. Finally, the host defensive responses are markedly influenced by environmental factors such as genetic background and smoking, which could also contribute to the development of periodontitis[13]. The pathogenesis of periodontitis is shown in Figure 1.

Figure 1. Pathogenesis of periodontitis. Bacteria invade the gingival tissues and cause gingival inflammation (a). Bacteria are not controlled timely and gather to form the unattached subgingival plaque, causing alveolar bone absorption and attachment loss (AL) (b). Although neutrophils engulf bacteria via phagocytosis at the early phase of inflammation, excessive pro-inflammatory cytokines produced during defense response accelerate the progress of periodontitis (c).
Non-coding RNAs (ncRNAs) are the RNAs that cannot be able to encode proteins. In the early stage, mRNAs are acted as a protein translation template while ncRNAs are considered as "transcription garbage"[14]. Heretofore, researchers found that around 98% of the human genome are transcribed into ncRNAs, extensively regulated the gene expression during the progress of diseases[15]. In inflammatory-related diseases like periodontitis, ncRNAs play the important role in regulating the homeostatic of cellular functions[16]. Studies about ncRNAs in the development of periodontitis mainly focus on microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs)[17]. However, how these ncRNAs interact and which specific mechanism involves in periodontitis remains to be explored[18]. Therefore, our review focuses on the dual role of miRNAs, lncRNAs and circRNAs related to inflammation, cell proliferation, apoptosis and osteogenic differentiation in periodontitis, aiming to explore more diagnostic markers and therapeutic targets of periodontitis.
-
MiRNAs have about 22 nucleotides in length and are first transcribed into primary RNAs (pri-miRNAs), then undergo sequential biogenesis, from pre-miRNAs and finally into mature miRNAs[19]. Later, mature miRNAs mostly silence the target genes by degrading or preventing translation[20]. Besides, the ceRNA network formed with miRNA- lncRNA/circRNA-mRNA was closely associate with the development of many diseases[21-24]. More importantly, miRNAs provide a powerful pathway for the diagnosis and treatments of disease[25,26].
-
LncRNAs, a class of ncRNAs with length greater than 200 nucleotides, were processed by RNA polymerase II (Pol II)[27]. Unlike mRNA, lncRNAs prefer to enrich in nucleus with the following possible mechanisms: 1) degradation or tethering on chromatin; 2) longer distance between the 3′ splice site and the branch point; 3) through nuclear retention elements[28,29]. In the nucleus, lncRNAs are closely related to chromatin, and directly or indirectly regulated chromatin organization by chromatin regulatory proteins (SWI/SNF or PRC subunits), thereby affecting the chromatin activity[30-32]. In addition, nuclear lncRNAs are also widely involved in transcriptional process through cis/trans regulation or transcription factors, in post-transcriptional process through alternative splicing or A-to-I editing, finally regulated the genes expression[33-36]. In the cytoplasm, diverse RNA binding proteins (RBPs) or miRNAs were interacted with lncRNAs to implement functions[37]. So far, many studies suggested the functional interactions among lncRNAs and miRNAs: 1) lncRNAs are degraded by miRNAs[38]; 2) lncRNAs serve as sponges/decoys for miRNAs to form ceRNAs, thereby derepressing miRNA target mRNAs[39]; 3) lncRNAs compete with miRNAs for degradation of target mRNAs[40]; 4) lncRNAs produce miRNAs[41]. In general, lncRNAs are widely involved in many cell processes and thus to regulate the progress of disease[42].
-
CircRNAs belong to a special class of ncRNAs with a covalent closed loop structure formed by trans-splicing. Up to now, the biological functions of circRNAs are summarized as follows: 1) as a ceRNA or miRNA sponge; 2) binding to proteins; 3) as a potential translation template for proteins; 4) regulating the pre-RNA splicing and parental gene expression[43]. As more molecular biology techniques were addressed to RNA studies, circRNAs with biological functions related to diseases has gradually been discovered, especially in various cancers[44,45]. Importantly, numerous circRNAs have been demonstrated to be potential biomarkers, such as circCHST15, circ-PTPDC1 and hsa_circRNA_0039480[46-48].
The formation process of miRNAs, lncRNAs, and circRNAs are shown in Figure 2.

Figure 2. The forming process of miRNAs (a), lncRNAs (b), and circRNAs (c). The genes of ncRNAs are firstly transcribed into linear RNAs by DNA polymerase in nucleus. Then, (a) miRNAs were processed from pri-miRNAs to pre-miRNAs through RNA polymerase II (Pol II) and Drosha, thereby transporting into the nucleus by export5. In the cytoplasm, mature miRNAs are formed by Dicer and subsequently bind to the silent effector complex (RISC), finally inhibited the expression of target gene. (b) Most lncRNAs are remained in the nucleus under the inefficient processing of Pol II and added a cap at the 5' end and a poly (A) tail at 3'. A small part of lncRNAs is then transported into the nucleus through NFX1 and further combined with miRNAs to release the inhibitory effects of miRNAs on target genes. (c) Four circRNAs are formed by trans-splicing, and subsequently act as miRNAs sponge or combine with RBP to regulate gene expression.
-
The protective effects of miRNAs on periodontitis are mainly concentrated on inhibiting pro-inflammatory cytokine production, improving cell functions and preventing the bone loss (Table 1). Table 1. Protective ncRNAs in the development of periodontitis
Functions MiRNAs LncRNAs CircRNAs Suppress Apoptosis, Inflammation, Proliferation miR-218 (target Mmp9)[49] miR-205-5p (target XBP1/ IL6ST)[50,51] miR-543-3P (target KLF6)[52] miR-212-5p (target Myd88)[53] miR-17-5p (target VEGFA)[54] miR-28-5p (target SPHK1)[55] miR-223-3p (target NLRP3-Caspase 1-GSDMD)[56] miR-466l (regulate IL-10)[57] lncRNA H19 (regulate PI3K/AKT)[58] IncRNA TUG1 (sponge miR-498/RORA)[59] lncRNA GAS5 (regulate NF-κB pathway)[60] IncZFY-AS1 (sponge miR-129-5p/DDX3X)[61] lncRNA PTCSC3 (target TLR4)[62] lncRNA MAFG-AS1 (target TLR4)[63] lncRNA LOXL1-AS1 (target IL-1β)[64] Iinc-RAM (target FGF2)[65] lncRNA JHDMID-AS1 (target DNAJC10)[66] IncRNA TUG1 (sponge miR-132)[67] IncRNA FGD5-AS1 (sponge miR-142-3p/SOCS6)[68] lncRNA DCST1 AS1 (sponge miR-21precursor/PLAP-1)[69] lncRNA MALAT1 (sponge miR-155-5p/ETS1)[70] lncRNA DLX2-AS1 (sponge miR-330-3p)[71] Circ 0062491 (sponge miR-498/SOCS6)[72] Circ_0085289 (sponge let-7f-5p/SOCS6)[73] Circ_0081572 (sponge miR-378h/RORA)[74] CircLRP6 (sponge miR-145a-5p/Zeb2)[75] hsa_circ_0003948 (sponge miR-144-3p/NR2F2)[76] Circ_00668819 (sponge miR-144-5p/RORA)[77] Bone Protection miR-21 (target Spry1)[78] miR-383-5p (target HDAC9)[79] miR-10a (Treg differentiation)[80] miR-17 (target HDAC9)[81] miR-203 (regulate AP-1/ICAM-1)[82] miR-200c (target IL-6, IL-8, and CCL-5)[83-85] miR-101 (regulate EZH2 /VCAM‐1)[86] miR-335-5p (unclear)[87] miR-543 (target TOB2)[88] miRNA-26a-5p (target Wnt5a)[89] IncRNA Nron (regulate NKRF)[90] lncRNA OIP5-AS1 (sponge miR-92a-3p)[91] lncRNA GAS5 (regulate GDF5 and p38/JNK)[92] lncRNA ANRIL (sponge miR-7/ NF-κB; miR-7-5p/IGF-1R)[93,94] IncRNA TWIST1 (regulate TWIST1)[95] IncRNA TUG1 (sponge Lin28A/miR-222-3p)[96] lncRNA XIST (sponge miR-214-3p) [97] lncRNA MEG3 (sponge miR-27a-3p/IGF1)[98] LINC00707 (sponge miR-490-3p/FOXO1)[99] CircMAP3K11 (sponge miR-511/ TLR4)[100] Circular RNA CDR1as (sponge miR-7/GDF5/SMAD0)[101] CircRNA FAT1 (sponge miR-4781-3p/SMAD5)[102] Circ 0062491 (sponge miR-142-5p/IGF1 axis)[103] Note. Mmp9: matrix metallopeptidase 9, XBP1: X-box binding protein 1, KLF6: kruppel-like factor 6, Myd88: myeloid differentiation primary response gene 88, IL6ST: interleukin 6 signa transducer, VEGFA: vascular endothelial growth factor A, SPHK1: sphingosine kinase 1, PI3K/AKT: phosphatidylinositol3-kinase/threonine protein kinase, TLR4: toll-like receptor 4, RORA: RAR-related orphan receptor A, NF-kB: Nuclear factor-kappa B, DDX3X: DEAD box polypeptide 3, IL-1β: β interleukin 1, FGF2: fibroblast growth factor 2, DNAJC10: a heat shock protein 40 family member, SOCS6: suppressor of cytokine signaling 6, PLAP-1: periodontal ligament-associated protein 1, ETS1: v-ets erythroblastosis virus E26 oncogene homolog 1, HIF-1α: hypoxia inducible factor 1 alpha, Zeb2: zinc finger E-box binding homeobox 2, NR2F2: nuclear receptor subfamily 2 group F member 2, Spry1: sprouty homolog 1, HDAC9: histone deacetylase 9, AP-1: activator protein 1, IGF1: insulin like growth facto 1, SMAD5: small mothers against decapentaplegic 5, GDF5: growth differentiation factor 5, SMAD: small mothers against decapentaplegic, TLR4: toll-like receptor 4, FOXO1: forkhead Box 1, IGF1: insulin-like growth factor-1, TWIST1: twist-related protein 1, NKRF: NF-κB repressing factor, JNK: jun N-terminal kinase. -
Burgeoning findings have revealed that miRNAs expression was dysregulated in periodontal tissues with periodontitis, resulting in abnormal expression of target genes[104]. For example, 91 miRNAs were differentially expressed in periodontal inflamed gingival tissues by gene sequencing technology, in which hsa-miR-126* and hsa-miR-20a were among the miRNAs with the most significant alteration[105]. The same conclusion was showed in a mouse study of periodontitis, which demonstrated that the miRNA expression was significant changed between healthy and periodontitis animals[106].
Emerging convincing studies suggested that miRNAs could control the progression of inflammation[16,107]. For example, exosomal miR-205-5p derived from PDLSCs relieved the inflammation by directly targeting XBP1 to influence the Th17/Treg balance in periodontitis rat model[50]. In periodontitis transgenic mice overexpressing miR-335-5p (335-Tg), 335-Tg group successfully relieved gingiva inflammation compared with wild-type mice[87]. Moreover, miR-17-5p suppressed abnormal angiogenesis by targeting vascular specific markers (CD31 and VEGFA), thereby controlling inflammatory infiltrates[54]. Besides, miRNAs could delay the development of periodontitis by elevating cell viability, inhibiting apoptosis and abnormal proliferation. For example, in LPS-induced cell apoptosis and viability inhibition, the overexpression of miR-543-3p, miR-212-5p and miR-203 reversed the destructive effects of LPS on PDLSCs functions by targeting KLF6, Myd88 and AP-1/ICAM-1 respectively[52].
PDLSCs possess multilineage differentiation potential and could differentiate into osteoblasts, thus initiating bone repair[108]. Many miRNAs including miR-383-5p, miR-543, and miR-21 were upregulated during the osteogenic differentiation of PDLSCs and interacted with HDAC9, TOB2, and Spry1, respectively, promoting the osteogenic differentiation of PDLSCs[78,79,88]. Similarly, numerous researchers used carriers in animal model to intuitively prove the ability of miRNAs in facilitating bone formation. For example, the PDLSCs transfected with miR-26a-5p mimics, which were subsequently loaded with hydroxyapatine-tricalcium phosphate (HA-TCT) and implanted into mice, promoted bone formation mechanically by targeting wnt5a[89]. Tadkamol Krongbaramee et al. found the local injection of PEI-plasmid miR-200 nanoplexes into the gingiva in periodontitis rat model effectively reduced alveolar bone loss[85]. In the subsequent study of miR-200c, Liu Hong et al. found that the bone protective effects of miR-200c were fulfilled by precisely targeting the 3’UTR of IL-6, IL-8, and CCL-5[83]. In addition, miR-10a combined the application of IL-2 and TGF-β could also prevent bone loss in a mouse periodontal disease model via nanofibrous spongy microspheres[80]. Some studies also indirectly proved the protective roles of miRNAs in bone loss. For example, in a rat model of periodontitis, miR-17 and miR-101 which separately targeted HDAC9 or EZH2/VCAM-1 ameliorated crestal bone loss[81,86].
-
Recent studies have confirmed the necessity of lncRNAs in the development of periodontitis and the interaction with miRNAs in regulating the cell functions of PDLSCs[109,110]. For example, 8925 lncRNAs were found to be differentially expressed in patients with periodontitis by microarray analysis[111]. Another study predicted that there were 84 lncRNAs involved in inflammatory processes, among them, AC000120.7, MZF1-AS1, FGD5-AS1, HOTAIR, and OIP5-AS1 were verified to downregulate in periodontitis patients[112]. Furthermore, the ceRNA networks conducted by lncRNA-miRNA-mRNAs also regulated the inflammatory process in periodontitis[113]. Those studies all proved that lncRNAs were central for the periodontitis development.
Similar to miRNAs, lncRNAs also account for the pathophysiological alteration of periodontitis. Several lncRNAs are negatively correlated with the degree of inflammatory state of periodontitis. For example, lncRNA GAS5 was downregulated in a mouse ligature-induced periodontitis model and its overexpression alleviated the expression of cytokines by regulating the NF-κB signaling pathway[60]. In LPS-stimulated inflammatory environments, the overexpression of lncRNA TUG1 and lncRNA DLX2-AS1 decreased the production of inflammatory cytokines by separately targeting miR-498 or miR-330-3p, thereby modulating downstream targets genes RORA or RO60[59,71]. Several lncRNAs are negatively correlated with the physiological functions of cells. For example, in PDLSCs from rats under oxidative stress, lncRNA JHDM1D-AS1 overexpression significantly blocked the promoting effect of H2O2 on apoptosis by negatively regulating DNAJC10[66]. In periodontal tissue from periodontitis, the knockdown of lncRNA FGD5-AS1 and lncRNA TUG1 that respectively targeted miR-142-3p and miR-132 exacerbated the inhibitory effects of LPS on PDLSCs, whereas overexpression rescued cell viability, blocked apoptosis and cytokine secretion[67,68]. lncRNA H19 was found to be upregulated in the mouse periodontitis model and closely associated with the activation of autophagy by inhibiting PI3K/AKT pathway, thereby achieving self-renewal of cells[58].
The abnormally increased proliferation of PDLSCs is considered to be associated with the disease progression of periodontitis[114]. The role of lncRNAs in proliferation inhibition was fullfed by targeting different genes. For example, lncRNA LOXL1-AS1, lncRNA DCST1-AS1 and Linc-RAM lncRNA could target TLR4, miR-21 precursor and FGF2 respectively, inhibiting the proliferation of PDLSCs[64,65,69]. Similarly, lncRNA PTCSC and lncRNA MAFG-AS1 could target TLR4 to inhibit the proliferation of PDLSCs as well[62,63]. Interestingly, the likelihood of periodontitis recurrence and proliferation rates of PDLSCs was inversely related to lncRNA MORT expression[115].
Many reports to date have indicated that lncRNAs could promote the expression of osteogenic transcription factors, thereby initiating bone repair[116]. For example, the overexpression of lncRNA-TWIST1 and LncRNA TUG1 upregulated the expression of Runx2, ALP, and OCN by targeting its downstream genes TWIST1 or lin-28 homolog A (Lin28A) respectively, consequently accelerating the formation of mineralized nodules[95,117].
The role of ceRNA formed by lncRNA-miRNA-mRNA in bone repair have been demonstrated by several studies. For example, in inflammatory environments, LncRNA OIP5-AS1 targeted miR-92a-3p directly to promote the osteogenic differentiation of PDLSCs[91]. In non-inflammatory environments under osteogenic induction, the silence of lncRNA TUG1, lncRNA XIST, lncRNA MALAT1, LIN00707 and lncRNA MEG3 decreased the osteogenic differentiation ability by separately sponging miRNA-222-3p, miR-214-3p, miR-155-5P, miR-490-3p and miR-27a-3p[70,96-99]. Similarly, lncRNA ANRIL could target several mechanisms simultaneously to promote the osteogenic differentiation of PDLSCs including miR-7/ NF-κB and miR-7-5p/IGF-1R[93,94]. Another two experiments validated the bone protective effects of lncRNAs at animal level. One of them was periodontitis model using lncRNA Nron knockout transgenic mice, in which TRAP staining results showed lncRNA Nron knockout significantly decreased the number of osteoclasts[90]. In another in vivo transplantation experiment, the HE staining results showed the silence of FoxO1 which negatively regulated lncRNA-POIR inhibited the formation of osteoids, indirectly suggesting the protective role of lncRNA-POIR in bone absorption[118]. Although lncRNAs play key role in PDLSCs osteogenic differentiation, studies on the role in inflammatory environments are rare, which requires further exploration.
-
The latest research has found that circRNAs involved in cell apoptosis, cell migration and oxidative stress are also differentially expressed in periodontitis[119]. For example, 70 differentially expressed circRNAs were identified in gingival tissues from periodontitis patients, among which circPTP4A2, chr22:23101560-23135351+, circARHGEF28, circBARD1 and circRASA2 played crucial role in the diagnosis of periodontitis[119]. Fifty-three circRNAs were differentially expressed in PDLSCs under hypoxia, in which lncRNA-FTX/circRNA-FAT1-hsa-miR-4781-3p-SMAD5 and circRNA LPAR1-hsa-miR-342-3p-ADAR might be involved in the progression of periodontitis[120].
circRNAs usually acted as miRNAs sponges to inhibit inflammation and improve cellular functions[121]. For example, Jie Li et al. first identified circ_0062491 as the sponge of miR-584, and they found it was downregulated in periodontitis patients[122]. Later, the roles of circ_0062491 in periodontitis were further studied. circ_0062491 was first described to directly regulate miR-498/SOCS6 to prevent PDLSCs from LPS-induced apoptosis and inflammation[72]. Another researcher found miR-142-5p/IGF1 axis was directly regulated by circ_0062491, promoting osteogenic differentiation of PDLSCs[103]. In periodontal tissues from periodontitis, the expression of Circ_0081572 and Circ_0066881 were downregulated and they were found to provide a sponge effect on miR-378h and miR-144-5p through targeting 3’ -UTR of RORA, which in turn restored the PDLSCs viability and inhibit apoptosis[74,77]. Likewise, Circ_ 0085289 and hsa_circ_0003948 enhanced cell viability and delayed progression of periodontitis by upregulating the expression of SOCS6 or NR2F2 through forming a competitive ring with let-7f-5p and miR-144-3p respectively, thereby achieving protective effects[73,76]. In periodontitis mouse model, tissues with si-circMAP3K11 accelerated the apoptosis and promoted the proliferation of PDLSCs by negatively regulating miR-511-3p[100]. Nowadays, the efficient bioinformatics prediction greatly helped investigators figuring out the potential role of circRNAs in promoting osteogenic differentiation. For example, Liangkun Xie et al. found 69–557 differentially expressed exosomal circRNAs during osteogenesis induction at day 0, 3, 7 by RNA sequencing. In the following qRT-PCR experiments, hsa_circ_0087960 and hsa_circ_0000437 were validated to be upregulated in the osteogenic differentiation of PDLSCs, meaning the upregulation of these two circRNAs could promote the osteogenic differentiation of PDLSCs[123]. In addition, 139 circRNAs were differentially expressed at day 14 of osteogenesis induction, VENN analysis further revealed that there were 52 circRNAs showed the same trends in all osteogenic stages[124]. Except for the bioinformatics analyses, Yu Ye et al. found the silencing of circFAT1 inhibited the bone formation in rat skull defect model mechanically through competitively binding miR-4781-3p and blocking SMAD5 expression[102]. What’s more, another animal experiment has shown that Circular RNA CDR1as deficiency could also inhibit the bone formation[101]. Consistently, the overexpression of circRNA CDR1as was proportionally related to the stemness of PDLSCs, which greatly enhanced the cell ability of self-renewal[125].
-
Some ncRNAs are positively correlated with the manifestation and pathophysiology of periodontitis. (Table 2).
Table 2. Destructive ncRNAs in the development of periodontitis
Functions MiRNA LncRNA CircRNA Promote Apoptosis, Inflammation, Proliferation miR-143-3p (through droving M1 macrophage polarization or targeting KLF5)[126,127] IncRNA 01126 (sponge miR-518a-5p/HIF-1α/MAP or regulate MEK/ERK signaling pathway)[128,129] lncRNA MALA1(target miR-20a/TLR4 or miR-769-5p/HIF3A)[130, 131] lncRNA AWPPH (unclear)[132] lncRNA NEAT1(sponge miR-200c-3p/TRAF6)[133] LINC00616 (sponge miR-370/TFRC)[134] Circ_0138959(sponge miR-527/caspase-5)[135] Circ_0097010 (sponge miR-769-5p/KLF6)[136] Circ_0138960 (sponge miR-518a-5p/HDAC6)[137] Circ_0099630(sponge miR-212-5p/SPRY1 or miR-940/TRAF6)[138-140] Bone Destruction miR-130a (Unclear)[141] miR-325-3 (target Runx2)[142] miR-25a-3p (target CD69 mRNA)[143] miR-138(osteocalcin OC)[144] miR-30a-5p (target Runx2) [145] miR-23a (BMPR1B)[146] miR-214 (target ATF4)[147] miR-6512-3p (target SNHG7)[148] miR-223 (target FGFR2/ TGFβR2)[149] miR-302b (unclear)[150] lncRNA-ANCR (sponge miRNA-758/ Notch2 or regulate canonical WNT signaling pathway) [151, 152] lncRNA DANCR (unclear)[153] Circ_0138959(sponge miR-495-3p/TRAF6)[154] Circ CDK8(regulate ER stress/autophagy)[155] Note. KLF5: Kruppel-like factor 5, HIF-1α: Hypoxia-inducible factor-1alpha, TLR4: Toll-like receptor 4, HIF3A: hypoxia-inducible factor (HIF) 3A, TRAF6: TNF receptor associated factor 6, TLR4: Toll like receptor 4, HDAC6: histone deacetylase 6, SPRY1: sprouty homolog 1, KLF6: Kruppel-like factor 6, ATF4: activating transcription factor 4, TFRC: transferrin receptor, SNHG7: Small nucleolar RNA hostgene 7, FGFR2: fibroblast growth factor receptor 2, TGFβR2: transforming growth factor beta receptor 2. -
The upregulation of a certain miRNA in periodontitis sometimes exerted destructive role in disease progress. For example, Young Hwa Lee et al. found miR-181b, miR-19b, miR-23a, miR-30a, miR-let7a, and miR-301a in periodontitis tissue were upregulated, which positively related with disease progression. MiR-143-3p was first found to be the most highly expressed miRNA through next generation sequencing (NGS) technology, which was validated to be positively correlated to clinical symptoms[156]. In the subsequent functional experiments, miR-143-3p was demonstrated to inhibit osteogenic differentiation of PDLSCs by targeting KLF5[127]. Recently, exosomal miR-143-3p was proved to enrich in inflammation-affected PDLSCs and could exacerbated inflammatory infiltrates by modulating macrophage polarization in periodontitis mouse model[126]. In LPS-induced inflammation environment, Tetramethylpyrazine protected PDLSCs from apoptosis and cell viability inhibition, however, miR-302b mimic reversed the protective effects[150].
Some miRNAs could inhibit bone formation by directly targeting osteogenic factors (Runx2, OPG, OCN and BMP), thereby limiting the effects of target gene[157]. For example, miR-30a-5p and miR-325-3p weakened bone formation by directly targeting Runx2, and miR‐325‐3p inhibitor rescued the formation of alveolar bone in rats[142,145]. Likewise, miR-23a and miR-138 inhibited osteogenesis of PDLSCs by individually targeting BMPR1B and OCN[144,146]. Additionally, miR-155-3p and miR-223 targeted Kctd1 and GFβR2/FGFR2 respectively, resulting in reduced differentiation of cementoblast[149,158]. Last but not least, miR-214 overexpression leads to a massive inhibition of osteogenic differentiation by directly targeting ATF4 and CTNNB1[147,159].
-
Five lncRNAs are involved in impairing the biological activities of periodontal cells by sponging miRNAs. Specifically, the upregulated expression level of lncRNA NEAT1 in LPS-treated PDLSCs caused the disordered expression of miR-200c-3p and TRAF6. This study further reported lncRNA NEAT1 silencing enhanced cell viability and repressed apoptosis in PDLSCs through miR-200c-3p/TRAF6[133]. In hypoxia environment, LIN01126 promoted apoptosis and inflammatory cytokines release of PDLSCs via sponging miR-518a-5p and inactivating MAPK signaling pathway[128]. Recently, one study reported the relationship between LINC00616 and ferroptosis, and found LINC00616 could promote ferroptosis by interacting with miR-370/TFRC, thereby speeding up the development of periodontitis[134]. LncRNA MALAT1 upregulation in LPS-stimulated human gingival fibroblasts (HGFs) and PDLSCs enhanced inflammatory cytokine production through interacting with miR-20a/TLR4 or miR-769-5p/HIF3A, which was reported by two studies[130,131]. Like lncRNA MALAT1, lncRNA ANCR could also block the osteogenic differentiation of PDLSCs through multiple pathways including activated Wnt signaling pathway and sponged miRNA-758[151,152].
Three lncRNAs was demonstrated to play destructive role in human bio-samples and animal model. For example, lncRNA AWPPH was positively correlated with the recurrence rate of periodontitis according to the analysis of blood samples from 80 patients with periodontitis[132]. LncZFY-AS1, which was elevated in Porphyromonas gingivali oral infected periodontitis mice, knock-down refrained oxidative stress and inflammation in mice with periodontitis by modulating miRNA-129-5p/DDX3X[61]. LncRNA-01126 was the most significantly increased lncRNA in periodontitis patients through lncRNA microarray, which was verified in the gingival tissues of periodontitis mice as well. Further functional experiments proved that lncRNA-01126 overexpression reduced the migration of PDLSCs by inhibiting MEK/ERK signal pathway[129].
-
The destructive role of circRNAs in periodontitis concentrated in disrupting the normal physiological functions of PDLSCs. For example, Jingjing Zheng et al. explored the regulation mechanism of CircCDK8 on osteogenic differentiation under hypoxia, and found CircCDK8 activated endoplasmic reticulum stress, autophagy, and apoptosis in PDLSCs through mTOR signaling[155].
Surely, the critical role of ceRNA formed by circRNA and miRNA is indisputable. Circ_0138959 was identified as a functional sponge of miR-527 that directly targeted the apoptosis proteins caspase-5 to block the effect of apoptotic protein on cells, ultimately promoting the apoptosis[135]. Wenjuan Deng et al. found circ_0138959 overexpression inhibited osteogenic differentiation, promoting apoptosis and cytokine release by interacting with miR-495-3p and TRAF6, leading to devastating outcomes[154]. Circ_0097010 and circ_0138959 inhibition could block LPS-mediated apoptosis and reduce inflammation by forming ceRNA with miR-769-5P/KLF6 and miR-518a-5p/HDAC6 respectively[136,154]. Moreover, silencing of circ_0099630 absorbed miR-940 consequently increased the mRNA level of TRAF6, promoting cell viability and inhibiting apoptosis and inflammatory response in LPS-induced PDLCs[139]. Consistent with a bioinformatics analysis, has_circ_0099630 overexpression played destructive role in the osteogenic differentiation of human periodontal ligament fibroblasts probably through interacting with miR-182, miR-1200, miR-338, miR-576 and miR-623[140]. Those experiments all proved the potential mechanism of circRNAs in accelerating the development of periodontitis. -
At present, the incidence of periodontitis is extremely high, and its pathogenesis remain ambiguous. Many ncRNAs have been proved to be differentially expressed in periodontitis patients and considered as biomarkers to predict the prognosis and recurrence of periodontitis[115,132,141]. Currently, to enhance the therapeutic effects, researchers have been investigating to implant functional ncRNAs into animals through scaffold or adenovirus or directly inject into gingiva[80,89,142,160]. However, there are still critical problems we are facing: Firstly, the expression of ncRNAs is easily affected by external stimuli, which makes it difficult to exert the therapeutic effects. For example, miR-146a significantly increased the activity of ALP and the expression of OCN and Runx2 on day7 and day14 after LPS stimulation[161]. Contrarily, miR-146a suppressed the activity of ALP and inhibited osteogenic differentiation of PDLSCs under mechanical forces[162]. Secondly, periodontitis is a complex infection caused by multiple bacteria. Up to now, research has shown that bacterial pathogens could suppress host miRNA expression for their survival[163]. Whether interaction between periodontal pathogens influence the expression of ncRNAs and whether there are bacteria-related biomarkers that can be used to diagnose periodontitis remain unclarified. Thirdly, ncRNAs are differentially expressed in divergent periodontal tissues. For example, the expression of miR-199a-5p was up-regulated in salivary level and decreased in plasma level among patients with periodontitis[164]. The miR-146a upregulation and miR-155 downregulation occurred only in gingiva after LPS stimulation instead of dental pulp and periodontal ligament fibroblasts from the same donor[165]. These uncertainties in ncRNAs expression led to challenges including the specificity, delivery and tolerability of ncRNA-based therapies. More importantly, many studies only focused on in vitro experiment, especially lncRNAs and circRNAs, which is different from the actual in vivo environmental state at the time of onset.
However, the future applications of ncRNAs still have great potential in periodontitis. We believe that ncRNAs could be applied as biomarkers in diagnose and typing of periodontitis. Additionally, ncRNAs could also be used as indicators for the treatment decision in periodontitis. In periodontitis treatment, the functional ncRNAs that processed into tablets or thin films could combine with classic periodontal surgery to enhance the therapeutic effectiveness. Surly, for genetic-related periodontitis, it is also a promising choice to develop vaccines based on ncRNAs.
Altogether, research on ncRNAs in periodontitis contains not only challenges but also opportunities, more preclinical and clinical studies are deserved to be done in this field, contributing to elucidate the underlying mechanisms in periodontitis as early as possible.
doi: 10.3967/bes2023.079
-
Abstract: This review aims to sum up how Non-coding RNAs (ncRNAs) regulate the development of periodontitis and provides a new perspective for understanding the pathogenesis of periodontitis. We explored the ncRNA's dual role in the development of periodontitis by summarizing evidence from previous in vivo and in vitro studies as well as clinical samples. In our review, the downregulation of 18 miRNAs, 22 lncRNAs and 10 circRNAs demonstrates protective roles in periodontitis. In contrast, the expression of other 11 miRNAs, 7 lncRNAs and 6 circRNAs are upregulated in periodontitis, which promote the progression of periodontitis. These dysregulated ncRNAs exert their protective or destructive roles by mainly influencing cell proliferation, differentiation and apoptosis via cross-talking with various molecules or signaling pathways. Our findings suggested which and how ncRNAs promote or delay the progression of periodontitis, which may greatly contribute to diagnose and therapy development of periodontitis based on ncRNAs in the future.
-
Key words:
- Periodontitis /
- MicroRNA /
- Long non-coding RNA /
- Circular RNA /
- Non-coding RNA
注释:1) AUTHOR CONTRIBUTION: -
Figure 1. Pathogenesis of periodontitis. Bacteria invade the gingival tissues and cause gingival inflammation (a). Bacteria are not controlled timely and gather to form the unattached subgingival plaque, causing alveolar bone absorption and attachment loss (AL) (b). Although neutrophils engulf bacteria via phagocytosis at the early phase of inflammation, excessive pro-inflammatory cytokines produced during defense response accelerate the progress of periodontitis (c).
Figure 2. The forming process of miRNAs (a), lncRNAs (b), and circRNAs (c). The genes of ncRNAs are firstly transcribed into linear RNAs by DNA polymerase in nucleus. Then, (a) miRNAs were processed from pri-miRNAs to pre-miRNAs through RNA polymerase II (Pol II) and Drosha, thereby transporting into the nucleus by export5. In the cytoplasm, mature miRNAs are formed by Dicer and subsequently bind to the silent effector complex (RISC), finally inhibited the expression of target gene. (b) Most lncRNAs are remained in the nucleus under the inefficient processing of Pol II and added a cap at the 5' end and a poly (A) tail at 3'. A small part of lncRNAs is then transported into the nucleus through NFX1 and further combined with miRNAs to release the inhibitory effects of miRNAs on target genes. (c) Four circRNAs are formed by trans-splicing, and subsequently act as miRNAs sponge or combine with RBP to regulate gene expression.
Table 1. Protective ncRNAs in the development of periodontitis
Functions MiRNAs LncRNAs CircRNAs Suppress Apoptosis, Inflammation, Proliferation miR-218 (target Mmp9)[49] miR-205-5p (target XBP1/ IL6ST)[50,51] miR-543-3P (target KLF6)[52] miR-212-5p (target Myd88)[53] miR-17-5p (target VEGFA)[54] miR-28-5p (target SPHK1)[55] miR-223-3p (target NLRP3-Caspase 1-GSDMD)[56] miR-466l (regulate IL-10)[57] lncRNA H19 (regulate PI3K/AKT)[58] IncRNA TUG1 (sponge miR-498/RORA)[59] lncRNA GAS5 (regulate NF-κB pathway)[60] IncZFY-AS1 (sponge miR-129-5p/DDX3X)[61] lncRNA PTCSC3 (target TLR4)[62] lncRNA MAFG-AS1 (target TLR4)[63] lncRNA LOXL1-AS1 (target IL-1β)[64] Iinc-RAM (target FGF2)[65] lncRNA JHDMID-AS1 (target DNAJC10)[66] IncRNA TUG1 (sponge miR-132)[67] IncRNA FGD5-AS1 (sponge miR-142-3p/SOCS6)[68] lncRNA DCST1 AS1 (sponge miR-21precursor/PLAP-1)[69] lncRNA MALAT1 (sponge miR-155-5p/ETS1)[70] lncRNA DLX2-AS1 (sponge miR-330-3p)[71] Circ 0062491 (sponge miR-498/SOCS6)[72] Circ_0085289 (sponge let-7f-5p/SOCS6)[73] Circ_0081572 (sponge miR-378h/RORA)[74] CircLRP6 (sponge miR-145a-5p/Zeb2)[75] hsa_circ_0003948 (sponge miR-144-3p/NR2F2)[76] Circ_00668819 (sponge miR-144-5p/RORA)[77] Bone Protection miR-21 (target Spry1)[78] miR-383-5p (target HDAC9)[79] miR-10a (Treg differentiation)[80] miR-17 (target HDAC9)[81] miR-203 (regulate AP-1/ICAM-1)[82] miR-200c (target IL-6, IL-8, and CCL-5)[83-85] miR-101 (regulate EZH2 /VCAM‐1)[86] miR-335-5p (unclear)[87] miR-543 (target TOB2)[88] miRNA-26a-5p (target Wnt5a)[89] IncRNA Nron (regulate NKRF)[90] lncRNA OIP5-AS1 (sponge miR-92a-3p)[91] lncRNA GAS5 (regulate GDF5 and p38/JNK)[92] lncRNA ANRIL (sponge miR-7/ NF-κB; miR-7-5p/IGF-1R)[93,94] IncRNA TWIST1 (regulate TWIST1)[95] IncRNA TUG1 (sponge Lin28A/miR-222-3p)[96] lncRNA XIST (sponge miR-214-3p) [97] lncRNA MEG3 (sponge miR-27a-3p/IGF1)[98] LINC00707 (sponge miR-490-3p/FOXO1)[99] CircMAP3K11 (sponge miR-511/ TLR4)[100] Circular RNA CDR1as (sponge miR-7/GDF5/SMAD0)[101] CircRNA FAT1 (sponge miR-4781-3p/SMAD5)[102] Circ 0062491 (sponge miR-142-5p/IGF1 axis)[103] Note. Mmp9: matrix metallopeptidase 9, XBP1: X-box binding protein 1, KLF6: kruppel-like factor 6, Myd88: myeloid differentiation primary response gene 88, IL6ST: interleukin 6 signa transducer, VEGFA: vascular endothelial growth factor A, SPHK1: sphingosine kinase 1, PI3K/AKT: phosphatidylinositol3-kinase/threonine protein kinase, TLR4: toll-like receptor 4, RORA: RAR-related orphan receptor A, NF-kB: Nuclear factor-kappa B, DDX3X: DEAD box polypeptide 3, IL-1β: β interleukin 1, FGF2: fibroblast growth factor 2, DNAJC10: a heat shock protein 40 family member, SOCS6: suppressor of cytokine signaling 6, PLAP-1: periodontal ligament-associated protein 1, ETS1: v-ets erythroblastosis virus E26 oncogene homolog 1, HIF-1α: hypoxia inducible factor 1 alpha, Zeb2: zinc finger E-box binding homeobox 2, NR2F2: nuclear receptor subfamily 2 group F member 2, Spry1: sprouty homolog 1, HDAC9: histone deacetylase 9, AP-1: activator protein 1, IGF1: insulin like growth facto 1, SMAD5: small mothers against decapentaplegic 5, GDF5: growth differentiation factor 5, SMAD: small mothers against decapentaplegic, TLR4: toll-like receptor 4, FOXO1: forkhead Box 1, IGF1: insulin-like growth factor-1, TWIST1: twist-related protein 1, NKRF: NF-κB repressing factor, JNK: jun N-terminal kinase. Table 2. Destructive ncRNAs in the development of periodontitis
Functions MiRNA LncRNA CircRNA Promote Apoptosis, Inflammation, Proliferation miR-143-3p (through droving M1 macrophage polarization or targeting KLF5)[126,127] IncRNA 01126 (sponge miR-518a-5p/HIF-1α/MAP or regulate MEK/ERK signaling pathway)[128,129] lncRNA MALA1(target miR-20a/TLR4 or miR-769-5p/HIF3A)[130, 131] lncRNA AWPPH (unclear)[132] lncRNA NEAT1(sponge miR-200c-3p/TRAF6)[133] LINC00616 (sponge miR-370/TFRC)[134] Circ_0138959(sponge miR-527/caspase-5)[135] Circ_0097010 (sponge miR-769-5p/KLF6)[136] Circ_0138960 (sponge miR-518a-5p/HDAC6)[137] Circ_0099630(sponge miR-212-5p/SPRY1 or miR-940/TRAF6)[138-140] Bone Destruction miR-130a (Unclear)[141] miR-325-3 (target Runx2)[142] miR-25a-3p (target CD69 mRNA)[143] miR-138(osteocalcin OC)[144] miR-30a-5p (target Runx2) [145] miR-23a (BMPR1B)[146] miR-214 (target ATF4)[147] miR-6512-3p (target SNHG7)[148] miR-223 (target FGFR2/ TGFβR2)[149] miR-302b (unclear)[150] lncRNA-ANCR (sponge miRNA-758/ Notch2 or regulate canonical WNT signaling pathway) [151, 152] lncRNA DANCR (unclear)[153] Circ_0138959(sponge miR-495-3p/TRAF6)[154] Circ CDK8(regulate ER stress/autophagy)[155] Note. KLF5: Kruppel-like factor 5, HIF-1α: Hypoxia-inducible factor-1alpha, TLR4: Toll-like receptor 4, HIF3A: hypoxia-inducible factor (HIF) 3A, TRAF6: TNF receptor associated factor 6, TLR4: Toll like receptor 4, HDAC6: histone deacetylase 6, SPRY1: sprouty homolog 1, KLF6: Kruppel-like factor 6, ATF4: activating transcription factor 4, TFRC: transferrin receptor, SNHG7: Small nucleolar RNA hostgene 7, FGFR2: fibroblast growth factor receptor 2, TGFβR2: transforming growth factor beta receptor 2. -
[1] Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nat Rev Dis Primers, 2017; 3, 17038. doi: 10.1038/nrdp.2017.38 [2] Luo WP, Li H, Ye F. Clinical therapeutic effects of probiotics in combination with antibiotics on periodontitis: a protocol for systematic review and meta-analysis. Medicine (Baltimore), 2021; 100, e23755. doi: 10.1097/MD.0000000000023755 [3] Chapple ILC, Van Der Weijden F, Doerfer C, et al. Primary prevention of periodontitis: managing gingivitis. J Clin Periodontol, 2015; 42 Suppl 16, S71-6. [4] Catunda RQ, Levin L, Kornerup I, et al. Prevalence of periodontitis in young populations: a systematic review. Oral Health Prev Dent, 2019; 17, 195−202. [5] Buset SL, Walter C, Friedmann A, et al. Are periodontal diseases really silent? A systematic review of their effect on quality of life. J Clin Periodontol, 2016; 43, 333−44. doi: 10.1111/jcpe.12517 [6] Liccardo D, Cannavo A, Spagnuolo G, et al. Periodontal disease: a risk factor for diabetes and cardiovascular disease. Int J Mol Sci, 2019; 20, 1414. doi: 10.3390/ijms20061414 [7] Page RC, Kornman KS. The pathogenesis of human periodontitis: an introduction. Periodontol 2000, 1997; 14, 9−11. doi: 10.1111/j.1600-0757.1997.tb00189.x [8] Bartold PM, Van Dyke TE. An appraisal of the role of specific bacteria in the initial pathogenesis of periodontitis. J Clin Periodontol, 2019; 46, 6−11. [9] Zhang JX, Yu JL, Dou JG, et al. The impact of smoking on subgingival plaque and the development of periodontitis: a literature review. Front Oral Health, 2021; 2, 751099. doi: 10.3389/froh.2021.751099 [10] Cekici A, Kantarci A, Hasturk H, et al. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2000, 2014; 64, 57−80. doi: 10.1111/prd.12002 [11] Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol, 2014; 35, 3−11. doi: 10.1016/j.it.2013.09.001 [12] Van Dyke TE, Bartold PM, Reynolds EC. The nexus between periodontal inflammation and dysbiosis. Front Immunol, 2020; 11, 511. doi: 10.3389/fimmu.2020.00511 [13] Kornman KS. Commentary: periodontitis severity and progression are modified by various host and environmental factors. J Periodontol, 2014; 85, 1642−5. doi: 10.1902/jop.2014.140430 [14] Palazzo AF, Koonin EV. Functional long non-coding RNAs evolve from junk transcripts. Cell, 2020; 183, 1151−61. doi: 10.1016/j.cell.2020.09.047 [15] Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet, 2006; 15, R17−29. doi: 10.1093/hmg/ddl046 [16] Marques-Rocha JL, Samblas M, Milagro FI, et al. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J, 2015; 29, 3595−611. doi: 10.1096/fj.14-260323 [17] Wang JH, Yan SS, Yang JH, et al. Non-coding RNAs in rheumatoid arthritis: from bench to bedside. Front Immunol, 2020; 10, 3129. doi: 10.3389/fimmu.2019.03129 [18] Qasim SSB, Al-Otaibi D, Al-Jasser R, et al. An evidence-based update on the molecular mechanisms underlying periodontal diseases. Int J Mol Sci, 2020; 21, 3829. doi: 10.3390/ijms21113829 [19] Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol, 2005; 6, 376−85. [20] Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol, 2019; 20, 5−20. [21] Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov, 2013; 3, 1113−21. doi: 10.1158/2159-8290.CD-13-0202 [22] Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell, 2011; 146, 353−8. [23] Acunzo M, Romano G, Wernicke D, et al. MicroRNA and cancer–a brief overview. Adv Biol Regul, 2015; 57, 1−9. doi: 10.1016/j.jbior.2014.09.013 [24] Wojciechowska A, Braniewska A, Kozar-Kaminska K. MicroRNA in cardiovascular biology and disease. Adv Clin Exp Med, 2017; 26, 865−74. [25] Kara G, Calin GA, Ozpolat B. RNAi-based therapeutics and tumor targeted delivery in cancer. Adv Drug Deliv Rev, 2022; 182, 114113. doi: 10.1016/j.addr.2022.114113 [26] Mori MA, Ludwig RG, Garcia-Martin R, et al. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab, 2019; 30, 656−73. doi: 10.1016/j.cmet.2019.07.011 [27] Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell, 2013; 154, 996−1009. doi: 10.1016/j.cell.2013.07.047 [28] Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res, 2012; 22, 1775−89. doi: 10.1101/gr.132159.111 [29] Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol, 2021; 22, 96−118. doi: 10.1038/s41580-020-00315-9 [30] Wang YY, He L, Du Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell, 2015; 16, 413−25. doi: 10.1016/j.stem.2015.03.003 [31] Werner MS, Ruthenburg AJ. Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep, 2015; 12, 1089−98. doi: 10.1016/j.celrep.2015.07.033 [32] Sun QY, Hao QY, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet, 2018; 34, 142−57. doi: 10.1016/j.tig.2017.11.005 [33] O'Leary VB, Ovsepian SV, Carrascosa LG, et al. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep, 2015; 11, 474−85. doi: 10.1016/j.celrep.2015.03.043 [34] Li XL, Subramanian M, Jones MF, et al. Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep, 2017; 20, 2408−23. doi: 10.1016/j.celrep.2017.08.041 [35] Gonzalez I, Munita R, Agirre E, et al. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol, 2015; 22, 370−6. doi: 10.1038/nsmb.3005 [36] Salameh A, Lee AK, Cardó-Vila M, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci USA, 2015; 112, 8403−8. doi: 10.1073/pnas.1507882112 [37] Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics, 2016; 14, 73−80. doi: 10.1016/j.gpb.2016.03.005 [38] Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell, 2012; 47, 648−55. doi: 10.1016/j.molcel.2012.06.027 [39] Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell, 2011; 147, 358−69. doi: 10.1016/j.cell.2011.09.028 [40] Franklin JL, Rankin CR, Levy S, et al. Malignant transformation of colonic epithelial cells by a colon-derived long noncoding RNA. Biochem Biophys Res Commun, 2013; 440, 99−104. doi: 10.1016/j.bbrc.2013.09.040 [41] Rogler LE, Kosmyna B, Moskowitz D, et al. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia. Hum Mol Genet, 2014; 23, 368−82. doi: 10.1093/hmg/ddt427 [42] Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol, 2021; 220, e202009045. doi: 10.1083/jcb.202009045 [43] Shan C, Zhang YF, Hao XD, et al. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer, 2019; 18, 136. doi: 10.1186/s12943-019-1069-0 [44] Meng SJ, Zhou HC, Feng ZY, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer, 2017; 16, 94. doi: 10.1186/s12943-017-0663-2 [45] Xue C, Li GL, Zheng QX, et al. The functional roles of the circRNA/Wnt axis in cancer. Mol Cancer, 2022; 21, 108. doi: 10.1186/s12943-022-01582-0 [46] Jiang B, Zhang JF, Sun XB, et al. Circulating exosomal hsa_circRNA_0039480 is highly expressed in gestational diabetes mellitus and may be served as a biomarker for early diagnosis of GDM. J Transl Med, 2022; 20, 5. doi: 10.1186/s12967-021-03195-5 [47] Gui CP, Liao B, Luo CG, et al. circCHST15 is a novel prognostic biomarker that promotes clear cell renal cell carcinoma cell proliferation and metastasis through the miR-125a-5p/EIF4EBP1 axis. Mol Cancer, 2021; 20, 169. doi: 10.1186/s12943-021-01449-w [48] Li ZX, Cheng Y, Fu K, et al. Circ-PTPDC1 promotes the progression of gastric cancer through sponging Mir-139-3p by regulating ELK1 and functions as a prognostic biomarker. Int J Biol Sci, 2021; 17, 4285−304. doi: 10.7150/ijbs.62732 [49] Guo J, Zeng XM, Miao J, et al. MiRNA-218 regulates osteoclast differentiation and inflammation response in periodontitis rats through Mmp9. Cell Microbiol, 2019; 21, e12979. [50] Kang LX, Miao YB, Jin Y, et al. Exosomal miR-205-5p derived from periodontal ligament stem cells attenuates the inflammation of chronic periodontitis via targeting XBP1. Immun Inflamm Dis, 2023; 11, e743. doi: 10.1002/iid3.743 [51] Li J, Li L, Wang XP, et al. Porphyromonas gingivalis inhibition of MicroRNA-205-5p expression modulates proinflammatory cytokines in gingival epithelial cells. Biochem Genet, 2020; 58, 566−79. doi: 10.1007/s10528-020-09957-y [52] Li W, Wang JW, Hao WJ, et al. MicroRNA-543-3p down-regulates inflammation and inhibits periodontitis through KLF6. Biosci Rep, 2021; 41, BSR20210138. doi: 10.1042/BSR20210138 [53] Hua B, Xiang JB, Guo L, et al. MicroRNA-212-5p regulates the inflammatory response of periodontal ligament cells by targeting myeloid differentiation factor 88. Arch Oral Biol, 2020; 118, 104831. doi: 10.1016/j.archoralbio.2020.104831 [54] Zhang Z, Shuai Y, Zhou F, et al. PDLSCs regulate angiogenesis of periodontal ligaments via VEGF transferred by exosomes in periodontitis. Int J Med Sci, 2020; 17, 558−67. doi: 10.7150/ijms.40918 [55] Huang PC, Jia LH. MicroRNA-28-5p as a potential diagnostic biomarker for chronic periodontitis and its role in cell proliferation and inflammatory response. J Dent Sci, 2022; 17, 1501−9. doi: 10.1016/j.jds.2022.04.031 [56] Xia YR, Zhou KC, Sun MJ, et al. The miR-223-3p regulates pyroptosis through NLRP3-caspase 1-GSDMD signal axis in periodontitis. Inflammation, 2021; 44, 2531−42. doi: 10.1007/s10753-021-01522-y [57] Liu Y, Yang J, Sun WB. Upregulation of IL-10 expression inhibits the proliferation of human periodontal ligament stem cells. Braz Oral Res, 2020; 34, e030. doi: 10.1590/1807-3107bor-2020.vol34.0030 [58] Guo RZ, Huang YP, Liu H, et al. Long non-coding RNA H19 participates in periodontal inflammation via activation of autophagy. J Inflamm Res, 2020; 13, 635−46. doi: 10.2147/JIR.S276619 [59] Huang NN, Li CX, Sun WJ, et al. Long non-coding RNA TUG1 participates in LPS-induced periodontitis by regulating miR-498/RORA pathway. Oral Dis, 2021; 27, 600−10. doi: 10.1111/odi.13590 [60] Yang QL, Liu P, Han YN, et al. Long noncoding RNA GAS5 alleviates the inflammatory response of human periodontal ligament stem cells by regulating the NF-κB signalling pathway. Eur J Orthod, 2022; 44, 669−78. doi: 10.1093/ejo/cjac030 [61] Cheng L, Fan YL, Cheng J, et al. Long non-coding RNA ZFY-AS1 represses periodontitis tissue inflammation and oxidative damage via modulating microRNA-129-5p/DEAD-Box helicase 3 X-linked axis. Bioengineered, 2022; 13, 12691−705. doi: 10.1080/21655979.2021.2019876 [62] Liu W, Zheng YY, Chen B, et al. LncRNA papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) regulates the proliferation of human periodontal ligament stem cells and toll-like receptor 4 (TLR4) expression to improve periodontitis. BMC Oral Health, 2019; 19, 108. doi: 10.1186/s12903-019-0802-9 [63] Wangzhou K, Gong L, Liu C, et al. LncRNA MAFG-AS1 regulates human periodontal ligament stem cell proliferation and Toll-like receptor 4 expression. Oral Dis, 2020; 26, 1302−7. doi: 10.1111/odi.13330 [64] Ruan DP, Wu CY, Zhang Y, et al. LncRNA LOXL1-AS1 inhibits proliferation of PDLSCs and downregulates IL-1β in periodontitis patients. J Periodontal Res, 2022; 57, 324−31. doi: 10.1111/jre.12962 [65] Wu XY, Cao ZY, Chen H, et al. Downregulation of Linc-RNA activator of myogenesis lncRNA participates in FGF2-mediated proliferation of human periodontal ligament stem cells. J Periodontol, 2020; 91, 422−7. doi: 10.1002/JPER.19-0317 [66] Shi B, Shao BY, Yang C, et al. Upregulation of JHDM1D-AS1 protects PDLSCs from H2O2-induced apoptosis by decreasing DNAJC10 via phosphorylation of eIF2α. Biochimie, 2019; 165, 48−56. doi: 10.1016/j.biochi.2019.06.018 [67] Han Y, Wang F, Shao LQ, et al. LncRNA TUG1 mediates lipopolysaccharide-induced proliferative inhibition and apoptosis of human periodontal ligament cells by sponging miR-132. Acta Biochim Biophys Sin (Shanghai), 2019; 51, 1208−15. doi: 10.1093/abbs/gmz125 [68] Chen H, Lan ZD, Li QM, et al. Abnormal expression of long noncoding RNA FGD5-AS1 affects the development of periodontitis through regulating miR-142-3p/SOCS6/NF-κB pathway. Artif Cells Nanomed Biotechnol, 2019; 47, 2098−106. doi: 10.1080/21691401.2019.1620256 [69] Wang XY, Wang YH. LncRNA DCST1-AS1 inhibits PDLCs' proliferation in periodontitis and may bind with miR-21 precursor to upregulate PLAP-1. J Periodontal Res, 2021; 56, 256−64. doi: 10.1111/jre.12809 [70] Hua L, Zhang XH. MALAT1 regulates osteogenic differentiation of human periodontal ligament stem cells through mediating miR-155-5p/ETS1 axis. Tissue Cell, 2021; 73, 101619. doi: 10.1016/j.tice.2021.101619 [71] Wang ZH, Wang DZ, Guo S, et al. Long noncoding RNA distal-less homeobox 2 antisense 1 restrains inflammatory response and apoptosis of periodontal ligament cells by binding with microRNA-330-3p to regulate Ro60, Y RNA binding protein. Arch Oral Biol, 2022; 133, 105298. doi: 10.1016/j.archoralbio.2021.105298 [72] Wang L, Li YL, Hong FF, et al. Circ_0062491 alleviates LPS-induced apoptosis and inflammation in periodontitis by regulating miR-498/SOCS6 axis. Innate Immun, 2022; 28, 174−84. doi: 10.1177/17534259211072302 [73] Du WW, Wang L, Liao Z, et al. Circ_0085289 alleviates the progression of periodontitis by regulating let-7f-5p/SOCS6 pathway. Inflammation, 2021; 44, 1607−19. doi: 10.1007/s10753-021-01445-8 [74] Wang J, Du CC, Xu LL. Circ_0081572 inhibits the progression of periodontitis through regulating the miR-378h/RORA axis. Arch Oral Biol, 2021; 124, 105053. doi: 10.1016/j.archoralbio.2021.105053 [75] Li MY, Du MY, Wang YL, et al. CircRNA Lrp6 promotes cementoblast differentiation via miR-145a-5p/Zeb2 axis. J Periodontal Res, 2021; 56, 1200−12. doi: 10.1111/jre.12933 [76] Li W, Zhang Z, Li YZ, et al. Abnormal hsa_circ_0003948 expression affects chronic periodontitis development by regulating miR-144-3p/NR2F2/PTEN signaling. J Periodontal Res, 2022; 57, 316−23. doi: 10.1111/jre.12961 [77] Li Q, Hu ZP, Yang F, et al. Circ_0066881 targets miR-144-5p/RORA axis to alleviate LPS-induced apoptotic and inflammatory damages in human periodontal ligament cells. Innate Immun, 2022; 28, 164−73. doi: 10.1177/17534259221079812 [78] Yang N, Li Y, Wang G, et al. Tumor necrosis factor-α suppresses adipogenic and osteogenic differentiation of human periodontal ligament stem cell by inhibiting miR-21/Spry1 functional axis. Differentiation, 2017; 97, 33−43. doi: 10.1016/j.diff.2017.08.004 [79] Ma L, Wu D. MicroRNA-383-5p regulates osteogenic differentiation of human periodontal ligament stem cells by targeting histone deacetylase 9. Arch Oral Biol, 2021; 129, 105166. doi: 10.1016/j.archoralbio.2021.105166 [80] Liu ZN, Chen X, Zhang ZP, et al. Nanofibrous spongy microspheres to distinctly release mirna and growth factors to enrich regulatory T cells and rescue periodontal bone loss. ACS Nano, 2018; 12, 9785−99. doi: 10.1021/acsnano.7b08976 [81] Li LY, Liu WJ, Wang H, et al. Mutual inhibition between HDAC9 and miR-17 regulates osteogenesis of human periodontal ligament stem cells in inflammatory conditions. Cell Death Dis, 2018; 9, 480. doi: 10.1038/s41419-018-0480-6 [82] Yang Y, Ren DP, Zhao D, et al. MicroRNA-203 mediates Porphyromonas gingivalis LPS-induced inflammation and differentiation of periodontal ligament cells. Oral Dis, 2023; 29, 1715−25. doi: 10.1111/odi.14132 [83] Hong L, Sharp T, Khorsand B, et al. MicroRNA-200c represses IL-6, IL-8, and CCL-5 expression and enhances osteogenic differentiation. PLoS One, 2016; 11, e0160915. doi: 10.1371/journal.pone.0160915 [84] Akkouch A, Zhu M, Romero-Bustillos M, et al. MicroRNA-200c attenuates periodontitis by modulating proinflammatory and osteoclastogenic mediators. Stem Cells Dev, 2019; 28, 1026−36. doi: 10.1089/scd.2019.0027 [85] Krongbaramee T, Zhu M, Qian QW, et al. Plasmid encoding microRNA-200c ameliorates periodontitis and systemic inflammation in obese mice. Mol Ther Nucleic Acids, 2021; 23, 1204−16. doi: 10.1016/j.omtn.2021.01.030 [86] Wang L, He YY, Ning WC. Role of enhancer of zeste homolog 2 in osteoclast formation and periodontitis development by downregulating microRNA-101-regulated VCAM-1. J Tissue Eng Regen Med, 2021; 15, 534−45. doi: 10.1002/term.3187 [87] Lian JX, Wu XW, Liu Y, et al. Potential roles of miR-335-5p on pathogenesis of experimental periodontitis. J Periodontal Res, 2020; 55, 191−8. doi: 10.1111/jre.12701 [88] Ge Y, Li J, Hao Y, et al. MicroRNA-543 functions as an osteogenesis promoter in human periodontal ligament-derived stem cells by inhibiting transducer of ERBB2, 2. J Periodontal Res, 2018; 53, 832−41. doi: 10.1111/jre.12572 [89] Zhang KK, Geng YD, Wang SB, et al. MicroRNA-26a-5p targets Wnt5a to regulate osteogenic differentiation of human periodontal ligament stem cell from inflammatory microenvironment. Chin J Stomatol, 2019; 54, 662−9. (In Chinese [90] Li J, Jin F, Cai M, et al. LncRNA nron inhibits bone resorption in periodontitis. J Dent Res, 2022; 101, 187−95. doi: 10.1177/00220345211019689 [91] Wang SW, Duan Y. LncRNA OIP5-AS1 inhibits the lipopolysaccharide-induced inflammatory response and promotes osteogenic differentiation of human periodontal ligament cells by sponging miR-92a-3p. Bioengineered, 2022; 13, 12055−66. doi: 10.1080/21655979.2022.2067291 [92] Yang QL, Han YN, Liu P, et al. Long noncoding RNA GAS5 promotes osteogenic differentiation of human periodontal ligament stem cells by regulating GDF5 and p38/JNK signaling pathway. Front Pharmacol, 2020; 11, 701. doi: 10.3389/fphar.2020.00701 [93] Liu XW, Zhou Y. Downregulation of lncRNA ANRIL inhibits osteogenic differentiation of periodontal ligament cells via sponging miR-7 through NF- κB pathway. Anal Cell Pathol, 2021; 2021, 7890674. [94] Bian MX, Yu Y, Li YZ, et al. Upregulating the expression of LncRNA ANRIL promotes osteogenesis via the miR-7-5p/IGF-1R axis in the inflamed periodontal ligament stem cells. Front Cell Dev Biol, 2021; 9, 604400. doi: 10.3389/fcell.2021.604400 [95] Xu YR, Qin W, Guo DH, et al. LncRNA-TWIST1 promoted osteogenic differentiation both in PPDLSCs and in HPDLSCs by inhibiting TWIST1 expression. Biomed Res Int, 2019; 2019, 8735952. [96] Wu D, Yin L, Sun DG, et al. Long noncoding RNA TUG1 promotes osteogenic differentiation of human periodontal ligament stem cell through sponging microRNA-222-3p to negatively regulate Smad2/7. Arch Oral Biol, 2020; 117, 104814. doi: 10.1016/j.archoralbio.2020.104814 [97] Feng YM, Wan PB, Yin LL. Long noncoding RNA X-Inactive Specific Transcript (XIST) promotes osteogenic differentiation of periodontal ligament stem cells by sponging MicroRNA-214-3p. Med Sci Monit, 2020; 26, e918932. [98] Liu Y, Liu CP, Zhang AK, et al. Down-regulation of long non-coding RNA MEG3 suppresses osteogenic differentiation of periodontal ligament stem cells (PDLSCs) through miR-27a-3p/IGF1 axis in periodontitis. Aging (Albany NY), 2019; 11, 5334−50. [99] Guo JB, Zheng MQ. The regulation mechanism of LINC00707 on the osteogenic differentiation of human periodontal ligament stem cells. J Mol Histol, 2022; 53, 13−26. doi: 10.1007/s10735-021-10029-7 [100] Yu BH, Hu JH, Li Q, et al. CircMAP3K11 contributes to proliferation, apoptosis and migration of human periodontal ligament stem cells in inflammatory microenvironment by regulating TLR4 via miR-511 sponging. Front Pharmacol, 2021; 12, 633353. doi: 10.3389/fphar.2021.633353 [101] Li XB, Zheng YF, Zheng Y, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther, 2018; 9, 232. doi: 10.1186/s13287-018-0976-0 [102] Ye Y, Ke Y, Liu L, et al. CircRNA FAT1 regulates osteoblastic differentiation of periodontal ligament stem cells via miR-4781-3p/SMAD5 pathway. Stem Cells Int, 2021; 2021, 5177488. [103] Wang CL, Gong JX, Li D, et al. circ_0062491 alleviates periodontitis via the miR-142-5p/IGF1 axis. Open Med (Wars), 2022; 17, 638−47. doi: 10.1515/med-2022-0442 [104] Luan XH, Zhou XF, Fallah P, et al. MicroRNAs: harbingers and shapers of periodontal inflammation. Semin Cell Dev Biol, 2022; 124, 85−98. doi: 10.1016/j.semcdb.2021.05.030 [105] Xie YF, Shu R, Jiang SY, et al. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int J Oral Sci, 2011; 3, 125−34. doi: 10.4248/IJOS11046 [106] Luan XH, Zhou XF, Naqvi A, et al. MicroRNAs and immunity in periodontal health and disease. Int J Oral Sci, 2018; 10, 24. doi: 10.1038/s41368-018-0025-y [107] Kebschull M, Papapanou PN. Mini but mighty: microRNAs in the pathobiology of periodontal disease. Periodontol 2000, 2015; 69, 201−20. doi: 10.1111/prd.12095 [108] Mert S, Malyaran H, Craveiro RB, et al. Comparative analysis of proliferative and multilineage differentiation potential of human periodontal ligament stem cells from maxillary and mandibular molars. J Periodontol, 2023; 94, 882−95. doi: 10.1002/JPER.22-0706 [109] Xu JC, Yin YY, Lin Y, et al. Long non-coding RNAs: emerging roles in periodontitis. J Periodontal Res, 2021; 56, 848−62. doi: 10.1111/jre.12910 [110] Sayad A, Mirzajani S, Gholami L, et al. Emerging role of long non-coding RNAs in the pathogenesis of periodontitis. Biomed Pharmacother, 2020; 129, 110362. doi: 10.1016/j.biopha.2020.110362 [111] Zou YG, Li C, Shu FP, et al. lncRNA expression signatures in periodontitis revealed by microarray: the potential role of lncRNAs in periodontitis pathogenesis. J Cell Biochem, 2015; 116, 640−7. doi: 10.1002/jcb.25015 [112] Sánchez-Muñoz F, Martínez-Coronilla G, Leija-Montoya AG, et al. Periodontitis may modulate long-non coding RNA expression. Arch Oral Biol, 2018; 95, 95−9. doi: 10.1016/j.archoralbio.2018.07.023 [113] Li S, Liu X, Li H, et al. Integrated analysis of long noncoding RNA-associated competing endogenous RNA network in periodontitis. J Periodontal Res, 2018; 53, 495−505. doi: 10.1111/jre.12539 [114] Zheng W, Wang S, Wang JG, et al. Periodontitis promotes the proliferation and suppresses the differentiation potential of human periodontal ligament stem cells. Int J Mol Med, 2015; 36, 915−22. doi: 10.3892/ijmm.2015.2314 [115] Wang YH, Sun YY, Zheng P, et al. Long non-coding RNAs mortal obligate RNA transcript regulates the proliferation of human periodontal ligament stem cells and affects the recurrence of periodontitis. Arch Oral Biol, 2019; 105, 1−4. doi: 10.1016/j.archoralbio.2019.04.013 [116] Khotib J, Marhaeny HD, Miatmoko A, et al. Differentiation of osteoblasts: the links between essential transcription factors. J Biomol Struct Dyn, 2022, 1−20. [117] He Q, Yang SY, Gu XG, et al. Long noncoding RNA TUG1 facilitates osteogenic differentiation of periodontal ligament stem cells via interacting with Lin28A. Cell Death Dis, 2018; 9, 455. doi: 10.1038/s41419-018-0484-2 [118] Wang L, Wu F, Song Y, et al. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis, 2016; 7, e2327. doi: 10.1038/cddis.2016.125 [119] Yu WJ, Gu QS, Wu D, et al. Identification of potentially functional circRNAs and prediction of circRNA-miRNA-mRNA regulatory network in periodontitis: bridging the gap between bioinformatics and clinical needs. J Periodontal Res, 2022; 57, 594−614. doi: 10.1111/jre.12989 [120] Ye Y, Fu L, Liu L, et al. Integrative analysis of ceRNA networks in human periodontal ligament stem cells under hypoxia. Oral Dis, 2023; 29, 1197−213. doi: 10.1111/odi.14096 [121] Jiao KX, Walsh LJ, Ivanovski S, et al. The emerging regulatory role of circular RNAs in periodontal tissues and cells. Int J Mol Sci, 2021; 22, 4636. doi: 10.3390/ijms22094636 [122] Li J, Xie RY. Circular RNA expression profile in gingival tissues identifies circ_0062491 and circ_0095812 as potential treatment targets. J Cell Biochem, 2019; 120, 14867−74. doi: 10.1002/jcb.28748 [123] Xie LK, Chen JZ, Ren XB, et al. Alteration of circRNA and lncRNA expression profile in exosomes derived from periodontal ligament stem cells undergoing osteogenic differentiation. Arch Oral Biol, 2021; 121, 104984. doi: 10.1016/j.archoralbio.2020.104984 [124] Zheng YF, Li XB, Huang YP, et al. The circular RNA landscape of periodontal ligament stem cells during osteogenesis. J Periodontol, 2017; 88, 906−14. doi: 10.1902/jop.2017.170078 [125] Gu XG, Li XY, Jin Y, et al. CDR1as regulated by hnRNPM maintains stemness of periodontal ligament stem cells via miR-7/KLF4. J Cell Mol Med, 2021; 25, 4501−15. doi: 10.1111/jcmm.16541 [126] Wang YZ, Zhang XG, Wang JJ, et al. Inflammatory periodontal ligament stem cells drive M1 macrophage polarization via exosomal miR-143-3p-mediated regulation of PI3K/AKT/NF-κB signaling. Stem Cells, 2023; 41, 184−99. doi: 10.1093/stmcls/sxac087 [127] Wangzhou K, Lai ZY, Lu ZS, et al. MiR-143-3p inhibits osteogenic differentiation of human periodontal ligament cells by targeting KLF5 and inactivating the Wnt/β-catenin pathway. Front Physiol, 2021; 11, 606967. doi: 10.3389/fphys.2020.606967 [128] Zhou M, Hu H, Han YN, et al. Long non-coding RNA 01126 promotes periodontitis pathogenesis of human periodontal ligament cells via miR-518a-5p/HIF-1α/MAPK pathway. Cell Prolif, 2021; 54, e12957. doi: 10.1111/cpr.12957 [129] Zhu YT, Ai RS, Ding ZQ, et al. LncRNA-01126 inhibits the migration of human periodontal ligament cells through MEK/ERK signaling pathway. J Periodontal Res, 2020; 55, 631−41. doi: 10.1111/jre.12749 [130] Li JS, Wang MW, Song LT, et al. LncRNA MALAT1 regulates inflammatory cytokine production in lipopolysaccharide-stimulated human gingival fibroblasts through sponging miR-20a and activating TLR4 pathway. J Periodontal Res, 2020; 55, 182−90. doi: 10.1111/jre.12700 [131] Chen QC, Cao M, Ge HY. Knockdown of MALAT1 inhibits the progression of chronic periodontitis via targeting miR-769-5p/HIF3A axis. Biomed Res Int, 2021; 2021, 8899863. [132] Wang XF, Ma F, Jia PZ. LncRNA AWPPH overexpression predicts the recurrence of periodontitis. Biosci Rep, 2019; 39, BSR20190636. doi: 10.1042/BSR20190636 [133] Zhang L, Lv H, Cui YX, et al. The role of long non-coding RNA (lncRNA) nuclear paraspeckle assembly transcript 1 (NEAT1) in chronic periodontitis progression. Bioengineered, 2022; 13, 2336−45. doi: 10.1080/21655979.2021.2018387 [134] Wang HW, Qiao XT, Zhang C, et al. Long non-coding RNA LINC00616 promotes ferroptosis of periodontal ligament stem cells via the microRNA-370 / transferrin receptor axis. Bioengineered, 2022; 13, 13070−81. doi: 10.1080/21655979.2022.2076508 [135] Pan JX, Zhao L, Liu J, et al. Inhibition of circular RNA circ_0138959 alleviates pyroptosis of human gingival fibroblasts via the microRNA-527/caspase-5 axis. Bioengineered, 2022; 13, 1908−20. doi: 10.1080/21655979.2021.2020396 [136] Sun DD, Wu X, Lin SC, et al. Anti-apoptosis and anti-inflammation activity of circ_0097010 downregulation in lipopolysaccharide-stimulated periodontal ligament cells by miR-769-5p/Krüppel like factor 6 axis. J Dent Sci, 2023; 18, 310−21. doi: 10.1016/j.jds.2022.04.024 [137] Li SS, Xu HL, Li YY, et al. Circ_0138960 contributes to lipopolysaccharide-induced periodontal ligament cell dysfunction. Immun Inflamm Dis, 2022; 10, e732. doi: 10.1002/iid3.732 [138] Wang J, Wang ZN, Huang M, et al. Circ_0099630 participates in SPRY1-mediated repression in periodontitis. Int Dent J, 2023; 73, 136−43. doi: 10.1016/j.identj.2022.06.025 [139] Zhao XQ, Ao CB, Yan YT. The circular RNA circ_0099630/miR-940/receptor-associated factor 6 regulation cascade modulates the pathogenesis of periodontitis. J Dent Sci, 2022; 17, 1566−76. doi: 10.1016/j.jds.2022.04.005 [140] Wei YR, Peng ZJ. Hsa_circ_0099630 knockdown induces the proliferation and osteogenic differentiation and attenuates the apoptosis of porphyromonas gingivalis lipopolysaccharide-induced human periodontal ligament fibroblasts. Ann Transl Med, 2022; 10, 993. doi: 10.21037/atm-22-4209 [141] Yu M, Chi CY. lncRNA FGD5-AS1 and miR-130a can be used for prognosis analysis of patients with chronic periodontitis. Biomed Res Int, 2021; 2021, 8544914. [142] Wang YZ, Li Y, Shao P, et al. IL1β inhibits differentiation of cementoblasts via microRNA-325-3p. J Cell Biochem, 2020; 121, 2606−17. doi: 10.1002/jcb.29482 [143] Byun JS, Lee HY, Tian JW, et al. Effect of salivary exosomal miR-25-3p on periodontitis with insulin resistance. Front Immunol, 2022; 12, 775046. doi: 10.3389/fimmu.2021.775046 [144] Zhou XD, Luan X, Chen Z, et al. MicroRNA-138 inhibits periodontal progenitor differentiation under inflammatory conditions. J Dent Res, 2016; 95, 230−7. doi: 10.1177/0022034515613043 [145] Liu X, Yang B, Zhang Y, et al. miR-30a-5p inhibits osteogenesis and promotes periodontitis by targeting Runx2. BMC Oral Health, 2021; 21, 513. doi: 10.1186/s12903-021-01882-9 [146] Zhang YC, Li SY, Yuan SJ, et al. MicroRNA-23a inhibits osteogenesis of periodontal mesenchymal stem cells by targeting bone morphogenetic protein signaling. Arch Oral Biol, 2019; 102, 93−100. doi: 10.1016/j.archoralbio.2019.04.001 [147] Yao SQ, Zhao W, Ou QM, et al. MicroRNA-214 suppresses osteogenic differentiation of human periodontal ligament stem cells by targeting ATF4. Stem Cells Int, 2017; 2017, 3028647. [148] Chen WY. SNHG7 promotes the osteo/dentinogenic differentiation ability of human dental pulp stem cells by interacting with hsa-miR-6512-3p in an inflammatory microenvironment. Biochem Biophys Res Commun, 2021; 581, 46−52. doi: 10.1016/j.bbrc.2021.09.081 [149] Zhang Z, Wang MH, Zheng YL, et al. MicroRNA-223 negatively regulates the osteogenic differentiation of periodontal ligament derived cells by directly targeting growth factor receptors. J Transl Med, 2022; 20, 465. doi: 10.1186/s12967-022-03676-1 [150] Duan Y, An W, Wu YX, et al. Tetramethylpyrazine reduces inflammation levels and the apoptosis of LPS-stimulated human periodontal ligament cells via the downregulation of miR-302b. Int J Mol Med, 2020; 45, 1918−26. [151] Jia Q, Jiang WK, Ni LX. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch Oral Biol, 2015; 60, 234−41. doi: 10.1016/j.archoralbio.2014.10.007 [152] Peng W, Deng W, Zhang J, et al. Long noncoding RNA ANCR suppresses bone formation of periodontal ligament stem cells via sponging miRNA-758. Biochem Biophys Res Commun, 2018; 503, 815−21. doi: 10.1016/j.bbrc.2018.06.081 [153] Wang Z, Huang YL, Tan LJ. Downregulation of lncRNA DANCR promotes osteogenic differentiation of periodontal ligament stem cells. BMC Dev Biol, 2020; 20, 2. doi: 10.1186/s12861-019-0206-8 [154] Deng WJ, Wang XL, Zhang J, et al. Circ_0138959/miR-495-3p/TRAF6 axis regulates proliferation, wound healing and osteoblastic differentiation of periodontal ligament cells in periodontitis. J Dent Sci, 2022; 17, 1125−34. doi: 10.1016/j.jds.2022.01.010 [155] Zheng JJ, Zhu XM, He YN, et al. CircCDK8 regulates osteogenic differentiation and apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia. Ann N Y Acad Sci, 2021; 1485, 56−70. doi: 10.1111/nyas.14483 [156] Nisha KJ, Janam P, Harshakumar K. Identification of a novel salivary biomarker miR-143-3p for periodontal diagnosis: a proof of concept study. J Periodontol, 2019; 90, 1149−59. doi: 10.1002/JPER.18-0729 [157] Iaquinta MR, Lanzillotti C, Mazziotta C, et al. The role of microRNAs in the osteogenic and chondrogenic differentiation of mesenchymal stem cells and bone pathologies. Theranostics, 2021; 11, 6573−91. doi: 10.7150/thno.55664 [158] Wang X, Sun H, Liao H, et al. MicroRNA-155-3p mediates TNF-α-inhibited cementoblast differentiation. J Dent Res, 2017; 96, 1430−7. doi: 10.1177/0022034517718790 [159] Cao FD, Zhan JL, Chen XF, et al. miR-214 promotes periodontal ligament stem cell osteoblastic differentiation by modulating Wnt/β-catenin signaling. Mol Med Rep, 2017; 16, 9301−8. doi: 10.3892/mmr.2017.7821 [160] Tenkumo T, Rojas-Sánchez L, Vanegas Sáenz JR, et al. Reduction of inflammation in a chronic periodontitis model in rats by TNF-α gene silencing with a topically applied siRNA-loaded calcium phosphate paste. Acta Biomater, 2020; 105, 263−79. doi: 10.1016/j.actbio.2020.01.031 [161] Zhu DW, Xue D, Lai W, et al. microRNA-146a reverses the inhibitory effects of Porphyromonas gingivalis lipopolysaccharide on osteogenesis of human periodontal ligament cells. Chin J Stomatol, 2018; 53, 753−9. (In Chinese [162] Meng XM, Wang WJ, Wang XL. MicroRNA-34a and microRNA-146a target CELF3 and suppress the osteogenic differentiation of periodontal ligament stem cells under cyclic mechanical stretch. J Dent Sci, 2022; 17, 1281−91. doi: 10.1016/j.jds.2021.11.011 [163] Riahi Rad Z, Riahi Rad Z, Goudarzi H, et al. MicroRNAs in the interaction between host-bacterial pathogens: a new perspective. J Cell Physiol, 2021; 236, 6249−70. doi: 10.1002/jcp.30333 [164] Rovas A, Puriene A, Snipaitiene K, et al. Analysis of periodontitis-associated miRNAs in gingival tissue, gingival crevicular fluid, saliva and blood plasma. Arch Oral Biol, 2021; 126, 105125. doi: 10.1016/j.archoralbio.2021.105125 [165] Sipert CR, Morandini AC, Dionísio TJ, et al. MicroRNA-146a and microRNA-155 show tissue-dependent expression in dental pulp, gingival and periodontal ligament fibroblasts in vitro. J Oral Sci, 2014; 56, 157−64. doi: 10.2334/josnusd.56.157 -




 下载:
下载:



 Quick Links
Quick Links