-
Foodborne Campylobacter spp. are common causes of gastrointestinal infections worldwide. Campylobacter is the dominant pathogen in patients with diarrhea, according to surveillance data in the S district of Beijing[1,2]. Compared to other enteric pathogenic bacteria, such as Salmonella typhimurium and Escherichia coli (C. coli), Campylobacter jejuni (C. jejuni) does not utilize carbohydrates as a food source. Campylobacter have been labeled as asaccharalytic microorganisms because early biological investigations on substrate consumption demonstrated that C. jejuni is unable to utilize glucose and other six-carbon carbohydrates as a carbon source[3-5]. Recently, two campylobacteriosis outbreaks have been reported in this region[6-8]. However, the sources of these infections have not yet been identified.
Contaminated food was considered as the source of infection; however, identifying the source of contamination proved difficult. The food may have been contaminated by raw materials or became cross-contaminated from the kitchen environment. There were also a few reports about the food being possibly contaminated by infected food handlers.
On August 3, 2021, three students with diarrhea from the same school visited a local hospital in the S District of Beijing. According to an epidemic investigation, three other students with diarrhea were found at the same school. These students studied and lived in the school, and their three meals per day were in the school canteen, which did not have a food business license. There were no special processing rooms for cooked or cold dishes, and the sanitary conditions were poor. Daily food items mainly included all kinds of staple foods, cold dishes, fruits, soup gruel, etc., and samples were not reserved for any food items. A total of 10 stool samples, six from the students with diarrhea and four from asymptomatic food handlers, and five suspected food samples were collected on the day of pathogen detection.
Written consent for stool sample collection was obtained from the patients in this study and the data were analyzed anonymously. Ethics approval for this study was also obtained from the ethics committee of the Shunyi District Center for Disease Control and Prevention (CDC) and the academic committee of the National Institute for Communicable Disease Control and Prevention in China CDC.
Real-time PCR was performed for 12 major enteric pathogens (C. jejuni, C. coli, Salmonella, Shigella, diarrheagenic E. coli, Vibrio cholerae, Vibrio parahaemolyticus, Yersinia enterocolitica, Norovirus, Sapovirus, and Enteric Adenovirus). According to the manual, tests with cycle threshold (Ct) < 35 with a typical S curve is considered positive; whereas, tests with Ct > 40 or no typical S curve are considered negative. Furthermore, tests are considered weak positive if repeated tests provide 35
$ \leq $ Ct$ \leq $ 40. All suspected food samples were negative for all tested pathogens. The stool samples from six patients with diarrhea were positive for C. jejuni with Ct values of 19.81, 21.64, 27.45, 32.9, 28.96, and 28.08, respectively. One of the four asymptomatic food handlers tested positive for C. jejuni and C. coli, with Ct values of 25.35 and 24.04, respectively. Except for Campylobacter, all stool samples were negative for other tested pathogens.According to the results of the PCR screening, Campylobacter isolation from the stool samples was carried out using a Campylobacter isolation kit incorporating a membrane filter method (ZC-CAMPY-002, Qingdao Sinova Biotechnology Co. Ltd., Qingdao, China)[9]. In total, 35 isolates were selected from seven positive samples. Furthermore, 25 C. jejuni isolates were obtained from six patients, and six C. jejuni and four C. coli isolates were selected from the specimens of the asymptomatic cook.
The minimum inhibitory concentrations (MICs) of 11 antimicrobial compounds (erythromycin, azithromycin, nalidixic acid, ciprofloxacin, gentamicin, streptomycin, chloramphenicol, florfenicol, tetracycline, telithromycin, and clindamycin) were determined using the agar dilution method as previously described[9]. The breakpoints for resistance used in this study were those with a MIC value greater than or equal to the standard values used in the National Antimicrobial Resistance Monitoring System (NARMS-2014) for Campylobacter in the USA: erythromycin (≥ 32 μg /mL), azithromycin (≥ 8 μg/mL), nalidixic acid (≥ 64 μg/mL), ciprofloxacin (≥ 4 μg/mL), gentamicin (≥ 8 μg/mL), streptomycin (≥ 16 μg/mL), chloramphenicol (≥ 32 μg/mL), florfenicol (≥ 8 μg/mL), tetracycline (≥ 16 μg/mL), telithromycin (≥ 16 μg/mL), and clindamycin (≥ 8 μg/mL). All 31 C. jejuni isolates had the same susceptibility pattern: they were resistant to nalidixic acid and ciprofloxacin and sensitive to the other nine drugs. Whereas, four C. coli isolates were sensitive to florfenicol and clindamycin and resistant to the other nine drugs.
A total of seven C. jejuni strains from each of the six patients and the cook were selected for whole-genome sequencing. Draft genomes of the seven C. jejuni strains were sequenced using an Illumina HiSeq 2500 platform at the Beijing Genomics Institute. The contig number, full genome length, and GC% are listed in Table 1. The MLST sequence Type (ST) of the seven draft genome sequences were determined using the, Genome Comparator online tool from the PubMLST website (http://pubmlst.org/campylobacter). All seven C. jejuni strains belonged to ST11329 and clonal complex 22. The core genome single nucleotide polymorphisms (SNPs) were obtained using Snippy4.3.6 software (https://github.com/tseemann/snippy) and only seven SNPs were identified among these seven genomes (Figure 1). Whole-genome multilocus sequence typing (wgMLST) analysis was performed with fast-GeP (https://github.com/jizhang-nz/fast-GeP) using the genome of the strain from the cook (cook-cj) as a reference. The ad hoc wgMLST analysis found that among the 1665 reference coding sequences (CDSs), 1662 CDSs were complete and shared by the seven genome sequences. The difference matrix of the allele loci among the seven C. jejuni whole Genome Sequencing is presented in Figure 2. The neighbor-net phylogeny of the seven isolates was constructed using SplitsTree4[10] based on the shared loci, and is presented in Figure 3.
Table 1. General features of the Campylobacter jejuni genome sequences studied
Genome Contig Full genome length Maximum genome length Minimum genome length Mean genome length GC (%) Patient1-Cj 22 1,642,553 533,041 507 74,661.50 30.43 Patient2-Cj 20 1,643,663 533,040 507 82,183.15 30.44 Patient3-Cj 25 1,644,257 533,040 507 65,770.28 30.46 Patient4-Cj 20 1,643,664 533,040 507 82,183.20 30.44 Patient5-Cj 20 1,643,663 533,040 507 82,183.15 30.44 Patient6-Cj 33 1,652,616 533,040 507 50,079.27 30.51 Cook-Cj 20 1,643,666 533,043 507 82,183.30 30.44 
Figure 1. Difference in single nucleotide polymorphisms (SNPs) between core genomes of seven Campylobacter jejuni strains. The horizontal and vertical columns of the matrix represent the names of the isolates. The number in the matrix indicates the different SNP numbers between the isolates in the horizontal and vertical columns.
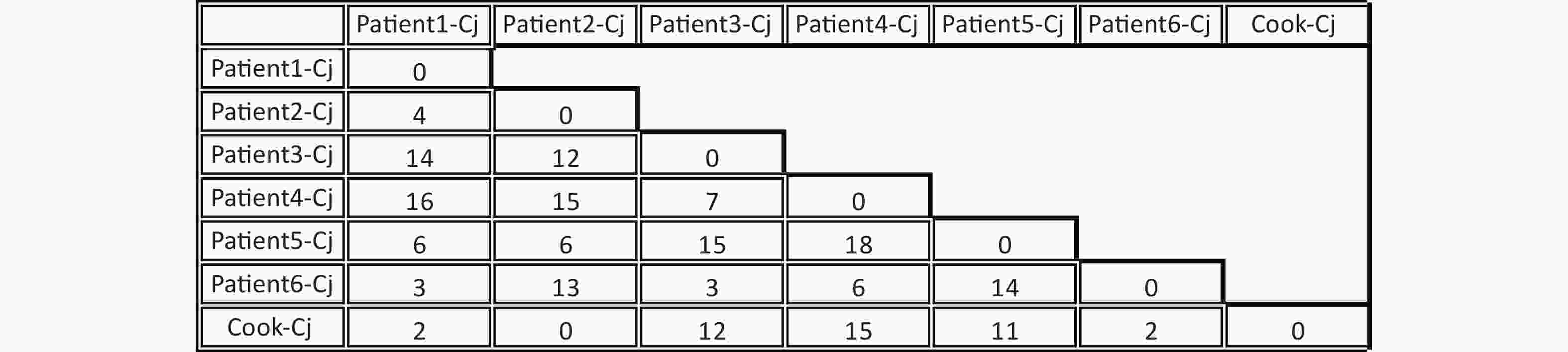
Figure 2. Difference matrix for alleles of the Whole-genome multilocus sequence typing (wgMLST) with Fast-GeP analysis. Allele sequences were searched with BLAST+(identity threshold ≥ 80) and cook-Cj was used as the reference genome. The horizontal and vertical columns of the matrix represent the names of the isolates. The number in the matrix indicates the different allele numbers between the isolates in the horizontal and vertical columns.
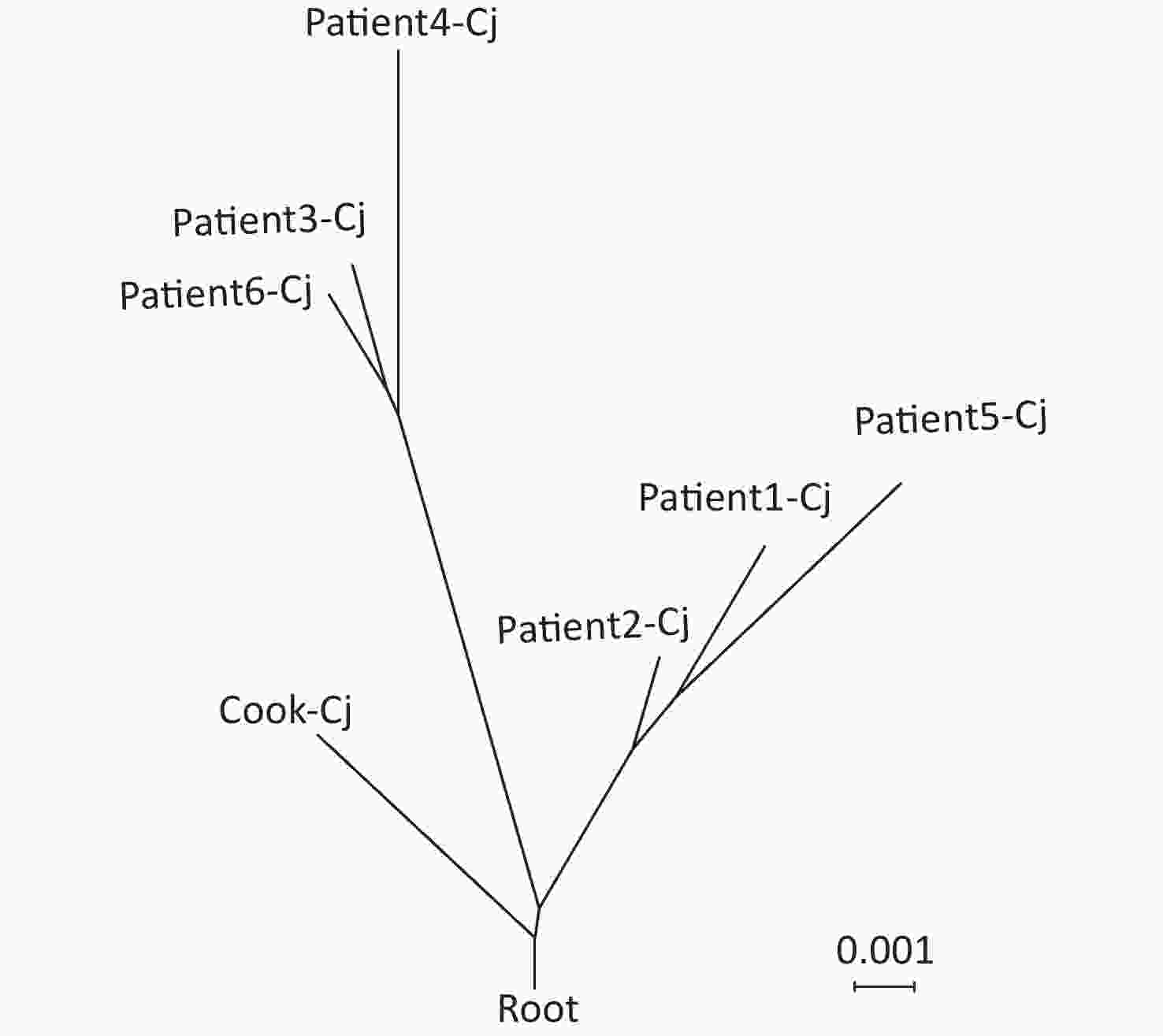
Figure 3. Neighbor-net phylogeny for alleles of core-genome multilocus sequence typing (cgMLST) loci of seven C. jejuni strains. The scale bar represents substitution per site. Cook-Cj represents the isolate from the cook, and patient-Cj represents the isolates from the patients.
The results of bacterial isolation, antimicrobial susceptibility testing, and genomic analysis support a campylobacteriosis outbreak in the school observed in this study. Through the phylogenomic tree, we found that the asymptomatic cook may have been the source of this outbreak. Foodhandler-associated foodborne outbreaks have been reported previously, and in one particular C. jejuni outbreak in Kansas in 1998, a cafeteria worker was identified as the source[11,12]. It is usually difficult to correctly identify the source of Campylobacter infections. In this study, we found that the asymptomatic food handler carried both C. jejuni and C. coli. However, patients with diarrhea only had C. jejuni infections. There was a possibility that both the asymptomatic cook and patients had the same contaminated food; however, there were two species colonization in the cook and only one species in the patients. The phylogenetic analysis based on the wgMLST indicated that the isolate carried by the cook was more similar to the ancestor of the investigated isolates from the patients. Based on our results, we speculated the possibility of the asymptomatic cook being the source of the infection.
Campylobacteriosis outbreaks observed in this study, and asymptomatic infections in cooks have been previously identified[4]. This study describes a campylobacteriosis outbreak in a school cafeteria, with an asymptomatic food handler as the most likely source. These findings emphasize that testing food-handling staff for Campylobacter, as well as for other foodborne pathogens, should be a part of the routine inspections of public food service establishments.
doi: 10.3967/bes2023.105
-
Abstract: In August 2021, three students with diarrhea from the same school visited a local hospital in the S district of Beijing. An epidemic investigation showed that there were more students with diarrhea in the same school and they had one meal together. Campylobacter jejuni was isolated from both patients with diarrhea and asymptomatic food handlers; however, the latter also carried Campylobacter coli. Phylogenomic analysis showed that there was a campylobacteriosis outbreak among the students, and the asymptomatic food handler may have been the source of the infection. Routine inspection and surveillance for Campylobacter is needed for the food producing staff, particularly those cooking in the cafeteria in schools or other public food services.
-
Key words:
- Campylobacter jejuni /
- Outbreak /
- Whole genome sequence /
- Asymptomatic food handler
注释: -
Figure 1. Difference in single nucleotide polymorphisms (SNPs) between core genomes of seven Campylobacter jejuni strains. The horizontal and vertical columns of the matrix represent the names of the isolates. The number in the matrix indicates the different SNP numbers between the isolates in the horizontal and vertical columns.
Figure 2. Difference matrix for alleles of the Whole-genome multilocus sequence typing (wgMLST) with Fast-GeP analysis. Allele sequences were searched with BLAST+(identity threshold ≥ 80) and cook-Cj was used as the reference genome. The horizontal and vertical columns of the matrix represent the names of the isolates. The number in the matrix indicates the different allele numbers between the isolates in the horizontal and vertical columns.
Table 1. General features of the Campylobacter jejuni genome sequences studied
Genome Contig Full genome length Maximum genome length Minimum genome length Mean genome length GC (%) Patient1-Cj 22 1,642,553 533,041 507 74,661.50 30.43 Patient2-Cj 20 1,643,663 533,040 507 82,183.15 30.44 Patient3-Cj 25 1,644,257 533,040 507 65,770.28 30.46 Patient4-Cj 20 1,643,664 533,040 507 82,183.20 30.44 Patient5-Cj 20 1,643,663 533,040 507 82,183.15 30.44 Patient6-Cj 33 1,652,616 533,040 507 50,079.27 30.51 Cook-Cj 20 1,643,666 533,043 507 82,183.30 30.44 -
[1] Li Y, Jia QL, Zhou GL, et al. Surveillance and analysis of etiology characteristics of campylobacter infection in adult diarrhea patients in Shunyi district of Beijing, 2016–2018. Dis Surveill, 2020; 35, 21−8. (In Chinese [2] Wang YY, Li Y, Zhang S, et al. Infection status and drug resistance of campylobacter in diarrhea patients in Shunyi district of Beijing, 2017. Dis Surveill, 2018; 33, 1048−53. (In Chinese [3] Parkhill J, Wren BW, Mungall K, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature, 2000; 403, 665−8. doi: 10.1038/35001088 [4] Ayna A, Moody PCE. Crystal structures of a dual coenzyme specific glyceraldehyde-3-phosphate dehydrogenase from the enteric pathogen Campylobacter jejuni. J Mol Struct, 2021; 1242, 130820. doi: 10.1016/j.molstruc.2021.130820 [5] Ayna A, Moody PCE. Activity of fructose-1, 6-bisphosphatase from Campylobacter jejuni. Biochem Cell Biol, 2020; 98, 518−24. doi: 10.1139/bcb-2020-0021 [6] Li Y, Gu YX, Lv JC, et al. Laboratory study on the gastroenteritis outbreak caused by a multidrug-resistant Campylobacter coli in China. Foodborne Pathog Dis, 2020; 17, 187−93. doi: 10.1089/fpd.2019.2681 [7] Li Y, Zhou GL, Gao P, et al. Gastroenteritis outbreak caused by Campylobacter jejuni - Beijing, China, August, 2019. China CDC Wkly, 2020; 2, 422−5. doi: 10.46234/ccdcw2020.108 [8] Zou L, Li Y, Zhou GL, et al. Laboratory investigation for one gastroenteritis outbreak caused by Campylobacter jejuni. Chin J Epidemiol, 2020; 41, 1692−6. (In Chinese [9] Li Y, Zhang S, He M, et al. Prevalence and molecular characterization of Campylobacter spp. isolated from patients with diarrhea in Shunyi, Beijing. Front Microbiol, 2018; 9, 52. doi: 10.3389/fmicb.2018.00052 [10] Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol, 2006; 23, 254−67. doi: 10.1093/molbev/msj030 [11] Ercoli L, Gallina S, Nia Y, et al. Investigation of a staphylococcal food poisoning outbreak from a Chantilly cream dessert, in Umbria (Italy). Foodborne Pathog Dis, 2017; 14, 407−13. doi: 10.1089/fpd.2016.2267 [12] Olsen SJ, Hansen GR, Bartlett L, et al. An outbreak of Campylobacter jejuni infections associated with food handler contamination: the use of pulsed-field gel electrophoresis. J Infect Dis, 2001; 183, 164−7. doi: 10.1086/317657 -




 下载:
下载:





 Quick Links
Quick Links