HTML
-
Legionella pneumophila (L. pneumophila ) is a gram-negative, facultative, intracellular pathogenic bacterium that widely exists in natural and man-made aquatic environments worldwide and causes Legionnaires' disease (LD)[1-2]. These bacteria are transmitted from the environment to humans via inhalation or aspiration of the aerosols they are present in[3-4]. There have been 61 known Legionella species and over 28 species reported to cause human disease[5]. L. pneumophila accounted for 90% was from the identified clinical cases, and over 75% of all LD cases of were caused due to the infection of L. pneumophila serogroup 1 (LP1); the remaining were caused by the other serogroups (LP2-LP15)[6-7].
Previous studies have described L. pneumophila pollution in public areas, including cooling towers, hot springs, foot spas, potable water systems in large facilities, hotels, hospitals, and public baths, and these bacteria can survive in various environments; they have also shown the relationship between the strains isolated from the environmental and biological specimens of LD patients[6, 8-14]. In China, since the first case of LD reported in 1982, the disease has been reported sporadically in many cities. However, there have been few reports of an outbreak, which could be attributed to the lack of a nationwide monitoring network[15]. The first LD case in Sichuan was reported in 1989, and the other case was reported in 2000. So, it is necessary to investigate the presence of L. pneumophila in the public environment and to take public health perspective in Sichuan.
Several subtyping techniques have been reported to identify and characterize L. pneumophila strains, such as Sequence-Based Typing (SBT), multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA), pulsed-field gel electrophoresis (PFGE), and amplified fragment length polymorphism (AFLP)[16] and so on. SBT is the current gold standard typing method for investigation of LD outbreaks and rapid identification of isolates that are closely related. While, MLVA is a rapid, reproducible and epidemiologically meaningful typing tool, which can be performed at low cost. In the present study, SBT and MLVA are used to identify L. pneumophila isolates, and the results can provide data for a specific geographic region and make it possible to perform phylogenetic analyses. The characteristics is important to explore the transmission routes of environmental and clinical isolates.
Several virulence genes have been proven to exist in L. pneumophila, including mip, ira, lvr, dot, lvh, and rtxA. The previous studies reported that lvh and rtxA regions were found more frequently in the strains associated with human disease[17-19]. The rtxA which encodes protein relating with adherence, cytotoxicity, and pore formation[18] is an important pathogenic factor involved in host cell invasion by L. pneumophila and other pathogenic bacteria[19]. The lvh derives proteins for type ⅣA secretion system that contributes to conjugation and virulence[20]. Specific genetic loci lvh and rtxA were chosen because they were suggested as appropriate indicators of L. pneumophila virulence. We examined the pathogenicity-specific genetic loci lvh and rtxA to survey whether the strains isolated in public environment had the virulence.
The invasion and intracellular replication of L. pneumophila within protozoa play major roles in the transmission of LD, and important relationships between the intracellular growth ability of L. pneumop-hila within protozoa or macrophages and human LD should been analysed. A variety of cell types, have been used as host cells to characterize the intracellular growth of L. pneumophila and to study the relationship between L. pneumophila and host cells. The J774 cell derived from the male BALB/c mouse has normal macrophage function. Therefore, we employed the J774 cell as the host to study the intracellular growth of 33 L. pneumophila strains with different STs.
The aim of the present study was to investigate the molecular genetic characteristics of L. pneumophila in Sichuan province, China. The isolates were identified to L. pneumophila including the 16S rRNA gene analysis and serum agglutination assay. Then we analyzed their molecular characteristics, the virulence genes (rtxA and lvh) and the intracellular growth ability.
-
The environmental isolates were collected based on the protocol according to ISO11731 and the National standard: Hygienic specification of central air conditioning ventilation system in public buildings (WS394-2012). A total of 79 L. pneumophila strains were included in this study: 64 strains were isolated during regular surveillance events which were performed for cooling towers and bath water in Sichuan Province. Thirteen strains were isolated from hot springs through an investigation regarding hot spring recreational areas in 2014, including two cities: Ya'an (10 strains) and Kang ding (3 strains). Ten strains were isolated from city fountains during fountain surveillance for L. pneumophila in 2010 in Chengdu. Lastly, 2 strains were isolated from the sputum of patients, of which one was from the case reported firstly in Sichuan province, in 1989 (Supplementary Table S1 available in www.besjournal.com).
Strain ID Year City Facility Type Source Strain ID Year City Facility Type Source 8931 1989 Chengdu Hospital Human XJ2010017 2010 Chengdu Hotel Cooling Tower 97008 1997 Chengdu Hotel Cooling Tower XJ2010025 2010 Chengdu Hotel Cooling Tower WZL 2000 Chengdu Hospital Human XJ2010041 2010 Chengdu Hotel Cooling Tower 00-02 2000 Chengdu Hotel Cooling Tower XJ2010044 2010 Chengdu Hotel Cooling Tower 00-03 2000 Chengdu Hotel Cooling Tower XJ2010051 2010 Neijiang Hotel Cooling Tower 2007026 2007 Chengdu Hotel Cooling Tower XJ2010098 2010 Chengdu Hotel Cooling Tower 2007091 2007 Chengdu Hotel Cooling Tower XJ20110891 2011 Luzhou Hotel Cooling Tower 2007094 2007 Chengdu Hotel Cooling Tower XJ20110895 2011 Zigong Hotel Cooling Tower 2007101 2007 Chengdu Hotel Cooling Tower XJ20111923 2011 Nanchong Hotel Cooling Tower 2007102 2007 Chengdu Hotel Cooling Tower XJ2011924 2011 Nanchong Hotel Cooling Tower 2007103 2007 Chengdu Hotel Cooling Tower XJ2011925 2011 Nanchong Hotel Cooling Tower 2007104 2007 Chengdu Hotel Cooling Tower XJ20120317 2012 Chengdu Hotel Bath Water 2007106 2007 Chengdu Hotel Cooling Tower XJ20130044 2013 Chengdu Hotel Cooling Tower 2007109 2007 Chengdu Hotel Cooling Tower XJ20130047 2013 Chengdu Hotel Bath Water 2007112 2007 Chengdu Hotel Cooling Tower XJ20130052 2013 Chengdu Hotel Cooling Tower 2008090 2008 Chengdu Hotel Cooling Tower XJ20130055 2013 Chengdu Hotel Cooling Tower 2008422 2008 Chengdu Hotel Cooling Tower XJ20130275 2013 Chengdu Hotel Bath Water 2008423 2008 Chengdu Hotel Cooling Tower XJ20130461 2013 Ya'an Hotel Cooling Tower 2009022 2009 Chengdu Hotel Cooling Tower XJ20140347 2014 Kangding Public Bath Hot Spring 2009029 2009 Chengdu Hotel Cooling Tower XJ20140348 2014 Kangding Public Bath Hot Spring 2009039 2009 Leshan Hotel Cooling Tower XJ20140360 2014 Kangding Public Bath Hot Spring 2009040 2009 Chengdu Hotel Cooling Tower XJ20140374 2014 Ya'an Public Bath Hot Spring 2009042 2009 Mianyang Hotel Cooling Tower XJ20140375 2014 Ya'an Public Bath Hot Spring 2009045 2009 Chengdu Hotel Cooling Tower XJ20140376 2014 Ya'an Public Bath Hot Spring 2009047 2009 Chengdu Hotel Cooling Tower XJ20140378 2014 Ya'an Public Bath Hot Spring 2009058 2009 Chengdu Hotel Cooling Tower XJ20140379 2014 Ya'an Public Bath Hot Spring 2009065 2009 Chengdu Hotel Bath Water XJ20140380 2014 Ya'an Public Bath Hot Spring 2009141 2009 Chengdu Hotel Cooling Tower XJ20140381 2014 Ya'an Public Bath Hot Spring 2009142 2009 Chengdu Hotel Bath Water XJ20140382 2014 Ya'an Public Bath Hot Spring CD1 2010 Chengdu City Center Fountain XJ20140386 2014 Ya'an Public Bath Hot Spring CD2 2010 Chengdu City Center Fountain XJ20140387 2014 Ya'an Public Bath Hot Spring CD3 2010 Chengdu City Center Fountain XJ20140450 2014 Neijiang Hotel Cooling Tower CD4 2010 Chengdu City Center Fountain XJ20140453 2014 Neijiang Hotel Cooling Tower CD5 2010 Chengdu City Center Fountain XJ20140454 2014 Nanchong Hotel Cooling Tower CD6 2010 Chengdu City Center Fountain XJ20140457 2014 Luzhou Hotel Cooling Tower CD7 2010 Chengdu City Center Fountain XJ20140469 2014 Neijiang Hotel Cooling Tower CD8 2010 Chengdu City Center Fountain 1607-GC-019A 2016 Luzhou Hotel Cooling Tower CD9 2010 Chengdu City Center Fountain 1609-GC-021A 2016 Luzhou Hotel Cooling Tower CD10 2010 Chengdu City Center Fountain 1609-GC-023A 2016 Luzhou Hotel Cooling Tower LP8 2010 Chengdu Hotel Cooling Tower Table Supplementary Table S1. Information of Strains Used in this Study
-
All isolates were identified using Legionella-specific PCR targeting the 16S rRNA gene[21] to determine the species and serogroup of L. pneumophila were determined by slide agglutination with polyclonal antisera (Tianjin Biochip Corporation).
Seventy-nine strains were lined on buffered charcoal yeast extract (BCYE) supplemented with glycine (3 g/L), vancomycin (1 mg/L), polymyxin B (80, 000 UI/L) and cycloheximide (80 mg/L) (GVPC) agar (BeiJing Land Bridge Technology Co., Ltd.), and the plates were incubated at 37 ℃ with 5% CO2 for 3-4 days, the bacterial colonies were collected to prepare for Genomic DNA. Genomic DNA was extr-acted with a QIAamp DNA minikit (Qiagen, Dusseldorf, Germany) according to the manufacturer's guidelines.
-
The genotyping was performed by means of the standard SBT method of the European Working Group for Legionella Infections (EWGLI) including 7 genes (flaA, pilE, asd, mip, mompS, proA, and neuA)[22-23]. PCR was performed with a reaction volume of 25 µL including 22 µL of the PCR mixture, 1 µL of each primer (10 µmol/L) (TsingKe Biological Technology Co., Ltd.), and 20 ng of the DNA template. PCRs were performed with an initial denaturation step at 95 ℃ for 5 min; followed by 35 cycles, each consisting of initial denaturation at 95 ℃ for 30 s, annealing at 55 ℃ for 30 s, and extension at 72 ℃ for 45 s; and final extension at 72 ℃ for 10 min. Then, each gene was sequenced by TsingKe Biological Technology Co., Ltd. The online Legionella SBT Quality Tool (http://www.hpa-bioinformatics.org.uk/cgi-bin/legionella/sbt-/seq_assemble_legionella1.cgi) was used to assign individual allele numbers (e.g., 1-4-3-1-1-1-1) for the 7 genes described previously and a sequence type (ST) represented by a number (e.g., ST1).
The BioNumerics software (Applied Maths, Kortrijk, Belgium) was used to create a maximum likelihood tree using the unweighted pair group method with arithmetic means (UPGMA), and this revealed the genetic distance among the 79 identified strains. The clonal complexes were also analyzed with this software to create minimum spanning trees (MST). In the MSTs, the clusters of related STs that descended from a common ancestor were defined as clonal groups (CGs)[9].
-
In this study, VNTR genotyping of the strains selected were conducted 8-locus MLVA (MLVA-8) including designated Lpms1b, Lpms3, Lpms13, Lpms17, Lpms19b, Lpms33, Lpms34, and Lpms35[24-25] (Supplementary Table S2 available in www.besjournal.com), according to which the primers were designed for PCR amplification of the markers. PCR was divided into three groups (VNTR1, VNTR2, VNTR3) and conducted in a reaction volume of 25 µL, respe-ctively, containing 12.5 µL of 2× PCR mixture, 1 µL of each fluorescent-labeled primer (10 µmol/L) (TsingKe Biological Technology Co., Ltd.), 20 ng of the DNA template, and 5.5 µL (VNTR1, VNTR2) or 7.5 µL (VNTR3) filtered sterile water. The PCR protocol was as follows: initial denaturation at 94 ℃ for 5 min; followed by 35 cycles, each consisting of initial denaturation at 94 ℃ for 30 s, annealing at 60 ℃ for 30 s, and extension at 72 ℃ for 45 s; and final extension at 72 ℃ for 10 min. PCR products were identified by dye-based capillary electrophoresis (TsingKe Biological Techn-ology Co., Ltd.)[26]. The number of repeats in the alleles was estimated by subtracting the invariable flanking region from the amplicon size divided by the repeat unit length, as determined for reference strain Philadelphia-1 according to a method described previously[24].
Loci Primer Sequences (5' to 3') Repeat Size/bp Panel Dye Lpms1b ACGAGCATATGACAAAGCCTTG 45 VNTR1 Fam CGGATCATCAGGTATTAATCGC Lpms3 CAACCAATGAAGCAAAAGCA 96 VNTR1 Hex AGGGGTTGATGGTCTCAATG Lpms13 CAATAGCATCGGACTGAGCA 24 VNTR1 TAMRA TGCCTGTGTATCTGGAAAAGC Lpms19 GAACTATCAGAAGGAGGCGAT 21 VNTR2 Fam GGAGTTTGACTCGGCTCAGG Lpms17 CAGCTCACCCCGTATCACTT 39 VNTR2 Hex TAACATCAATGACCGCGAAA Lpms33 ACCACAGCAGTTTGAACATAAT 125 VNTR2 TAMRA GGGAGAAGTTATAGATCTATTCG Lpms34 GAAAAGGAATAAGGCGCAGCAC 125 VNTR3 Hex AAACCTCGTTGGCCCCTCGCTT Lpms35 CTGAAACAGTTGAGGATGTGA 18 VNTR3 TAMRA TTATCAACCTCATCATCCCTG Table Supplementary Table S2. L. pneumophila MLVA-8 Setup for Capillary Electrophoresis
The BioNumerics software was used to create a maximum likelihood tree using UPGMA, which revealed the genetic distance among the 42 strains.
-
Two virulence genes lvh and rtxA, which were previously proven to have a strong correlation with LD in clinical cases, were chosen for examination in 79 strains. The primers and PCR reaction procedure were the same as those described previously[17].
-
Thirty-three L. pneumophila strains were selected to represent different 33 STs, sources, and sero-groups and subjected to an intracellular growth assay repeated three independent times, with the reference strain Philadelphia-1 strain (ATCC33152) as a positive control.
All the strains were added to J774 cells, and together to co-culture for 1 h, 24 h, 48 h, and 72 h, respectively, then the CFUs of intracellular bacteria were counted to determine growth[9]. J774 cells were incubated in RPMI 1640 tissue culture medium containing 10% calf serum at 37 ℃ with 5% CO2. Phosphate-buffered saline (PBS) was used to dilute the bacteria to 1 × 108 CFU/mL as the primary dilution, which was then diluted 10-fold with the culture medium containing the J774 cells (2 × 105 per well) to reach a multiplicity of infection (MOI) of approximately 10. The 24-well plates containing the culture medium and bacteria were incubated at 37 ℃ in 5% CO2 and with saturated humidity. To measure internalization, PBS was used to wash away extracellular bacteria and other potentially interfering substances. Then, 1 mL of sterile distilled water was added to the wells to release internal bacteria from the host cells, and the CFUs were counted by plating dilutions of the medium on BCYE agar plates (Oxoid Microbiological Products, UK).
Bacterial Isolates and Culture Conditions
Strain Identification
SBT and Data Analyses
MLVA and Data Analysis
Detection of Virulence Genes
Intracellular Growth Assay
-
Legionella-specific 16S rRNA gene was detected for identifying the 79 isolates included in the present study, by which all of the strains belonged to Legionella species, of which eleven serogroups were identified. Two serogroups (LP9 and LP14) were simultaneously identified in one strain (XJ20140386) isolated from a hot spring. The major serogroup was LP1 containing 43 strains (54.43%), the following was LP7 of 12 strains (15.19%), LP6 of 6 strains (7.59%), LP11 of 4 strains (5.06%), LP12 of 3 strains (3.80%), and 9 strains belonging to various serogroups: 2 LP3 strains (2.53%), 2 LP9 strains (2.53%), 2 LP13 strains (2.53%), 2 LP15 strains (2.53%), 1 LP5 (1.27%), and LP10 strain respectively (Table 1).
Serogroup Cooling Tower (n = 49) Bath Water (n = 5) Fountain (n = 10) Hot Spring (n = 13) Clinical (n = 2) Total (n = 79) No. Proportion (%) No. Proportion (%) No. Proportion (%) No. Proportion (%) No. Proportion (%) No. Proportion (%) LP1 30 61.22 2 40.00 4 40.00 6 46.15 1 50.00 43 54.43 LP3 0 0.00 0 0.00 0 0.00 2 15.38 0 0.00 2 2.53 LP5 0 0.00 1 20.00 0 0.00 0 0.00 0 0.00 1 1.27 LP6 4 8.16 0 0.00 0 0.00 1 7.69 1 50.00 6 7.59 LP7 10 20.41 0 0.00 2 20.00 0 0.00 0 0.00 12 15.19 LP9 2 4.08 0 0.00 0 0.00 0 0.00 0 0.00 2 2.53 LP10 1 2.04 0 0.00 0 0.00 0 0.00 0 0.00 1 1.27 LP11 0 0.00 0 0.00 3 30.00 1 7.69 0 0.00 4 5.06 LP12 1 2.04 2 40.00 0 0.00 0 0.00 0 0.00 3 3.80 LP13 0 0.00 0 0.00 0 0.00 2 15.38 0 0.00 2 2.53 LP15 1 2.04 0 0.00 1 10.00 0 0.00 0 0.00 2 2.53 LP9*LP14 0 0.00 0 0.00 0 0.00 1 7.69 0 0.00 1 1.27 Table 1. Serogroup Distribution of 79 Strains
-
For SBT, full 7-allele profile was obtained of 79 isolates, 22 STs could be found in the EWGLI SBT database (http://bioinf-ormatics.phe.org.uk/legionella/legionella_sbt/php/sbt_homepage.php), but the other 11 profiles could not be found in the database. Novel profiles were submitted to the ESGLI SBT database and acquired the STs.
As shown in Figure 1, the 79 strains were analyzed by SBT, and 33 different STs were found, of which 11 (ST2355-ST2365) were identified for the first time. A total of 17 STs were from cooling towers [index of discrimination (IOD), 0.763], 4 were from bath water (IOD, 0.900), 8 were from hot springs (IOD, 0.897), and 5 were from fountains (IOD, 0.756). ST1 have been found in both cooling towers, fountain and bath water. All these isolates had high IODs.
Ten STs (ST1, ST36, ST630, ST2359, ST1777, ST187, ST2363, ST328, ST641, and ST7) contained more than one strain, while the other 23 STs contained only a single strain, respectively, among which ST1 was the predominant type, accounting for 37.97% (30/79), followed by ST630 (7/79, 8.86%), ST2359 (4/79, 5.0%), and ST1777 (3/79, 3.8%). However, the strains with identical ST types belonged to different serogroups. This was observed in the case of ST1, which corresponded to three different serogroups-LP1, LP7, and LP9.
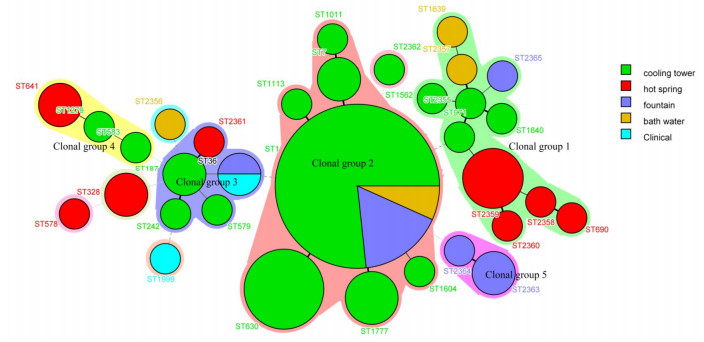
The results of MST analysis (Figure 2) showed that 33 STs were predicted to form five clonal groups, whereas 5 STs, which differed from every other ST in four or more genes, were identified as singletons. Clonal group 1 contained 11 STs, 4 were isolated from the hot spring (ST690, ST2358, ST2359, ST2360), 4 from cooling tower (ST2355, ST1640, ST1562, ST971), 2 from bath water (ST1639, ST2357), and 1 from fountain (ST2365). Clonal group 2 contained 7 STs, 6 STs were isolated from cooling towers (ST1011, ST7, ST630, ST1604, ST1113, and ST1777), 1 ST (ST1) was distributed in cooling towers, bath water, and fountain. Clonal group 3 contained 5 STs in all: 3 isolated from cooling towers (ST579, ST187, and ST242), 1 from a hot spring (ST2361), and 1 from a fountain and clinical patient (ST36). Clonal group 4 had only 3 STs, ST583 and ST1279 from cooling towers and 1 from a hot spring (ST641). Lastly, clonal group 5 had 2 STs, both of which were from the fountain (ST2363 and ST2364). The five singletons were from various sources, S1 (ST1562), S2 (ST2356), S3 (ST1999), and S5 (ST578) contained 1 strain, and were isolated from cooling tower, bath water, patient and hot spring, respectively. S4 contained 2 stains (ST328), and both were isolated from hot spring in the same city (Kang ding).
Figure 2. Minimum spanning tree analysis of the 79 L. pneumophila from Sichuan, China. The sequence types (STs) are displayed as circles. The size of each circle indicates the number of isolates within the particular type, and the ST are shown near the circle. The colors of the halo surrounding the STs denote types that belong to the same clonal group.
-
The 42 LP1 strains were selected for MLVA, from which 8 MLVA types were obtained. Using these, we created a maximum likelihood tree, demonstrated in Figure 3. Of the 8 MLVA types, M08 was the most principal type, accounting for 47.62% of the LP1 strains (20/42), the other major type was M07 (10/42, 23.81%), then the following was M01 and M04 (6/42, 14.29%; 2/42, 4.76%). Most of the MLVA were source specific. All but one of the 20 strains of the M08 were obtained from cooling towers; the exception was 1 strain from a fountain. Similarly, all 6 strains of the M01 were obtained from hot springs, except 1 that came from a fountain.
As shown in Figure 3 for of the 42 LP1 strains, 12 different STs and 8 different MLVA types were identified. Comparing these results, it seems that the strains of the same ST can could belong type into more than one MLVA type. For example, the strains of ST1 belonged were typed into 2 different MLVA (M07 and M08), as did the strains of ST630. In the same way, the strains of the same MLVA could belong to more than one ST, such as the strains of M01 were typed into three STs (ST2369, ST2360, and ST971).
-
The virulence genes rtxA and lvh of L. pneumophila were identified in all of 79 strains. Both loci were present in all strains, it was suggested that the strains isolated from Sichuan area might have the potential infectivity and pathogenicity to human being.
-
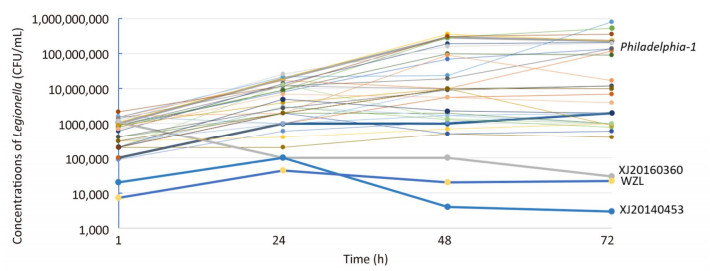
The mouse BALB/c macrophage cell line J774 was used for testing the intracellular growth ability of the 33 strains, and Philadelphia-1 (ATCC33152) was used as a positive control. To further evaluate the intracellular growth ability of the strains, plate culture counting was performed. Thirty-one isolates (including ATCC33152) had bacterial concentrations of 104-105 CFU/mL, 105-106 CFU/mL, 106-107 CFU/mL and 107-108 CFU/mL on 1 h, 24 h, 48 h, and 72 h of infection, respectively, and showed high proliferative ability in J774 macrophage cells. However, XJ20140360 (ST578), XJ20140453 (ST1604), and WZL (ST1999) exhibited bacterial concentrations of 1 × 105 CFU/mL, 4 × 103 CFU/mL, 2 × 104 CFU/mL on 48 h, and 3 × 104 CFU/mL, 3 × 103 CFU/mL, 2.2 × 104 CFU/mL on 72 h, respectively, and showed low proliferative ability in J774 macrophage cells (Figure 4).
Strain Identification
SBT Analysis
MLVA of LP1
Detection of Virulence Genes
Intracellular Growth Ability
-
Our findings showed that L. pneumophila was widespread in public environment, suggesting the high prevalence of L. pneumophila in Sichuan, China. The standardized SBT scheme for L. pneumophila presents an important advancement in the study of the molecular epidemiology of this bacterium. The results of ST analysis on the isolates of L. pneumophila showed that there was the high genetic diversity of L. pneumophila in Sichuan province from 1989 to 2016. There are 2703 STs in the EWGLI SBT Database (http://bioinformatics.phe.org.uk/legionella/legionella_sbt/php/sbt_homepage.php) among 12820 isolates (up to 22th Jan, 2019). According to our research, ST1, which is the most prevalent ST reported in several countries[7, 15, 27-29], is also the most common ST in Sichuan, accounting for 37.9% of the isolates (30/79). Interestingly, all ST1 strains were isolated from man-made aquatic environments, namely, cooling towers (76.6%), fountains (16.6%), and bath water (6.6%), rather than natural water (hot springs). These results were consistent with previous studies[6, 27, 30]. The reasons that ST1 strains seem to have a poor ability to survive in the natural environment, such as hot springs and soil, need further examination.
The 54 strains isolated from cooling towers and bath water were collected during environmental surveillance for L. pneumophila in a public water system, including a hotel and public bath. Seventeen STs were obtained from cooling towers (2 were unique STs) and 4 were collected from bath water (2 were unique STs). ST630 was the second most predominant ST in this study (8.86%). All strains of ST630 isolated from cooling towers were distributed in five cities (Chengdu, Ya'an, Nachong, Neijiang, and Leshan), suggesting the widespread prevalence of this type in Sichuan. According to the reports by Qin[6] and Guo[31], ST630 was isolated from cooling towers in Beijing and Guangzhou, China. Moreover, a case of clinical LD associated with ST630 was reported in France. The widespread distribution of this type around the world needs further investigation. According to the EWGLI SBT database, ST1113, ST1279, ST1562, ST1604, ST1640, ST1777, and ST971 have been isolated from the environment in many counties such as the Netherlands, Greece, Israel, and USA. However, ST1, ST187, ST242, ST579, ST583, ST7, and ST630 have been found during environmental surveillance and in clinical cases, which indicate that people are at a high risk of contracting L. pneumophila infection in hotels and public baths, and more stringent disinfection measures should be adopted in these places.
Ten strains were isolated from city fountains in Chengdu during fountain surveillance for L. pneumophila in 2010, which involved 50 water fountains from five collection points, making the positive rate high, at 20%. The fountain isolates, distributed across four serogroups (LP1, LP7, LP11, and LP15), were divided into 5 STs, including 3 novel STs (ST2363-ST2365), of which most of the isolates belonged to ST1 (50%). The strains of ST36, which is closely related to Philadelphia-1 (ATCC33152), were isolated from a fountain and a clinical case. Researches reported that ST36 was widely distributed throughout the world[7, 32-34] and had been responsible for multiple sporadic cases and outbreaks in the past, including the first described outbreak in Philadelphia, Pennsylvania, in 1976[34]. The first case of LD in Sichuan was reported in 1989 by the Sichuan Centers for Disease Control, and it was also caused by an LP1 strain of ST36; after 21 years, we isolated a strain, shared the same ST (ST36) as the clinical isolate, suggesting that L. pneumophila persists in the environment for a long time and have potential risk to spread and infect human beings. According to the previous studies on outbreaks, because the fountains were the suspected source, many people were affected[35-36], suggesting that human could be infected by exposing to Legionella-contaminated fountains. In environmental surveillance studies, the detection rates of Legionella in fountains varied from 2.20% (3/134) to 80.00% (4/5)[37]. Therefore, environmental monitoring should be improved, especially in public areas.
We selected nine hot springs in three cities (Ya'an, Emei, and Kangding) as study points for L. pneumophila in 2014, and the procedure for collection and pretreatment of these samples was based on ISO11731 (1998) protocols. Thirteen strains were isolated from all 40 water samples collected during the study period, which showed a high positive rate of L. pneumophila (13/40, 32.50%) in the hot spring water samples in Sichuan. L. pneumophila with temperatures ranging from 35 ℃ to 43 ℃ and pH ranging from 7.0 to 7.5. This isolation rate of Legionella positivity was higher than that reported in previous studies in Sivas, Turkey, in 2002 (11.5%)[38], Japan in 2004 (28.7%)[39], while lower than that in a report from Beijing in 2013 (51.9%)[9], and Wenzhou in 2016 (62.5%)[15]. It seems that contamination of hot springs with L. pneumophila in China is more serious than that in the other countries, indicating that some hot springs may be endemic origin for LD in China. Among the 8 STs identified from hot spring in Sichuan, 4 STs were the location-specific STs (ST2358-ST2361), and according to the SBT database, ST328, ST641, ST690, and ST578 were isolated from the environment and clinical cases worldwide.
The L. pneumophila MLVA database can be acquired online at . Until 22th Jan, 2019, 1140 L. pneumophila strains had been deposited in the MLVA bank, including 338 LP1 strains. In the present study, 8 MLVA types were identified. M04, belonging to MLVA Clone Complex 2 (VACC2), was identified by SBT as ST36 which is widespread across the world, including 2 strains: one strain was the first L. pneumophila isolated from a patient in Sichuan in 1989, the other was isolated from a central fountain in Chengdu (capital of Sichuan) in 2010. The results showed the high consistency between MLVA and SBT. M06 belongs to VACC6, which was isolated from a cooling tower in Chengdu, also reported isolated from environment or clinical case in France and Sweden by Pourcel C[40] and Visca P[41]. M01, M02, M03, and M05 were unique types found in Sichuan and have not been reported elsewhere. M07 and M08, which belong to VACC1, isolated from man-made aquatic environments, including cooling towers, bath water, and fountains, were the most prevalent MLVA types in Sichuan. These two MLVA types strains were also found in other countries[41] and may infect humans.
Both the SBT and MLVA methods have the advantages of good classification ability, easy operation, good repeatability, time saving, and digital standardization and can be applied to type LP1. However, SBT is non-economical for forward and reverse sequencing. Moreover, according to the results of the SBT analysis, ST1 corresponds to different serogroups, namely, LP1, LP7, and LP9, while the 42 LP1 strains were divided into 13 STs, suggesting the serogroups have no direct correlation with STs.
We found 3 isolates, that is, XJ20140360 (ST578), XJ20140453 (ST1604), and WZL (ST1999), did not grow from 24 h to 72 h, suggesting that these isolates have lower intracellular growth ability and maybe less likely to cause disease in humans. It seems that the strain WZL, although obtained from a patient's sputum, has lost its pathogenicity. A possible reason for this is that the strain developed some lethal mutations after frequently proliferating. However, the mechanism of how these mutations transfer requires further study. The remaining 30 isolates showed high growth ability in J774, indicating that these isolates, which were obtained from hot springs, cooling towers, fountains, and bath water in Sichuan, have the potential to cause LD.
-
In conclusions, this study has investigated the prevalence and genetic characteristics of L. pneumophila in Sichuan, China, from 1989 to 2016. Both SBT and MLVA can applied for molecular epidemiological investigation of L. pneumophila. There are the high genetic polymorphism and the regional specificity in Sichuan. Further, the results of virulence gene detection and the intracellular growth ability of the isolates suggest a high possible risk of contamination by this organism in aquatic environments, including cooling towers, hot springs, fountains, and bath water, and effective control and prevention strategies are urgently needed.
-
The authors declare no conflict of interest.
-
This article does not contain any studies with human participants or animals performed by any of the authors.










 Quick Links
Quick Links
 DownLoad:
DownLoad: