-
Pneumoconiosis, a fatal lung disease caused by dust inhalation and deposition, is one of the leading occupational diseases worldwide[1]. The dust is primarily inorganic particles, such as silica particles, coal dust, and asbestos fibers. If the diameter of dust is less than 5 μm, it is defined as ‘respirable dust’ that can easily reach the distal airways and alveoli[2]. The most common types of pneumoconiosis include silicosis, coal workers’ pneumoconiosis (CWP), and asbestosis. Pneumoconiosis is currently incurable, and is a serious public health threat. Recently, pneumoconiosis has not only arisen in traditional industries but also in emerging modern industries (e.g., biomedical applications, denim jeans production, and jewelry polishing) related to the use of new materials (e.g., silica nanoparticles, sandblast, and artificial stone). Many countries, both developing and developed, presently experience significant outbreaks, especially silicosis[3-5], thereby leading to an increase in pneumoconiosis morbidity and mortality worldwide[6].
Current treatment options for pneumoconiosis patients are extremely limited by a lack of effective diagnostic biomarkers and therapeutic targets[7] because critical molecular mechanisms underlying the disease remain poorly understood. For end-stage pneumoconiosis patients, lung transplantation is still the only means of prolonging survival[8]. Moreover, the current diagnosis of pneumoconiosis primarily depends on reliable occupational exposure history and chest radiographs. Although diagnostic technologies have been significantly improved in recent years, there is a lack of early diagnostic biomarkers for pneumoconiosis. Owing to strong compensation in lungs, most pneumoconiosis patients have already suffered severe irreversible damage by the time abnormalities appear in radiological images. Moreover, current therapeutic drugs cannot attenuate pneumoconiosis progression and mortality. Therefore, identifying effective diagnostic biomarkers and therapeutic targets of pneumoconiosis is urgently required.
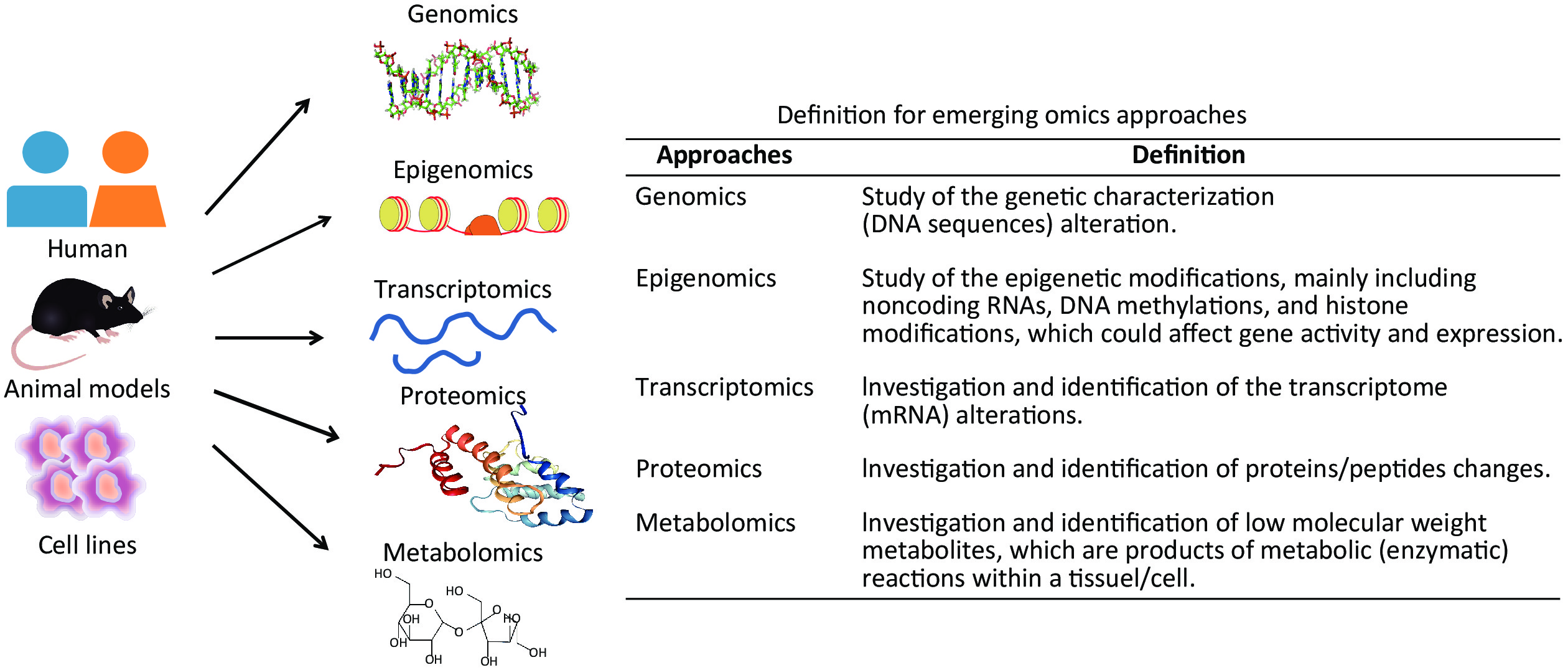
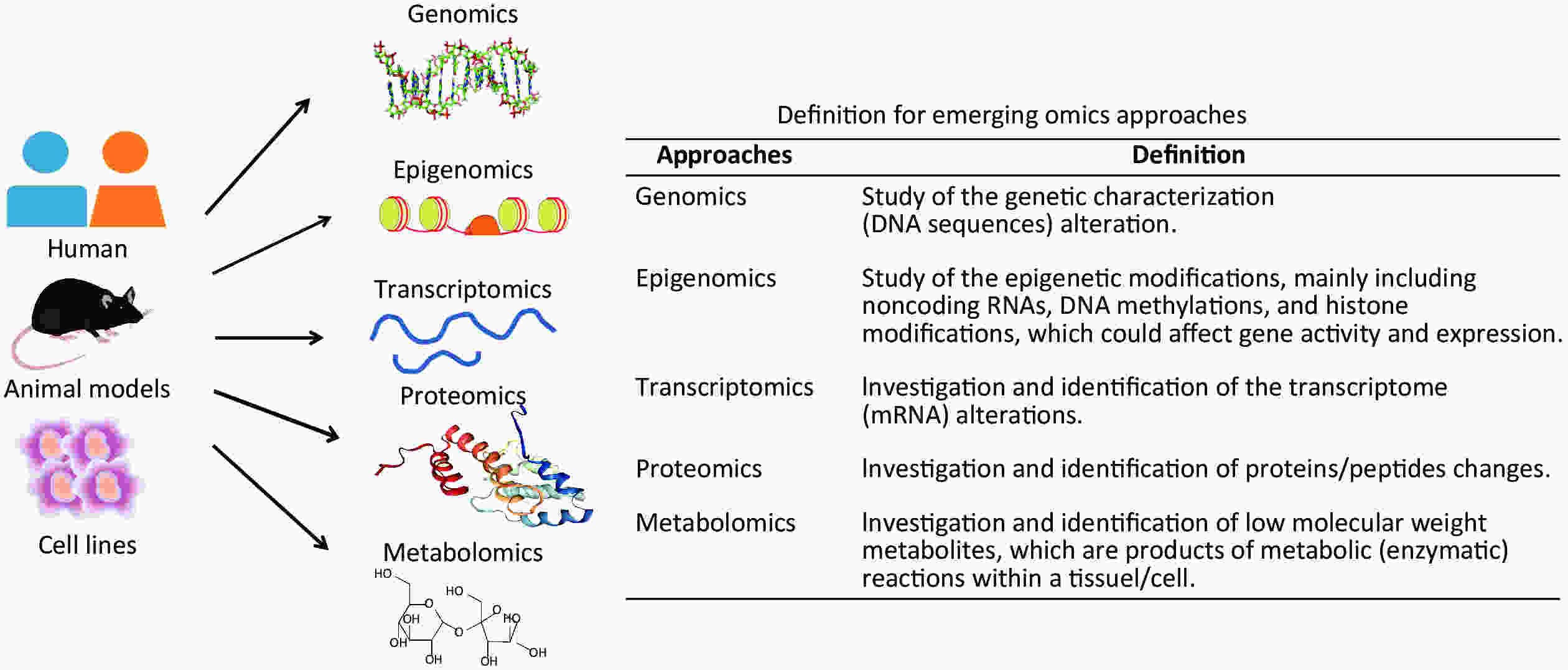
Recently, with the development of high-throughput technologies and bioinformatics, multiple omics approaches have bridged underlying molecular alterations with disease progression[9]. An overview of multiple omics approaches, including genetics, epigenomics, transcriptomics, proteomics, and metabolomics is shown in Figure 1. Compared with traditional hypothesis-driven methods, omics approaches are more comprehensive means of uncovering molecular alterations and facilitate deeper insights into disease pathogenesis[9]. Presently, omics approaches are an increasingly popular method for characterising pulmonary responses to occupational exposures. The omics studies have been widely conducted on pneumoconiosis patients and in vivo and in vitro experimental models for exploring underlying pneumoconiosis pathogenesis. Until now, omics approaches have made significant progress in revealing effective therapeutic targets for pneumoconiosis drug development and candidate biomarkers for early disease diagnosis.
This review is a unique first summary of pneumoconiosis omics studies. We describe putative omics-based diagnostic biomarkers and therapeutic targets and discuss the underlying pathogenesis mechanisms of pneumoconiosis. Moreover, we assess future prospects of single-cell omics and integrated multi-omics approaches for pneumoconiosis study. Additionally, future research challenges associated with these emerging omics techniques are clarified.
-
The cause of pneumoconiosis is industrial dust. It has been recognized that individual variation exists between subjects with similar occupational exposures, indicating a genetic susceptibility exists in pneumoconiosis patients[10]. Recently, genotyping technologies have significantly expanded our understanding of disease genetic factors, particularly single-nucleotide polymorphisms (SNPs) identified by genome-wide association studies (GWAS)[11]. Chu et al.[12] first conducted GWAS with a cohort of 400 Chinese coal workers, comprised of 202 pneumoconiosis patients and 198 healthy controls. The study successfully identified three novel SNPs, rs73329476, rs4320486, and rs117626015, which closely associated with mine-related pneumoconiosis risk[12]. Similarly, a case-control study involving a systematic screen of genes near rs73329476 found a strong association between silicosis susceptibility and rs12812500, which can regulate macrophages differentiation[13]. Here, we summarize several SNPs that have been reported to be significantly associated with CWP susceptibility (Table 1)[12,14,15]. Additionally, GWAS investigations have indicated that micro-RNAs (miRNAs) may play critical roles in CWP[16] and silicosis[17] pathogenesis and recommended miRNAs as putative diagnostic biomarkers and therapeutic targets.
Gene (SNPs) Chromosome Gene functions References CPM (rs73329476) 12q15 Regulates the differentiation of macrophages, and activates BKB1R signalling pathway Chu et al., 2014[12] LAMB1 (rs4320486) 7q31 Increases expression levels of the laminin and laminin-1 receptor Chu et al., 2014[12] PDZRN4 & CNTN1 (rs117626015) 12q12 Regulates Notch signalling pathway Chu et al., 2014[12] TICAM1 (rs8120, rs2292151) 19p13 Activates NF-κB, regulates the innate immune response and the biosynthesis of type I interferon Wang et al., 2015[14] HIST3H3 (rs2230656) 1q42 Participates in various biological processes, including gene expression and regulation, DNA repair, apoptosis, etc. Wang et al., 2015[14] ATP1A4 (rs10797062) 1q23 May alter the binding motifs of transcription factors. Wang et al., 2018[15] RBM26 (rs2274554) 13q31 Regulates TNF and NF-κB signalling pathways Wang et al., 2018[15] FNBP1P1 (rs1667614) 2q13 Involves in the peroxisome pathway Wang et al., 2018[15] Note. GWAS: genome-wide association studies, BKB1R: bradykinin B1 receptor, NF-κB: nuclear factor kappa-B, TNF: tumour necrosis factor. Table 1. SNPs identified by GWAS in pneumoconiosis
In summary, pneumoconiosis susceptibility is influenced by genetic and environmental factors, and genomics studies could significantly increase our understanding of the impact of genetic risk factors on pneumoconiosis pathogenesis. However, it will be necessary to validate and clarify underlying molecular mechanisms of identified gene variants. The genetic data could then be incorporated into pneumoconiosis clinical trials to better assess disease risk.
-
Epigenetics serves as a bridge connecting occupational exposures with underlying molecular alterations of disease progression and is currently drawing wide attentions from pneumoconiosis researchers[18-22]. Pneumoconiosis epigenomic studies are largely subdivided into the following three subjects: noncoding RNAs (ncRNAs), DNA methylation, and histone modification. ncRNAs dysfunction has been widely studied in pneumoconiosis, while histone modification and DNA methylation are investigated more rarely. ncRNAs, mainly including miRNAs, long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), regulate genetic expression and biological functions via RNA interference[21]. Here, we focus discussion on regulatory effects of ncRNAs, especially miRNAs, in pneumoconiosis.
-
To date, miRNAs studies have been conducted with lung tissues from silicosis animal models, using cell lines exposed to silica or transforming growth factor-β1 (TGF-β1), as well as with serum samples, peripheral blood leukocytes, and bronchoalveolar lavage fluid (BALF) from pneumoconiosis patients. The studies indicate that miRNAs are closely correlated with pneumoconiosis risk and disease progression and miRNAs have potential to serve as non-invasive biomarkers for early pneumoconiosis diagnosis and prognostic assessment (see Table 2). We recognize that a large number of miRNAs may contribute to early pneumoconiosis diagnosis, including miR-18a*, miR-149, miR-222, miR-671-3p, miR-16, miR-204, miR-19a, has-miR-4516, miR-29b-3p, and miR-34c-3p[16,23-26]. Furthermore, miR-146a, miR-18a, miR-29b-3p, and miR-34c-3p have been used to predict pulmonary severity and assess pneumoconiosis prognosis[26-28]. Additionally, a study conducted on serum samples from pneumoconiosis patients with pulmonary tuberculosis complications indicates that miR-155 and miR-29a are closely correlated with pulmonary tuberculosis risk in pneumoconiosis patients[29]. Because of their stable morphological characteristics, miRNAs are presently recognized as ideal biomarkers.
Micro-RNA Samples & Models/Patients Effects Reference miR-18a* ↑ Serum samples of CWP An early diagnostic biomarker Guo et al., 2013[16] miR-149 ↓ Serum samples of CWP An early diagnostic biomarker Guo et al., 2013[16] miR-222 ↓ Serum samples of CWP An early diagnostic biomarker Guo et al., 2013[16] miR-671-3p ↑ Serum samples of CWP An early diagnostic biomarker Guo et al., 2013[16] miR-16 ↑ Serum samples of CWP An early diagnostic biomarker Zhang et al., 2014[23] miR-204 ↓ Serum samples of CWP An early diagnostic biomarker Zhang et al., 2014[23] miR-19a ↓ Peripheral blood leukocytes from
patients with silicosisAn early diagnostic biomarker Yang et al., 2016[24] miR-146a ↑ BALF from silicosis patients A biomarker of silicosis severity Zhang et al., 2016[28] miR-155, miR-29a ↑, ↓ Serum samples from pneumoconiosis
with pulmonary TB complicationA biomarker related to pulmonary TB risk Liu et al., 2016[29] has-miR-4516 ↑ Serum samples from patients
with pneumoconiosisAn early diagnostic biomarker Huang et al., 2018[25] miR-18a ↑ Fibroblasts exposed to nano-SiO2 A biomarker of pulmonary injury Yang et al., 2018[27] miR-29b-3p,
miR-34c-3p↓, ↓ Lungs of rats and A549 cells
exposed to silicaAn indicator of early diagnosis and
prognosis for silicosisWang et al., 2019[26] Note. *: Indicates the passenger strand of miRNA, ↑: increased expression, ↓: decreased expression, CWP: coal workers’ pneumoconiosis, BALF: bronchoalveolar lavage fluid, TB: tuberculosis. Table 2. Studies of micro-RNAs as potential biomarkers of pneumoconiosis
-
Table 3 presents all miRNAs with anti-fibrotic effects or pro-fibrotic effects, which represent potential pneumoconiosis therapeutic targets. A brief summary follows.
Micro-RNA Samples & Models/Patients Effects Reference miRNAs with anti-fibrotic effects miR-181b ↓ Lungs of rats with silicosis Decreases TNF-α Wang et al., 2012[41]; Zhang et al., 2015[42] miR-486-5p ↓ Serum samples from silicosis patients Inhibits Smad2 Ji et al., 2015[30] miR-146a ↑ Lungs of silicosis rats Decreases IL-1β Zhang et al., 2015[42] miR-449a ↓ Lungs of silicosis mice Regulates autophagy and targets Bcl2 Han et al., 2016[43] miR-489 ↓ Mouse lungs and macrophages exposed to
silica and fibroblasts exposed to TGF-β1Inhibits Smad3 and MyD88 Wu et al., 2016[31] miR-27a-3p ↓ Fibroblasts exposed to TGF-β1 Inhibits α-SMA, Smad2 and Smad4 Cui et al., 2016[32] miR-503 ↓ Lungs of mice exposed to silica Inhibits EMT by targeting PI3K/p85 Yan et al., 2017[34] miR-29b ↓ Macrophages expose to silica;↓ RLE-6TN
cells exposed to silicaInhibits extracellular matrix synthesis;
Promotes METLian et al., 2017[36]; Sun et al., 2019[37] miR-18a ↓ Fibroblasts exposed to TGF-β1 Inhibits TGF-β1-induced lung fibrosis Li et al., 2018[39] miR-29c Overexpressed miR-29c via a lentiviral vector in lungs of silicosis mice Suppresses Wnt/β-catenin pathway Wang et al., 2018[46] miR-7 ↓ Lungs of mice and human bronchial epithelial cells exposed to silica Represses fibrogenesis of lung fibroblasts
and blocks EMTYao et al., 2018[35] miR-542-5p ↓ Lungs of mice expose to silica and fibroblasts exposed to TGF-β1 Attenuates fibroblast activation by targeting integrin α6 Yuan et al., 2018[45] miR-19a-19b-20a ↓ Lung fibroblasts exposed to silica Suppressed TGF-β-induced activation of fibroblasts Souma et al., 2018[40] miR-326 ↓ Lungs of mice exposed to silica Promotes autophagy activity of fibroblasts
by targeting PTBP1Xu et al., 2019[44] miR-411-3p ↓ Lungs of rats exposed to silicosis and fibroblasts exposed to TGF-β1 Blocks Smurf2/TGF-β pathway and decrease Smad7 degradation Gao et al., 2020[33] miRNAs with pro-fibrotic effects miR-26a ↓ Lungs of rats with silicosis Regulates the transcription of Smad1 Faxuan et al., 2012[41] miR-1224-5p ↑ Lungs of mice exposed to silica and fibroblasts exposed to TGF-β1 Mediates mitochondrial damage Wu et al., 2017[48] miR-200c ↓ Patients with impaired lung function induced
by silicaAssociated with abnormal ECM Rong et al., 2018[47] miR-155-5p The mice exposed to silicosis andtreated with
anti-miR-155-5pRegulates meprin α Chen et al., 2020[49] Note. miR: micro-RNA, ↑: increased expression, ↓: decreased expression, TNF-α: tumour necrosis factor-α, IL-1β: interleukin-1β, TGF-β1: transforming growth factor-β1, α-SMA: α-smooth muscle actin, EMT: epithelial -mesenchymal transition, MET: mesenchymal-epithelial transformation, PTBP1: polypyrimidine tract binding protein 1, ECM: extracellular matrix. Table 3. Recent micro-RNAs research related to pneumoconiosis pathogenesis
Firstly, we found that over 20 miRNAs participate in silica-induced pulmonary fibrosis, mainly by regulating Smad, TGF-β1, α-smooth muscle actin (α-SMA), endothelial-mesenchymal transition (EMT), extracellular matrix (ECM) synthesis in lung fibroblasts, autophagy activity in fibroblasts, and the production of inflammatory factors (e.g., tumour necrosis factor-α, interleukin-1β). These changes are clearly correlated with occurrence and progression of lung fibrosis under various particle exposures.
Secondly, most miRNAs had anti-fibrotic properties, and the expression levels of miRNAs are negatively correlated with the degree of lung fibrosis (Table 3). A series of studies with in vivo and in vitro experimental models or silicosis patients indicate that the expression of Smad, promoting silica-induced fibrosis, can be inhibited by several miRNAs, including miR-486-5p, miR-489, miR-27a-3p, and miR-411-3p[30-33]. Furthermore, dysregulated miRNAs may affect EMT. For example, a study conducted with lungs from silica-exposed mice indicates that miR-503 modulates EMT by targeting the PI3K/p85 signalling pathway[34]. Similarly, miR-7 and miR-29b, inhibited EMT by repressing fibrogenesis in lung fibroblasts and promoting mesenchymal epithelial transformation (MET)[35-37]. Further, TGF-β1, a well-known pro-fibrotic factor, plays a critical role in lung fibrosis by inducing fibroblast differentiation into myofibroblasts, promoting excessive ECM deposition[38]. We observe that many miRNAs, including miR-411-3p, miR-18a, and miR-19a-19b-20a, may be capable of alleviating silica-induced fibrosis by inhibiting TGF-β1 signalling[33, 39, 40]. Other investigations have demonstrated the powerful potential of miRNAs as pneumoconiosis therapeutic targets. For example, 1) miR-181b and miR-146a may attenuate lung fibrosis by decreasing lung inflammation in rats[41, 42]; 2) miR-449a and miR-326 exhibit anti-fibrotic effects by promoting fibroblast autophagy activity[43, 44]; 3) miR-542-5p attenuates fibroblast activation by targeting integrin α6[45]; and 4) miR-29c suppresses Wnt/β-catenin signalling, alleviating silica-induced lung fibrosis in mice[46].
Thirdly, there are a few miRNAs that have pro-fibrotic effects. A study performed with lungs of silica-exposed rats found that miR-26a regulates the transcription of Smad-1[41]. Further, miR-200c is associated with abnormal ECM deposition, playing a crucial role in silicosis progression[47]. Wu et al.[48] suggested that miR-1224-5p mediates mitochondrial damage, promoting silica-induced pulmonary fibrosis. Chen et al.[49] showed that miR-155-5p exerts a pro-fibrotic effect in silicosis by regulating meprin α. These studies indicate that targeting these miRNAs may alleviate lung fibrosis during silicosis. Although these miRNAs have shown promise for attenuating pneumoconiosis development, further studies are urgently needed to validate the safety and efficacy of these miRNAs and related inhibitors.
-
LncRNAs and circRNAs are playing crucial roles in the progression of many diseases, including lung cancer, acute respiratory distress syndrome, pulmonary hypertension, pulmonary tuberculosis, and pneumoconiosis[50]. Accumulating evidence shows that lncRNAs and circRNAs regulate corresponding miRNAs expression to affect downstream gene expression, playing vital pro-fibrosis or anti-fibrosis roles. These ncRNAs are considered potential therapeutic targets for silica-induced pulmonary fibrosis[51, 52].
A large number of studies indicate that lncRNAs could act as miRNA sponges, known as competing endogenous RNAs, to modulate the activity of miRNAs and thereby regulate pulmonary fibrosis[53]. For example, a recent study showed that pulmonary fibrosis-associated lncRNA (PFAL) could promote lung fibrosis by competitively binding miR-18a[39]. Moreover, studies by Liu et al.[54] and Yan et al.[34] indicate that lnRNA-ATB and lncRNA MALAT1 may promote silica-induced pulmonary fibrosis by competitively binding miR-200c and miR-503, respectively. In addition, another study identified plasma lncRNA-ATB as a potential biomarker for diagnosis of CWP patients[55]. For circRNAs, the latest investigations indicate that circHECTD1 can promote silica-induced fibroblast activation and lung EMT via HECTD1[56, 57]. Yang et al.[52] suggested that silica-induced initiation of a circular ZC3H4 RNA/ZC3H4 pathway promotes pulmonary macrophage activation, identifying a novel therapeutic target for silicosis. The circRNA CDR1AS is stimulated by silica and could regulate miR-7 to release TGFBR2, importantly promoting EMT in silicosis progression[35].
In summary, serum-specific miRNAs, with stable properties and levels closely correlated with fibrosis progression, could be ideal biomarkers for diagnosing pneumoconiosis. Moreover, many ncRNAs, especially miRNAs, show anti-fibrosis or pro-fibrosis effects and are promising putative targets for pneumoconiosis treatment. Because of the transient nature and biodegradability of ncRNAs, permanent adverse effects from ncRNAs could potentially be avoided. However, ncRNAs functions are extremely complicated, involving perplexing crosstalk between mRNAs, miRNAs, lncRNAs, and circRNAs. Furthermore, specific therapeutic targets and underlying regulatory mechanisms of many miRNAs remain unclear. Clearly, any single miRNA is capable of regulating a broad array of genes and one gene can be regulated by multiple miRNAs, which increases the potential for off-target effects that would limit clinical applications. To address these obstacles, further studies are urgently needed.
Additionally, due to a lack of lung tissues and other human samples, few epigenetic studies have been performed on pneumoconiosis patients. Considering large species differences between animals and humans in epigenetic studies, we hope that more future investigations with human samples can be performed.
-
Transcriptomics analysis uncovers differentially expressed genes (DEGs), largely using RNA sequencing (RNA-Seq) analysis, with advantages of a broad dynamic range, high specificity and sensitivity, and enhanced detection of biological isoforms[58]. Transcriptomics studies of pneumoconiosis using in vivo and in vitro experimental models have been conducted within the past four years. Transcript differences between model groups and control groups in lung tissues of silica-exposed animals or cell lines have led to the identification of DEGs, which may contribute mechanistic understanding or novel biomarkers for pneumoconiosis. We summarize pneumoconiosis transcriptomics studies in Table 4.
Samples & Models/Populations Profiling approach Candidate biomarkers/targets/
signalling pathwaysReferences Transcriptomics Lung epithelial cells exposed to silica RNA sequencing ID1, ID2, and ID3 Chan et al., 2017[60] Macrophages exposed to silica RNA sequencing Activating transcription factor 3 (ATF3) Chan et al., 2018[59] Lungs of a silicosis mouse model RNA sequencing Ccl2, Ccr2, and JAK-STAT pathway Chen et al., 2018[61] Lungs of a silicosis rat model RNA sequencing Long non-coding RNAs (lncRNAs) Sai et al., 2019[51] Proteomics Lungs of a silicosis rat model 2D gel with MALDI-TOF MS Cathepsin D precursor, SEC14-like
protein 3 and peroxiredoxin-1Chen et al., 2009[65];
Chen et al., 2012[66]Serum samples of asbestosis patients SELDI-TOF KIF18A and KIF5A Tooker et al., 2011[64] Serum samples of asbestosis patients 2D gel with MALDI-TOF MS Cytokines, α1-AT, L-ficolin Ju et al., 2012[63] Lungs of a silicosis rat model 2D gel with MALDI-TOF MS Alpha B-crystallin Chu et al., 2015[68] Macrophages exposed to silica iTRAQ-coupled 2D LC-MS NOD-RIP2-NF-κB pathway Fu et al., 2018[74] Lungs of a silicosis rat model LC-MS/MS VLDLR and syndecan-2 Na et al., 2019[73] Lungs of a silicosis rat model iTRAQ PTPN2, p-STAT3 and vimentin Zhu et al., 2020[69] Metabolomics Lungs of a silicosis mouse model 1 H NMR Membrane choline phospholipids
metabolism and glucose metabolismHu et al., 2008[82] Lungs and BALF of rats inhaled ZnO particles 1 H NMR Lung tissues: lipid metabolism;
BALF: amino acid metabolismLee et al., 2016[83] Note. ID: DNA-binding protein inhibitor, 2D: two-dimensional, MALDI-TOF: matrix-assisted laser desorption/ionization time-of-flight, MS: mass spectrometry, SELDI-TOF: the surface-enhanced laser desorption/ionization time of flight, iTRAQ: isobaric tags for relative and absolute quantitation, LC: liquid chromatography, 1 H NMR: proton nuclear magnetic resonance, VLDLR: very low density lipoprotein receptor, PTPN2: protein tyrosine phosphatase non-receptor type 2, p-STAT3: phospho-signal transducer and activator of transcription 3, BALF: bronchoalveolar lavage fluid, ZnO: zinc oxide. Table 4. Novel biomarkers/targets/signalling pathways in pneumoconiosis revealed by omics (transcriptomics, proteomics, or metabolomics) approaches
In vitro, an RNA-Seq analysis of silica-exposed macrophages found that activating transcription factor 3 (ATF3) is significantly upregulated, and small interfering RNA (siRNA) inhibition of ATF3 expression improved silica-induced lung inflammation[59]. DEGs uncovered in a study of lung epithelial cells exposed to silica for different time intervals showed that the DNA-binding protein inhibitor (ID) family, particularly ID1, ID2 and ID3, are potential biomarkers for early lung epithelial cell responses to silica[60]. In vivo, a recent RNA-Seq analysis conducted in a silicosis mouse model revealed a comprehensive profile of molecular alterations, and bioinformatic analyses indicate that cytokine-cytokine receptor interaction and downstream JAK-STAT signalling pathways were the most significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways[61]. Ccl2 and Ccr2, which participate in silica-induced pulmonary inflammation and fibrosis, were significantly up-regulated[61]. Previous research suggested that Dexamethasone attenuates bleomycin-induced lung fibrosis in mice through the JAK-STAT pathway[62], which suggests Dexamethasone treatment could potentially attenuate silica-induced lung fibrosis.
In summary, to date only a few studies using RNA-Seq methods have been completed to discover DEGs involved in pneumoconiosis progression. Unfortunately, transcriptomics studies using human pneumoconiosis lung samples have not yet been performed. To better understand underlying mechanisms of pneumoconiosis pathogenesis, it will be necessary to examine downstream alterations of proteins and metabolites.
-
Proteins levels reflect the instantaneous state of tissues or cells, vitally informing disease pathogenesis studies. Several proteomics studies of pneumoconiosis have been conducted within the past decade. Presently, proteomics approaches are being applied to quantitatively analyse protein changes in serum samples from asbestosis patients, lung tissues from silicosis animal models, and silica-exposed macrophage cells.
Proteomics has provided crucial information about pneumoconiosis, and critical biomarkers, targets, or signalling pathways are summarized in Table 4. A study of 37 patients with asbestosis, 254 workers exposed to asbestos, and 439 healthy controls identified differentially expressed serum proteins, such as cytokines and α1-AT, that could serve as biomarkers for early asbestosis diagnosis and intervention[63]. In a study of serum samples from 35 asbestosis patients who subsequently developed cancer and 35 patients who did not develop cancer, two kinesin proteins, KIF18A and KIF5A, showed potential as blood biomarkers to identify asbestosis patients at risk of developing lung cancer[64]. Analysis of differential protein expression in lungs of silica-exposed rats identified six differentially expressed proteins and indicated that these proteins may participate in silicosis progression[65]. The same researchers further validated these six differentially expressed proteins and indicated that cathepsin D precursor, SEC14-like protein 3, and peroxiredoxin-1 may participate in lung fibrosis[66]. Further, a previous study showed that a lack of peroxiredoxin-I can aggravate bleomycin-induced pulmonary fibrosis in mice[67]. However, the effects of Cathepsin D precursor and SEC14-like protein 3 have not yet been investigated in silicosis experimental models. Thus, it would be worthwhile to validate the roles of the two proteins in pneumoconiosis.
In a two-dimensional gel electrophoresis image of early lung injury in silica-exposed rats, alpha B-crystallin was one of 13 significantly changed proteins, suggesting an important role in silicosis progression[68]. Similarly, a recent investigation also conducted with lungs of silica-exposed rats indicated significantly increased expression of protein tyrosine phosphatase non-receptor type 2 (PTPN2)[69]. In the same study, expression of phospho-signal transducer and activator of transcription 3 (p-STAT3) was markedly reduced, and PTPN2 inhibited EMT in silicosis by dephosphorylating STAT3[69]. STAT3, a well-known TGF-β-related signalling factor, possesses pro-fibrosis effects[70]. A more recent in vivo murine model study of bleomycin induced pulmonary fibrosis also indicated that STAT3 inhibition exerts anti-fibrotic functions[71]. Moreover, another recent study showed that PTPN2 may improve renal fibrosis by repressing STAT-induced inflammation in early diabetic nephropathy[72]. Taken together, PTPN2 shows promise as a pneumoconiosis therapeutic target.
Two recent studies using in vitro experimental models identified very low density lipoprotein receptor (VLDLR) and syndecan-2 as novel molecular targets of activated myofibroblasts in silica-exposed rat lungs and showed that activation of the NOD-RIP2-NF-κB pathway in RAW264.7 silica-exposed macrophages may induce macrophage polarization[73,74]. Syndecan-2 overexpression has been observed in alveolar macrophages of IPF patients, and syndecan-2 can attenuate lung fibrosis by inhibiting TGF-β1-induced fibroblast-myofibroblast differentiation, migration, and proliferation[75,76]. Further, VLDLR has been associated with hepatic fibrosis[77]. Accordingly, it is reasonable and necessary to further study effects of syndecan-2 and VLDLR on silica-induced pulmonary fibrosis.
In summary, proteomics has provided us a large number of potential therapeutic targets for pneumoconiosis. Although there are numerous pneumoconiosis-related proteomics publications, challenges, including accurate quantification of protein changes, remain, and continual improvement of proteomics technologies will be necessary. Additionally, integration of multiple omics approaches shows promise for increasing insight into the underlying pathogenesis of pneumoconiosis.
-
Metabolomics systematically detects small molecular metabolites (weighing less than 1,000 Daltons) in an untargeted or targeted manner, revealing disease-induced metabolic changes in blood, urine, BALF, cells, and tissues, and their extracts[78]. Small molecular metabolites, such as peptides, amino acids, nucleic acids, and organic acids, reflect protein activity and serve as signalling molecules. Identified marker metabolites can also serve as therapeutic targets[79]. Metabolic profile characterisation is crucial for revealing combined effects of multiple pneumoconiosis factors, including occupational exposures, altered genetic expression, and associated proteomic changes.
The most commonly used metabolomic analytical techniques are: 1) proton (1 H) nuclear magnetic resonance (NMR); 2) LC-MS; and 3) gas chromatography (GC)–MS. Advances in metabolomic techniques permit unique insights into underlying pathophysiology and early biomarkers of pneumoconiosis, which could improve pneumoconiosis diagnosis and risk assessment. 1 H NMR-based metabolomics have the advantage of being rapid and reproducible. Currently, metabolomics studies of pulmonary disease are mainly focused on lung cancer, chronic obstructive pulmonary disease, asthma, and cystic fibrosis[80,81]. To date, the two metabolomics studies conducted with lungs or BALF from experimental animal models exposed to silica or zinc oxide (Table 4) both used 1 H NMR-based metabolomics methods. Hu et al.[82] authored the first metabolomics study, finding notably altered membrane choline phospholipids metabolism in lungs of silica-exposed mice. Additionally, the study suggested cellular energy metabolism pathways and collagen pathway activation were affected by silica exposure. Lee et al.[83] showed that metabolic alterations in lung tissues (lipid metabolism) and BALF (amino acid metabolism) were different. However, validation is critically needed to confirm the functions of pneumoconiosis metabolic biomarkers. Although many omics approaches have been applied, future studies need to address effects of environment on lung samples and multiple endogenous factors, such as mucus, epithelial, and inflammatory cells, in lung tissues[84]. Standardization of large pneumoconiosis population metabolomics and proteomics approaches across different platforms remains a significant challenge.
In summary, metabolomics is a novel method that can be used for earlier disease detection, therapy monitoring, and ultimately for understanding pneumoconiosis pathogenesis—a step towards realizing the goal of personalized healthcare. Additional effort will be required to improve metabolomics technologies for future clinical application.
-
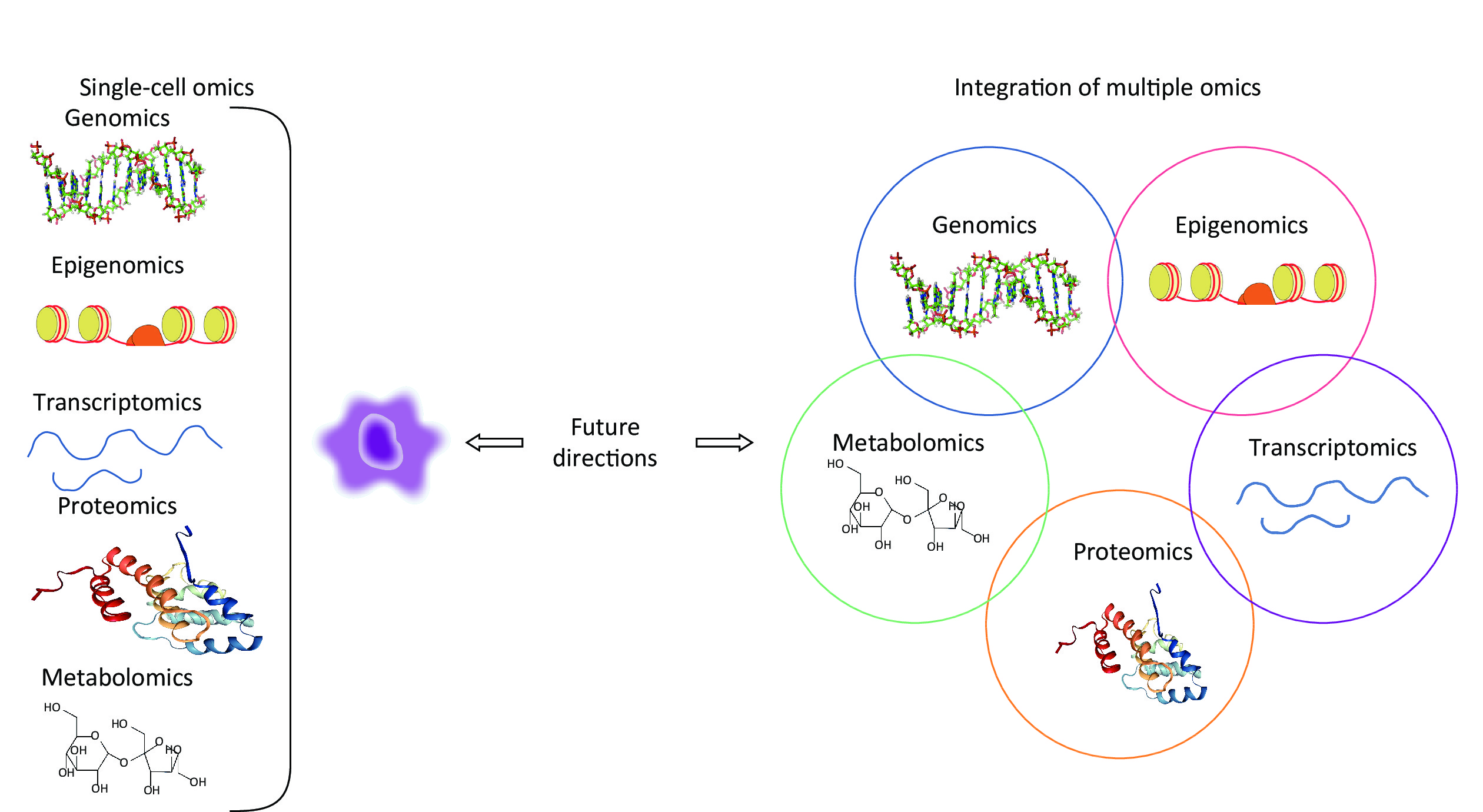
Emerging single-cell genomics approaches comprehensively measure omics at multiple levels, including genome, epigenome, transcriptome, protein, and metabolites in a single cell, providing increased sensitivity and accuracy over any single omics method[85]. Single-cell genomics plays a vital role in understanding various biology processes, contributing not only to a comprehensive picture of cellular phenotype and status, but also insight into cell interactions and spatial organization[86]. Furthermore, integrated multiple omics approaches will be required to fully understand underlying mechanisms of pneumoconiosis that are not readily observable with single omics assays. A scheme for single-cell omics and multi-omics approaches for future pneumoconiosis studies is shown in Figure 2.
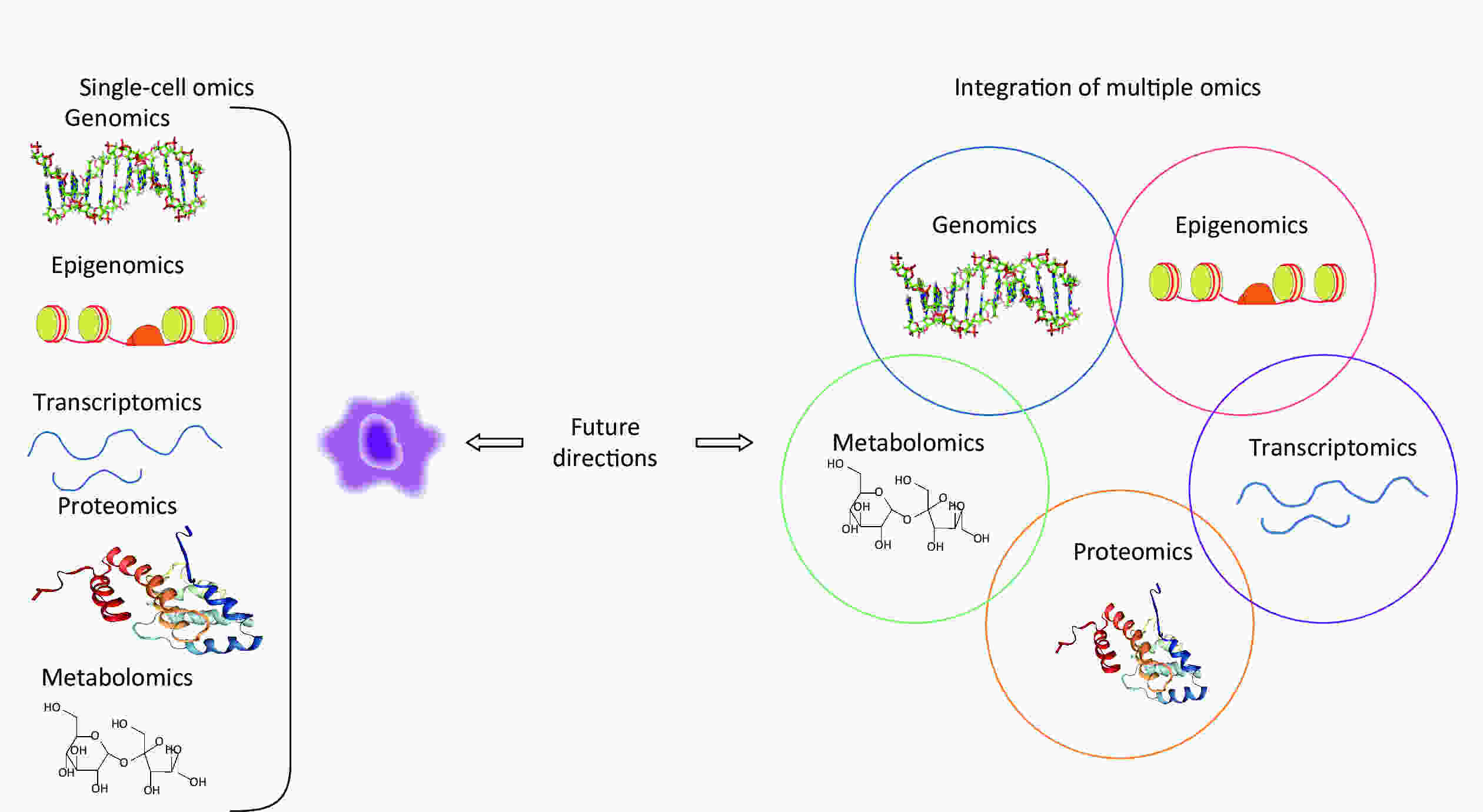
Figure 2. Schematic diagram of single-cell omics and multi-omics approaches for future pneumoconiosis studies.
It has become widely accepted that integrated omics approaches would reduce the sample sizes needed for single omics methods. Unfortunately, integrated multi-omics are not currently used to study pneumoconiosis. Identification and quantification of both peptides and metabolites remain barriers to integrating metabolomics and proteomics approaches into multi-omics studies of pneumoconiosis. Analysing ‘big data’ may reduce the feasibility of systems biology data integration from multiple sources. Moreover, as comprehensively summarized by Bowler et al.[84] in a recent review, many sample types possess inevitable weaknesses. Challenges for using emerging omics technologies in pneumoconiosis research are summarized in Table 5.
Single omics approach · Lung samples affected by multiple exogenous factors and endogenous components are difficult to collect. · Lack of standardization of different samples and analytical procedures. · Lack of standards for performance assessment and result interpretation. · Lack of power in single omics approach. · High costs. Single-cell omics and multi-omics approaches · Still under developing, sensitivity and throughput still need to improve. · The process of sample processing is still not ideal. · Improved quality control and standardization are necessary, but it is difficult to unify analytical standardization. · Enhanced multiplexity is required to reduce errors and bias. · Poor correlation between different omics approaches. · Standardization of large pneumoconiosis population omics approaches across different platforms and applying those technologies into routine clinical practice are challengeable. · High costs. Note. miRNAs: microRNAs, lncRNAs: long non-coding RNAs, circRNAs: circular RNAs. Table 5. Challenges for omics approaches in pneumoconiosis research
In summary, single-cell omics and multi-omics approaches have shown the potential to enhance precision medicine for pneumoconiosis. Moreover, omics approaches seem to be the optimal path for understanding pneumoconiosis pathogenesis. However, technical issues hinder application of these approaches in pneumoconiosis clinical practice.
-
Pneumoconiosis outbreaks create heavy burdens worldwide that seriously threaten public health. Although there still are no therapeutic drugs that effectively alleviate pneumoconiosis progression, emerging technologies are now providing new insights into pneumoconiosis pathogenesis and opening frontiers for both basic research and precision medicine. Innovative pneumoconiosis insights from omics research include genomics identification of pneumoconiosis susceptibility, transcriptomics detection of key genes involved in pneumoconiosis development, and indication that miRNAs may be ideal diagnostic biomarkers and promising therapeutic targets of pneumoconiosis. Proteomics, metabolomics, single-cell omics, and integrated multi-omics are powerful approaches for exploring potential diagnostic biomarkers, therapeutic targets, and key molecular mechanisms; however, many technological challenges need to be overcome. Moreover, it remains essential to validate findings and confirm the functions of key pneumoconiosis biomarkers or targets in order to move clinical applications forward. Thus, additional research depth is urgently warranted.
Author Contributions Conceived and designed the manuscript: WANG Jing, WANG Chen, and LUO Ya; Searched and read papers: LUO Ya and QI Xian Mei; Wrote the manuscript: LUO Ya, Qi Xian Mei, and PANG Jun Ling; Critically revised the manuscript: WANG Jing, WANG Chen, and LUO Ya; Provided final approval of manuscript to be published: WANG Jing and WANG Chen.
Competing Interests None of the authors have competing interests to declare.
Omics Approaches for Exploring Pneumoconiosis: A Review
doi: 10.3967/bes2021.010
- Received Date: 2020-04-26
- Accepted Date: 2020-09-09
| Citation: | LUO Ya, QI Xian Mei, PANG Jun Ling, WANG Jing, WANG Chen. Omics Approaches for Exploring Pneumoconiosis: A Review[J]. Biomedical and Environmental Sciences, 2021, 34(1): 71-82. doi: 10.3967/bes2021.010 |







 Quick Links
Quick Links
 DownLoad:
DownLoad: