-
Colorectal cancer (CRC) is a malignancy of the digestive system that poses a serious threat to human life and health. Approximately 1.93 million new CRC cases were identified in 2020, including 0.94 million CRC-related deaths worldwide, which accounted for 10% of the global cancer incidence and 9.4% of all cancer-related fatalities[1]. Although the cause of CRC has not been revealed, epidemiological studies have shown that nongenetic factors, such as smoking and alcohol consumption, and genetic factors, such as single nucleotide polymorphisms (SNPs) on CRC-related genes, play an important role in the occurrence of CRC[2,3]. Identifying CRC risk factors can help prevent the disease.
The phospholipase C epsilon 1 (PLCE1) gene on human chromosome 10q23.33 encodes a phospholipase enzyme that catalyzes the hydrolysis of phosphatidylinositol-4,5-bisphosphate to produce the second messengers, inositol 1,4,5-triphosphate and diacylglycerol. These second messengers govern several processes affecting cell growth, differentiation, and gene expression. Abnormal PLCE1 expression has been associated with multiple cancers, including CRC. The expression level of PLCE1 is significantly lower in CRC tissues than that in normal tissues[4]. The overexpression of PLCE1 could decrease the growth of colon cancer cells and reduce their malignant potential[5]. PLCE1 may act as a Ras receptor to promote apoptosis and thus act as a tumor suppressor[5]. These findings show that PLCE1 plays a suppressive role in the occurrence of CRC. Furthermore, some studies have focused on the role of several SNPs, such as rs753724, rs11187842, rs10882424, rs2274223, and rs3765524, on the CRC-related gene in the development of CRC. Among these SNPs, rs2274223 A > G is a missense variant (NP_057425.3: p.His1927Arg) that affects PLCE1 expression. The rs2274223 G allele is related to lower PLCE1 mRNA levels[6]. In addition, bioinformatics analysis revealed that PLCE1 has two important functional domains, a Ca2+ binding pocket related to protein activity and a catalytic binding pocket linked to catalytic efficiency[7]. The rs2274223 genetic polymorphism may alter the Ca2+ binding pocket, thereby deregulating its Ca2+-related bioactivity[7]. Changes in the structure and activity of PLCE1 further affect its cancer-suppressive function. Although several studies have reported the association between the rs2274223 polymorphism and CRC risk, the results are inconsistent. To clarify the relationship between this polymorphism and CRC risk, we performed a case-control study and meta-analysis.
In the case-control study, we included 706 patients with CRC and 775 healthy controls (Supplementary Table S1, available in www.besjournal.com). All patients were confirmed via pathological diagnosis. Patients with any other malignancy or a history of radiotherapy or chemotherapy before surgery were excluded. The control subjects were obtained from a pool of healthy volunteers. The exclusion criteria for the controls were no personal or family history of cancer or inflammatory diseases of the intestine. The Ethics Committee of Shanghai’s Xuhui District Central Hospital approved this study protocol (no. 047-001). We extracted genomic DNA from the peripheral blood of these individuals using TIANamp genomic DNA kits. Subsequently, DNA fragments containing the rs2274223 loci were amplified via polymerase chain reaction (PCR). The PCR reaction parameters and primer sequences were 95 ℃ for 5 min, 35 cycles (94 ℃ for 30 s, 56 ℃ for 30 s, 72 ℃ for 30 s), and 72℃ for 8 min; F: 5′-TTCATCATTCACTTTGTCCAT-3′; R: 5′-TTTCCTTTTGCTTCTTAATTC-3′. The PCR products were analyzed using direct sequencing. The results showed that the rs2274223 genetic polymorphism was associated with an increased risk of CRC under GG vs. AA [odds ratio (OR) = 1.80, 95% confidence interval (CI) = 1.15–2.83, P = 0.01], (AG + GG) vs. AA (OR = 1.25, 95% CI: 1.01–1.53, P = 0.04), GG vs. (AA + AG) (OR = 1.68, 95% CI: 1.08–2.61, P = 0.03), and G vs. A (OR = 1.24, 95% CI: 1.05–1.46, P = 0.01) (Table 1). In addition, patients with CRC carrying the rs2274223 GG genotype or the G allele were more likely to develop stage III+IV tumors (Table 2).
Genotype/Allele Cases (%) Controls (%) OR (95% CI)a Pa Power AA 362 (51.3) 439 (56.6) Reference AG 292 (41.4) 300 (38.7) 1.19 (0.96−1.47) 0.13 0.54 GG 52 (7.4) 36 (4.6) 1.80 (1.15−2.83) 0.01 0.50 AA 362 (51.3) 439 (56.6) Reference AG + GG 344 (48.7) 336 (43.4) 1.25 (1.01−1.53) 0.04 0.53 AA + AG 654 (92.6) 739 (95.4) Reference GG 52 (7.4) 36 (4.6) 1.68 (1.08−2.61) 0.03 0.56 A 1,016 (72.0) 1,178 (76.0) Reference G 396 (28.0) 372 (24.0) 1.24 (1.05−1.46) 0.01 0.49 Note. a: Adjusted for gender and age. CRC, colorectal cancer. Table 1. Association between the PLCE1 rs2274223 genetic polymorphism and CRC risk
Genotype/Allele I + II (%) III + IV (%) OR (95% CI)a Pa Power AA 215 (53.3) 147 (48.5) Reference AG 167 (41.4) 125 (41.3) 1.10 (0.81−1.51) 0.60 0.66 GG 21 (5.2) 31 (10.2) 2.17 (1.20−3.93) 0.01 0.48 AA 215 (53.3) 147 (48.5) Reference AG + GG 188 (46.7) 156 (51.5) 1.22 (0.91−1.65) 0.22 0.54 AA + AG 382 (94.8) 272 (89.8) Reference GG 21 (5.2) 31 (10.2) 2.09 (1.17−3.71) 0.02 0.55 A 597 (74.1) 419 (69.1) Reference G 209 (25.9) 187 (30.9) 1.28 (1.01−1.62) 0.04 0.50 Note. a: Adjusted for gender and age. CRC, colorectal cancer. Table 2. Association between the PLCE1 rs2274223 genetic polymorphism and CRC clinical stage
Characteristics CRC patients Healthy control subjects P Age (years, mean ± SD) 59.3 ± 6.7 59.9 ± 6.3 0.13 Gender, n (%) Male 421 (59.6) 431 (55.6) 0.12 Female 285 (40.4) 344 (44.4) Tumour stage, n (%) I + II 403 (57.1) III + IV 303 (42.9) Note. CRC, colorectal cancer. Table S1. Clinical and demographic characteristics of CRC patients and healthy control subjects
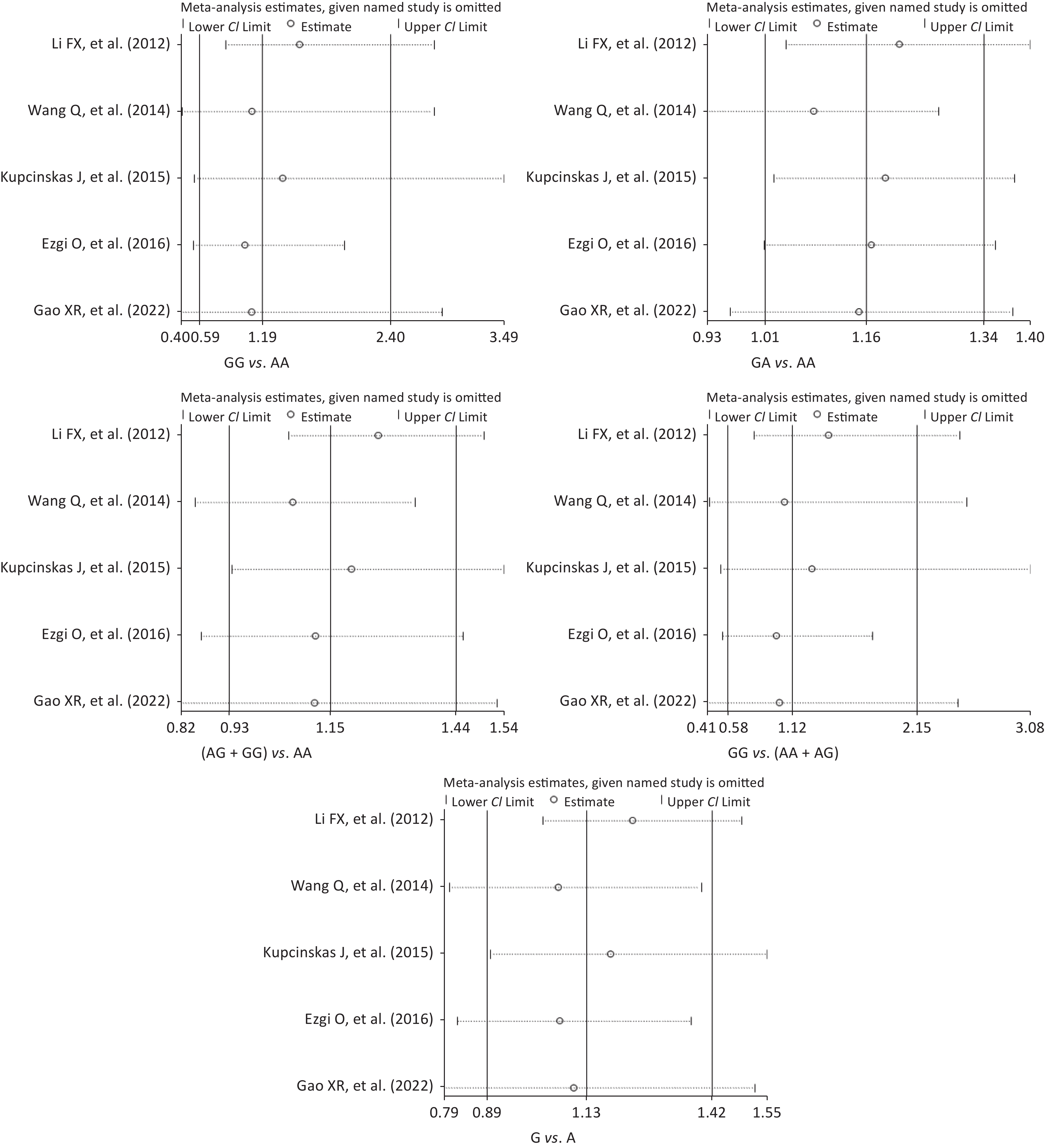
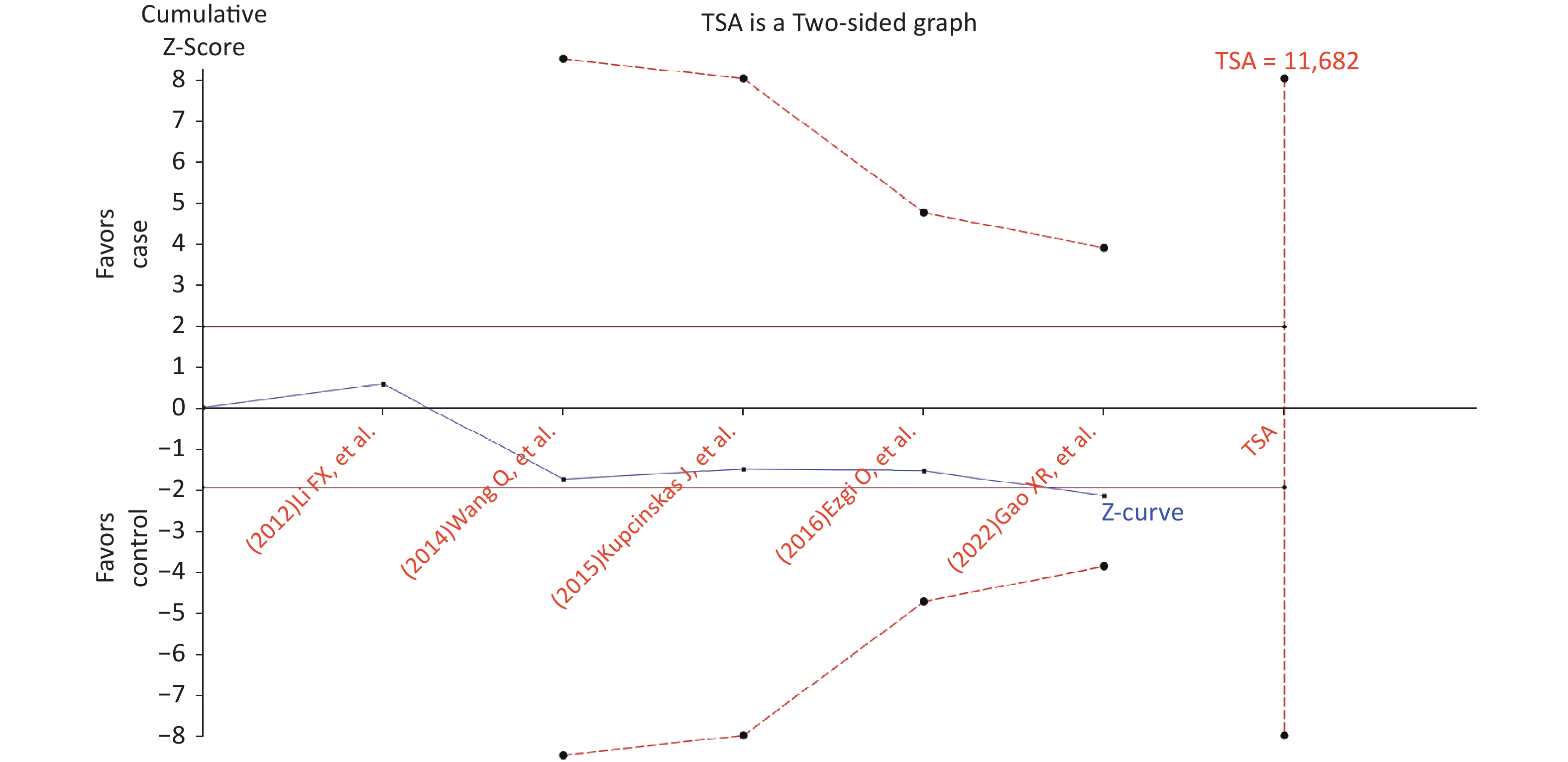
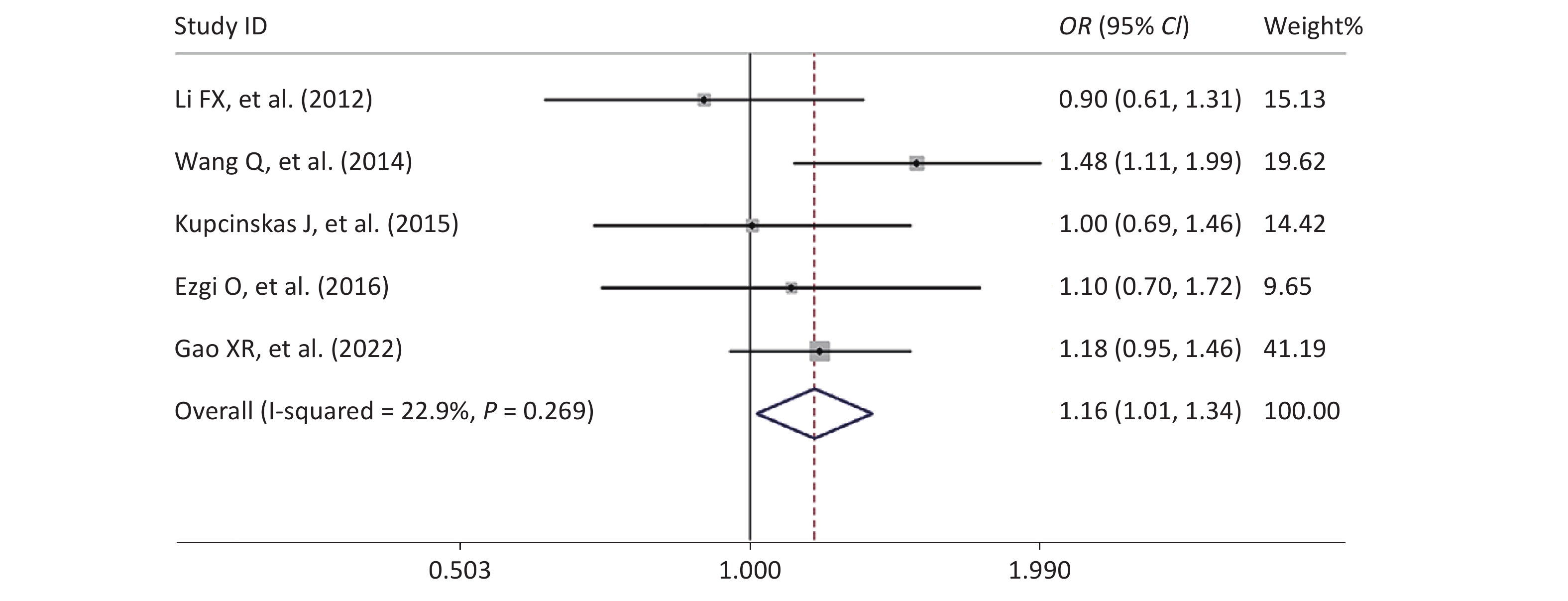
We searched Pubmed, Embase, and the Web of Science databases and obtained four case-control studies on the rs2274223 genetic polymorphism and CRC risk. Wang et al. reported that the AG and GG genotypes of PLCE1 rs2274223 are associated with an increased risk of CRC in the Chinese population[6]. Similarly, Ezgi et al. observed that Turkish subjects carrying the rs2274223 G allele had a significantly increased risk of developing CRC compared with Turkish subjects carrying the rs2274223 A allele[8]. However, a study by Li et al. showed that the rs2274223 G allele or GG genotype has a significant protective effect against CRC[9]. Kupcinskas et al. stated that the rs2274223 genetic polymorphism is not associated with the risk of CRC in European populations[10]. By combining the current case-control study with four previous studies, we included 1,746 patients with CRC and 2,089 healthy controls in the current meta-analysis (Supplementary Table S2, available in www.besjournal.com). The results of the combined analysis showed that the rs2274223 genetic polymorphism was significantly associated with an increased risk of CRC under AG vs. AA (Table 3 and Supplementary Figure S1, available in www.besjournal.com). No publication bias existed in the current meta-analysis (Supplementary Table S3 and Supplementary Figure S2, available in www.besjournal.com). Notably, although the current meta-analysis contained a relatively large sample size, sensitivity and trial sequential analyses suggested that the results of the current meta-analysis were unstable and needed to be further confirmed by more case-control studies (Supplementary Figures S3–S4, available in www.besjournal.com).
Comparison Test of heterogeneity Test of association Power I2 (%) P Model OR 95% CI P GG vs. AA 78 0.001 Random 1.19 0.59−2.40 0.64 0.84 AG vs. AA 23 0.27 Fixed 1.16 1.01−1.34 0.03 0.50 (AG + GG) vs. AA 59 0.04 Random 1.15 0.93−1.44 0.20 0.80 GG vs. (AA + AG) 76 0.002 Random 1.12 0.58−2.15 0.74 0.82 G vs. A 77 0.002 Random 1.13 0.89−1.42 0.32 0.91 Note. CRC, colorectal cancer. Table 3. Meta-analysis of the association between the PLCE1 rs2274223 genetic polymorphism and CRC risk
Author Year of
publicationCountry Cancer type Genotyping
methodCase group Control group PHWE value AA AG GG AA AG GG Li FX, et al.[9] 2012 China Colorectal cancer MassARRAY 155 71 5 180 92 20 0.09 Wang Q, et al.[6] 2014 China Colorectal cancer TaqMan 228 161 28 269 128 19 0.45 Kupcinskas J, et al.[10] 2015 Lithuania,Latvia Colorectal cancer TaqMan 77 91 24 147 173 56 0.66 Ezgi O, et al.[8] 2016 Turkey Colorectal cancer PCR-RFLP 142 48 10 176 54 0 0.04 Gao XR, et al. 2022 China Colorectal cancer Sequencing 362 292 52 439 300 36 0.09 Note. PCR-RFLP: Polymerase Chain Reaction-Restriction Fragment Length Polymorphism. Table S2. Characteristics of the case-control studies included in the current meta-analysis
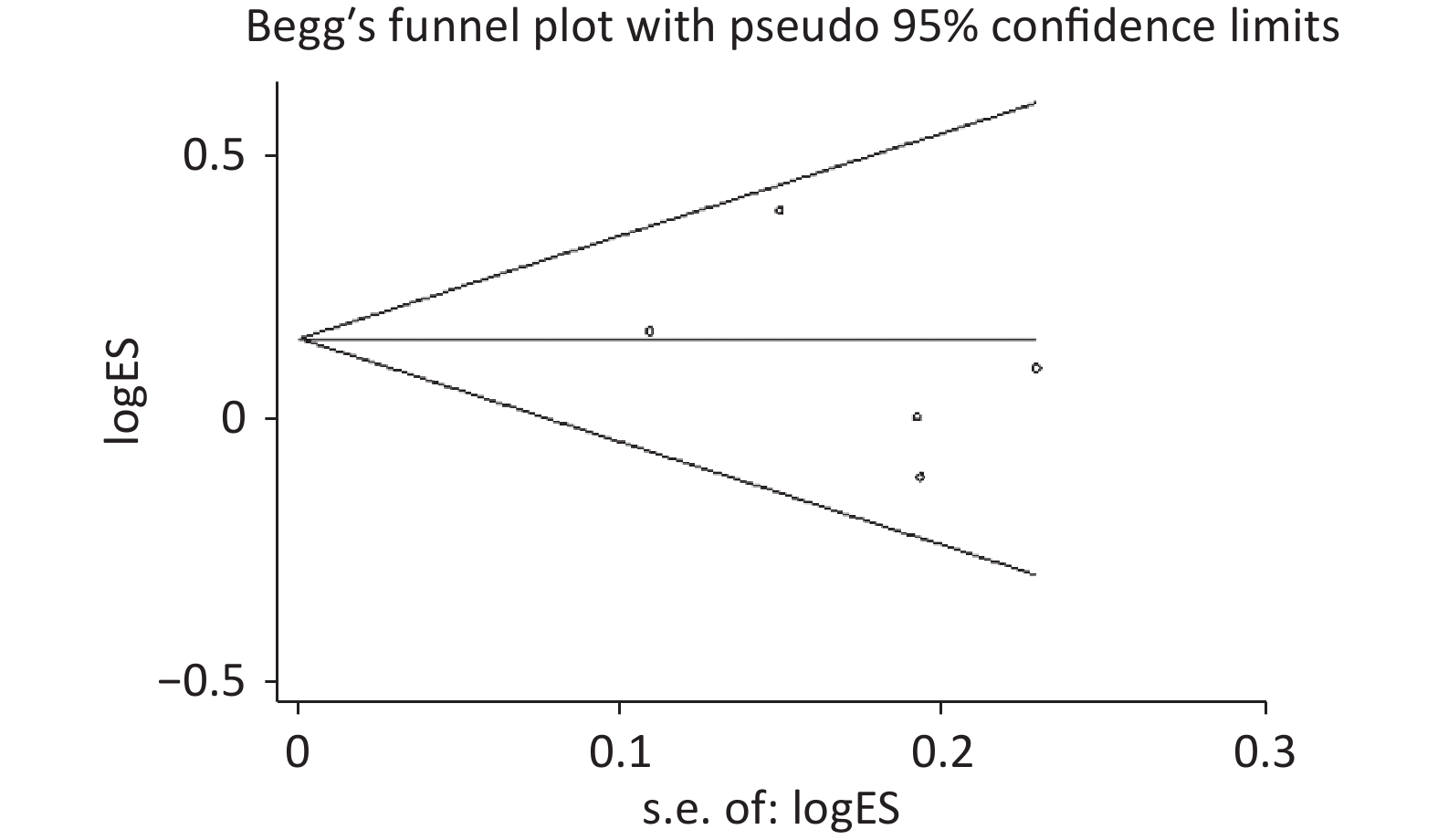
Comparison P Begg’s test Egger’s test GG vs. AA 1 0.75 AG vs. AA 0.46 0.45 (AG + GG) vs. AA 1 0.35 GG vs. (AA + AG) 1 0.88 G vs. A 0.81 0.76 Note. CRC, colorectal cancer. Table S3. Publication bias analysis of the relationship between the rs2274223 genetic polymorphism and CRC risk
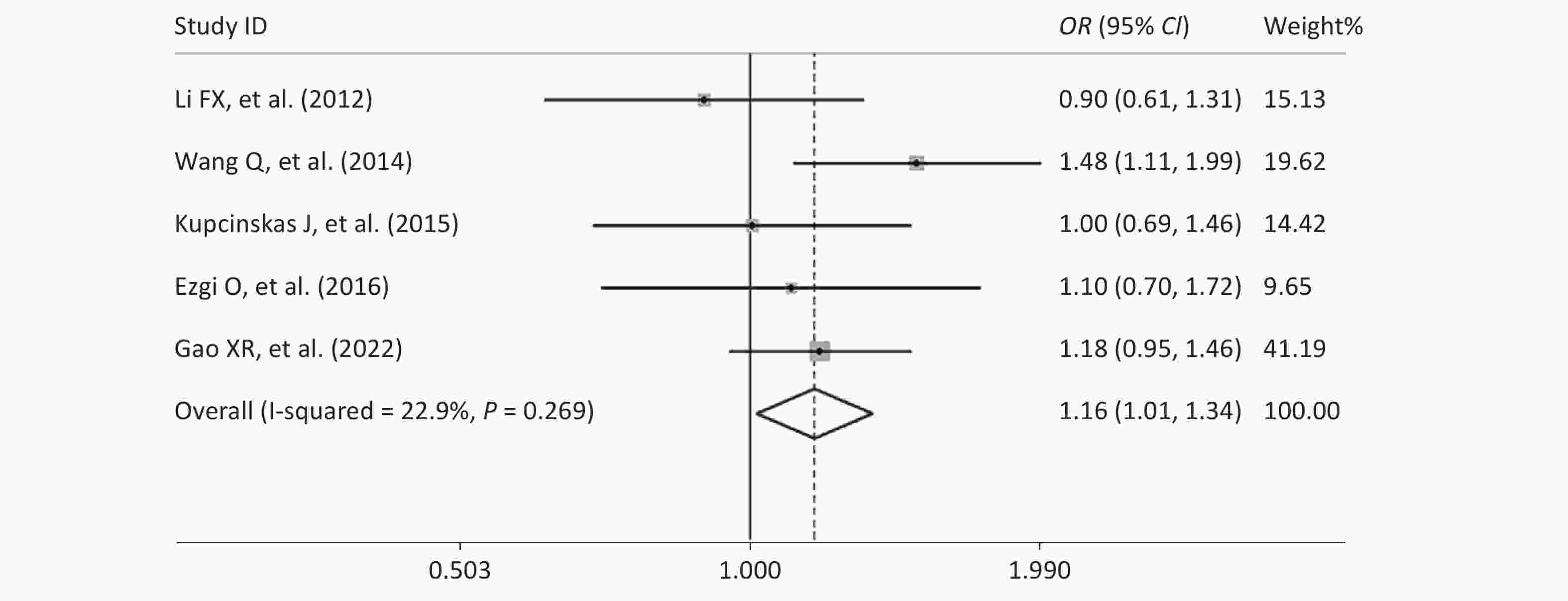
Figure S1. Forest plot of the relationship between the rs2274223 genetic polymorphism and CRC risk under AG vs. AA
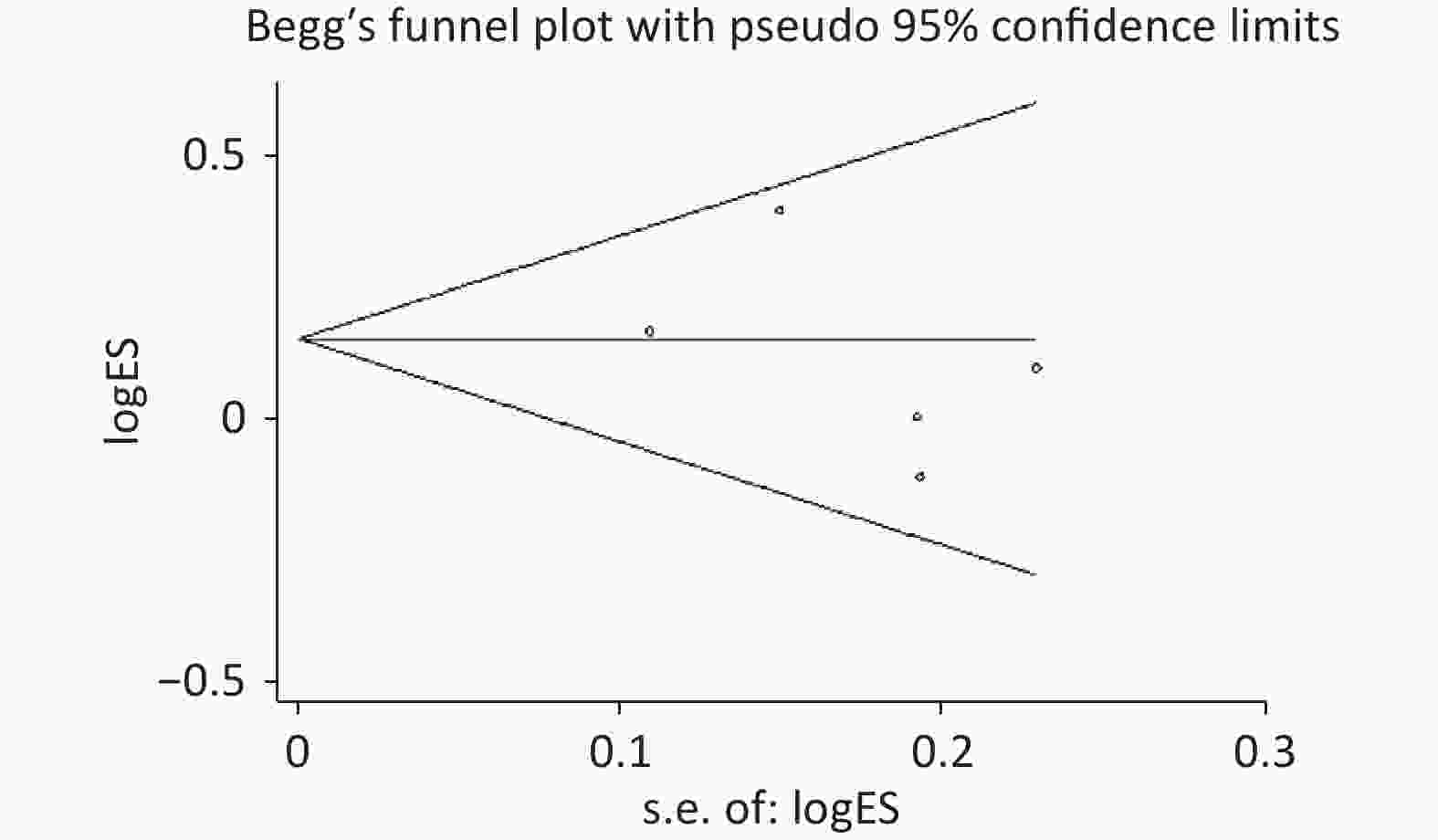
Figure S2. Begg’s funnel plot of the relationship between the rs2274223 genetic polymorphism and CRC risk under AG vs. AA
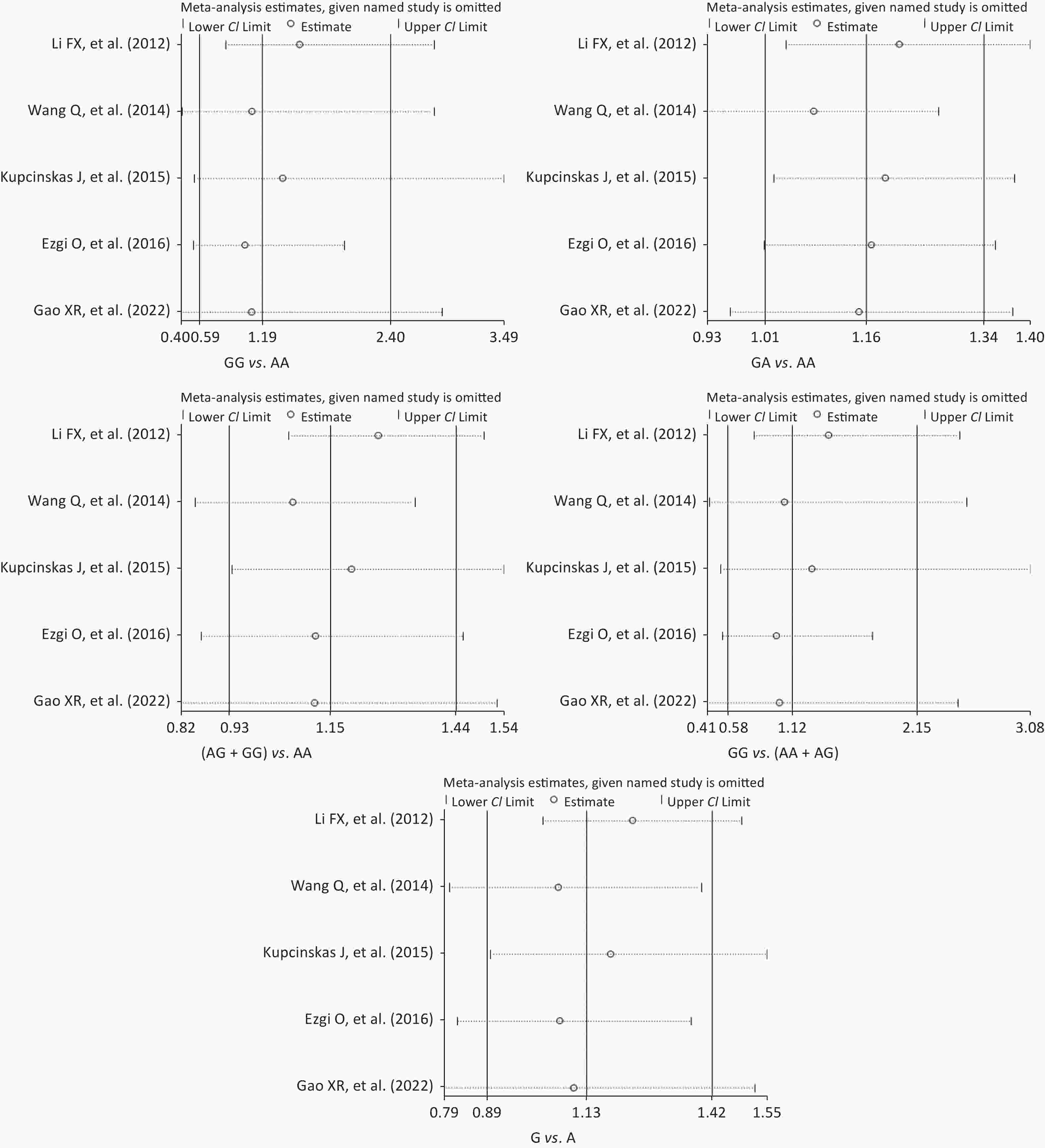
Figure S3. Sensitivity analysis of the relationship between the rs2274223 genetic polymorphism and CRC risk. CRC, colorectal cancer.
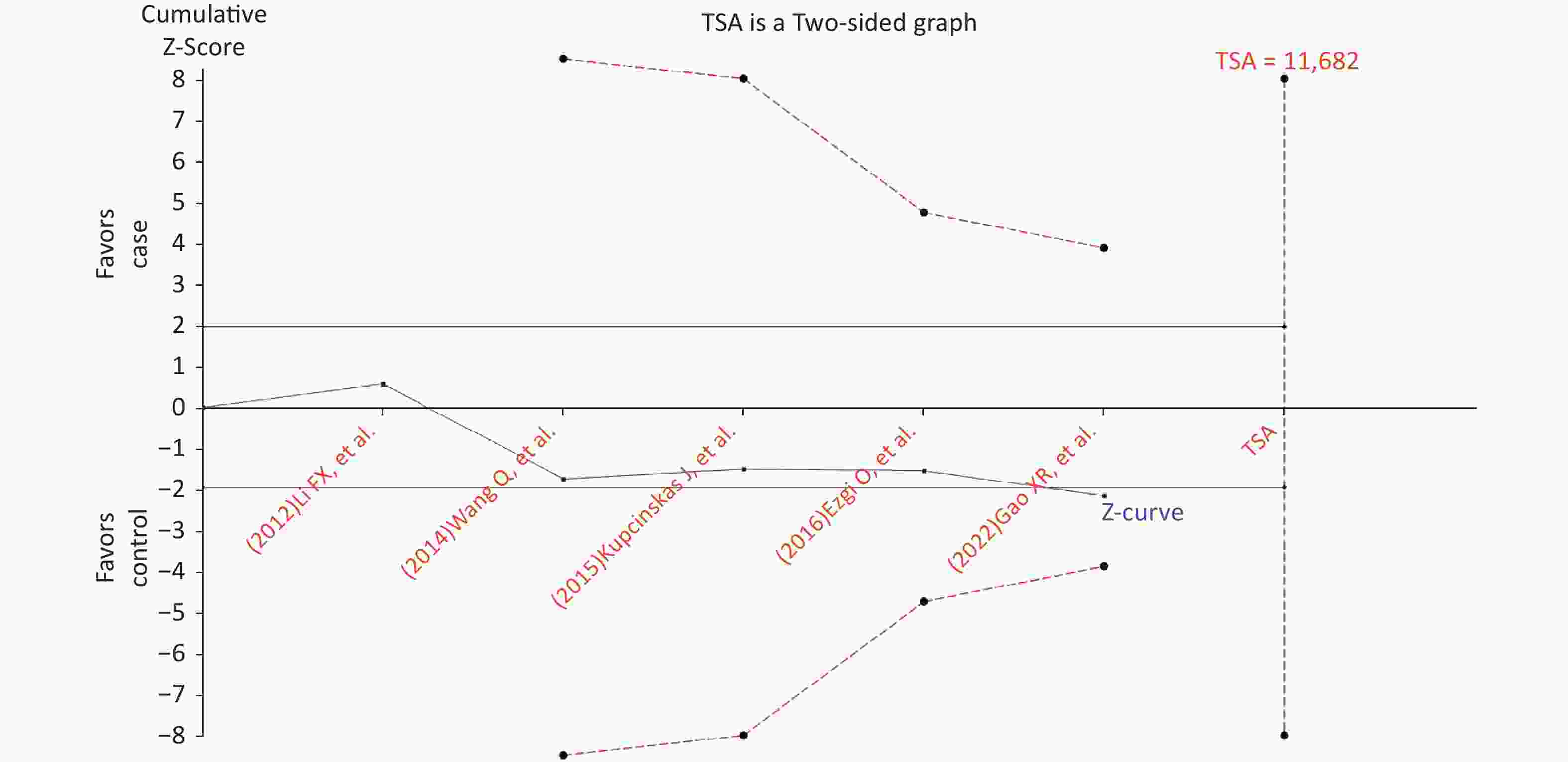
Figure S4. Trial sequential analysis for the relationship between the rs2274223 genetic polymorphism and CRC risk under AG vs. AA (The needed sample size is 11,682 samples, and the cumulative z-curve did not cross the trial sequential monitoring boundary before reaching the required sample size, indicating that our findings need to be confirmed further). CRC, colorectal cancer
In conclusion, the results of the current study suggest that the PLCE1 rs2274223 genetic polymorphism is associated with CRC risk and CRC tumor stage and can be useful as a biomarker.
HTML
 22305Supplementary Materials.pdf
22305Supplementary Materials.pdf
|

|







 Quick Links
Quick Links
 DownLoad:
DownLoad: