-
The overall incidence and burden caused by breast cancer is increasing, affecting the global socio-economic development and life expectancy [1]. According to “WHO Global Breast Cancer Initiative (GBCI) reports”, if the mortality rate due to breast cancer can be reduced by 2.4% annually, we may prevent 2.5 million breast cancer deaths between 2020–2040. Approximately 2.7 million new cases of breast cancer were reported globally in 2020, with 685,000 deaths. As per the study report of the National Cancer Institute-All India Institute of Medical Sciences (NIC-AIIMS), one out of every two women diagnosed with cancer in India dies because of late diagnosis. The first potential sign or symptom of breast cancer, such as a lump or thickening in the breast, is usually ignored and goes undetected until a later stage [2]. Existing screening methods like mammogram, fail to identify the disease in earlier stages as the interpretation of the mammogram result remains challenging for the clinicians. Incidence of false-positive mammographic reports can result in invasive clinical procedures, leading to long-term anxiety in patients. Meanwhile, false-negative reports result in the advancement of clinical stages [3]. Studies show that mammogram results are less sensitive among young women with higher breast density while mammographic screening precision is affected by varying tumour growth properties. Mammogram, biopsy and fine needle aspiration cytology are hospital-centric making it not accessible by certain groups of the society. Researchers are constantly investigating and developing modern screening techniques for the earlier identification of breast cancer. Before the actual onset of cancer, several underlying genetic changes take place leading to dysregulation of various biomarkers [4]. Among the various biomarker-based detection methods, serum-based miRNA biomarkers support non-invasive methods with more accurate bias-free results. miRNAs are 18–25 nucleotides long non-coding RNA that play an important role in regulating gene expression and metabolic pathways. miRNAs are the best biomarkers due to their substantially altered expression pattern in conjunction with cancer progression, stability, and ease of accessibility in multiple biofluids [5]. Under normal physiological conditions, miRNA plays an important role in post-translational gene regulation through translational inhibition and mRNA destabilisation. During the multistep transition of a normal cell to a cancerous cell, several genes involved in cell proliferation, differentiation, and death are impacted. The two most common types of genes involved in cancer progression are oncogenes (OncomiR in case of miRNA) and tumour suppressor genes. Overexpression of oncogenic gene products lead to tumour development, meanwhile, reduction or loss of function of tumour suppressor gene products lead to dysregulations associated with cancer progression [6]. It is well known that miRNA regulates carcinogenesis through its oncogenic and tumour suppressor activity. miRNA-21 being an oncomiR, was found to be over-expressed in breast cancer patients’ serum and was found to play a significant role as a non-invasive diagnostic biomarker for point-of-care (POC) application. A study on miRNA-195 along with three different target miRNAs in metastatic breast cancer has reportedly indicated downregulation in breast cancer. On the other hand, miRNA-195 was found to be under-expressed in benign and malignant groups. In our study we choose these two circulating miRNA biomarkers as a combination and evaluated their potential in screening breast cancer with increased specificity. In this work, we investigated the expression level of miRNA-21 and 195 using qPCR and studied the improvement in specificity when the two biomarkers come as a combination. This study is done towards developing a more specific POC screening instrument.
Ethical clearance was obtained from the Institutional Human Ethics Committee, Government of India (IHEC Approval No. PSG/IHEC/APPR/FB002 dated 8th Jan 2020 and renewal PSG/IHEC/Renew/052 dated 5th April 2021). The inclusion criterion is based on the specific factors such as, Age Group, consanguinity, medical history and family history. Pathologically confirmed breast cancer patients admitted to the Oncology department of PSG hospitals (PSGIMSR, Coimbatore) were included for the study with written consent. According to the clinical features and biopsy, breast cancer stages were confirmed. The desirable sample size for each group of diseased and non-diseased for an AUC ranging from 0.7 to 0.9 is 92 each with 95% of CI and 80% of power[7]. Since this study is using purposive sampling where the status of the participants is known, a sample size of 100 is chosen. A total of 93 blood samples were collected from the volunteers including 43 healthy and 50 clinically proven breast cancer patients. As per the inclusion criteria participants with a distinct age group 30–60 were included for breast cancer study. The samples were sorted based on the stage of the disease as well as Estrogen (ER), Progesterone (PR) and HER2/neu status. miRNA was isolated from serum using the miRNeasy Serum / Plasma Kit (Qiagen, Germany) according to the manufacturer's protocol and eluted with RNase-free water. Isolated miRNA was quantified using Nanodrop (SpectroMax, Quick Drop, Molecular Devices) and cDNA was synthesised using TaqMan miRNA cDNA synthesis kit and primer (ThermoFisher Scientific, USA) for miRNA-21, miRNA-195, and U6 snRNA as endogenous controls according to the manufacturer's protocol. The synthesised cDNA was used to perform quantitative real-time PCR (q-RT-PCR) with TaqManTM miRNA Assays (Thermo Fisher Scientific, USA) on a light cycler 96 (Roche, Switzerland). The data was normalised using U6 snRNA, and relative gene expression was calculated using the
$ {2}^{-\Delta \Delta Ct} $ method. Biostatistical analysis was done using MiniTab. Sensitivity and specificity were calculated for miRNA21 alone, miRNA195 alone and with ratio of expressions of the two chosen miRNAs at various cut off levels. Receiver operating characteristic (ROC) curves were plotted and areas under the curve (AUC) were calculated.Serum samples collected from 43 Healthy control and 50 breast cancer participants were sorted based on their age and clinical stage. Eight of the breast cancer samples were falling under triple negative (TNBC) category, out of which 2 had undergone surgery. All the samples were evaluated using q-RT-PCR and the expression patterns for both miRNAs were studied at different stages of breast cancer. miRNA-21 level was found elevated in breast cancer serum samples in comparison with healthy controls. On the other hand, miRNA-195 level was shown to be low in breast cancer serum. Clinical stages of 15 grade I (Invasive carcinoma), 14 grade II (Ductal carcinoma in situ), 7 grade III (Infiltrating ductal carcinoma) and 1 metastatic breast cancer grade IV and 13 were without the stage information.
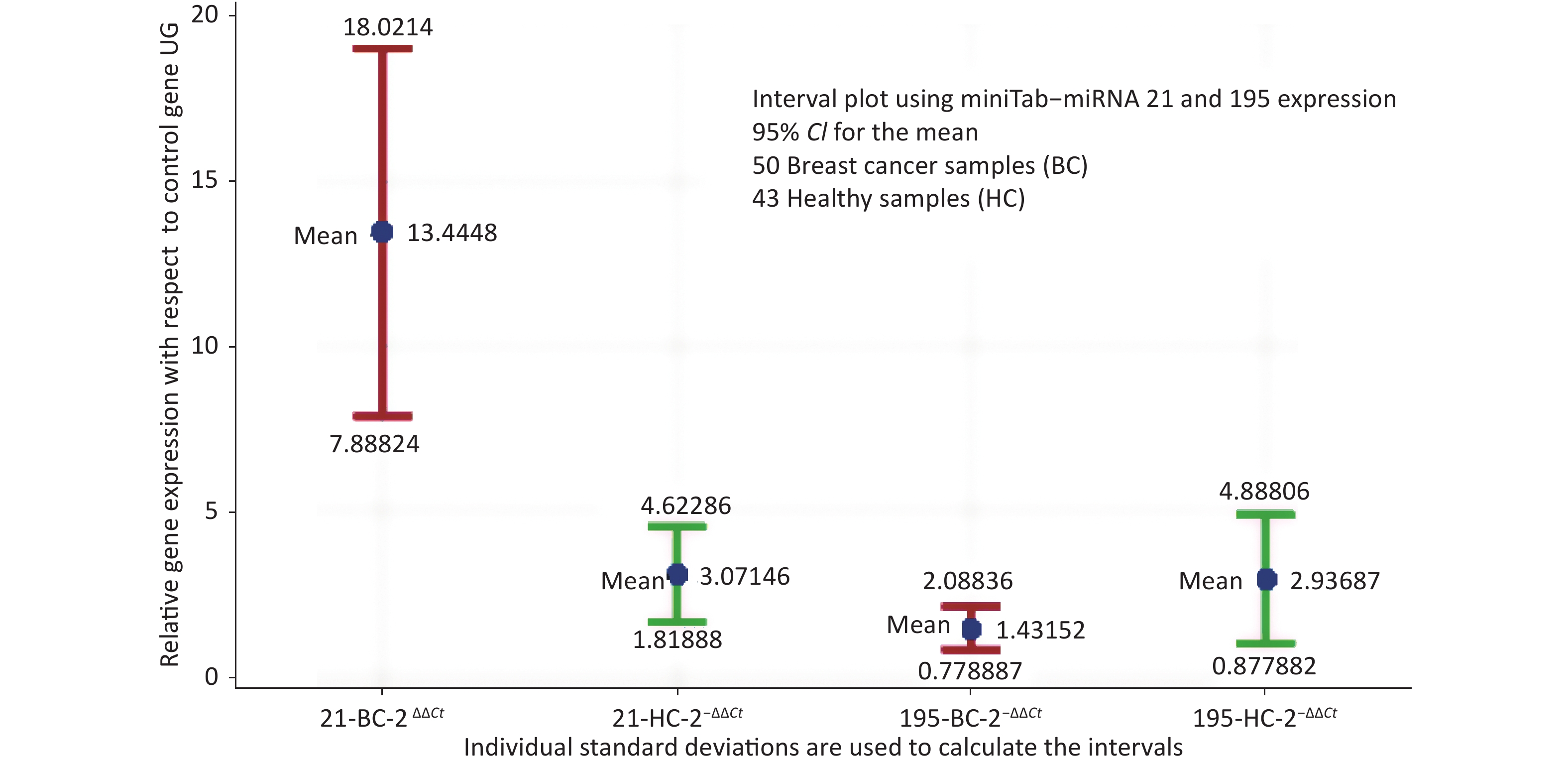
The descriptive statistics of miRNA-21 and miRNA-195 expression in healthy control and breast cancer cases were estimated and the normalcy of the data was analysed, where miRNA-21 represented normal distribution with P = 0.06, whereas miRNA-195 represented normalcy with P < 0.005. 50 BC samples presented a mean value of 13.44 and SD of 19.62 for
$ {2}^{-\Delta \Delta Ct} $ miRNA 21, while 43 HC presented 3.071 and 4.716 respectively. Similarly,$ {2}^{-\Delta \Delta Ct} $ miRNA 195 presented a mean of 1.432 and SD of 2.304 in BC and 2.937 and 6.366 respectively in HC. The mean values of miRNA-21 in healthy samples and breast cancer cases with respect to the control gene has a markable difference. miRNA-195 is at a higher expression level in healthy samples compared to breast cancer samples.The interval plot for the expression of miRNA-21 and 195 for breast cancer and healthy controls depicted in Figure 1 and it reveals that a combination of the two miRNAs will provide a better decision making logically. While screening using miRNA21 alone, since miRNA-21 is not only overexpressed in breast cancer serum, one more housekeeping gene like miRNA-195 expression will add to the specificity. The interval plot indicates that, miRNA 21 being low and miRNA 195 being high is indicative of the tested individual screened negative.
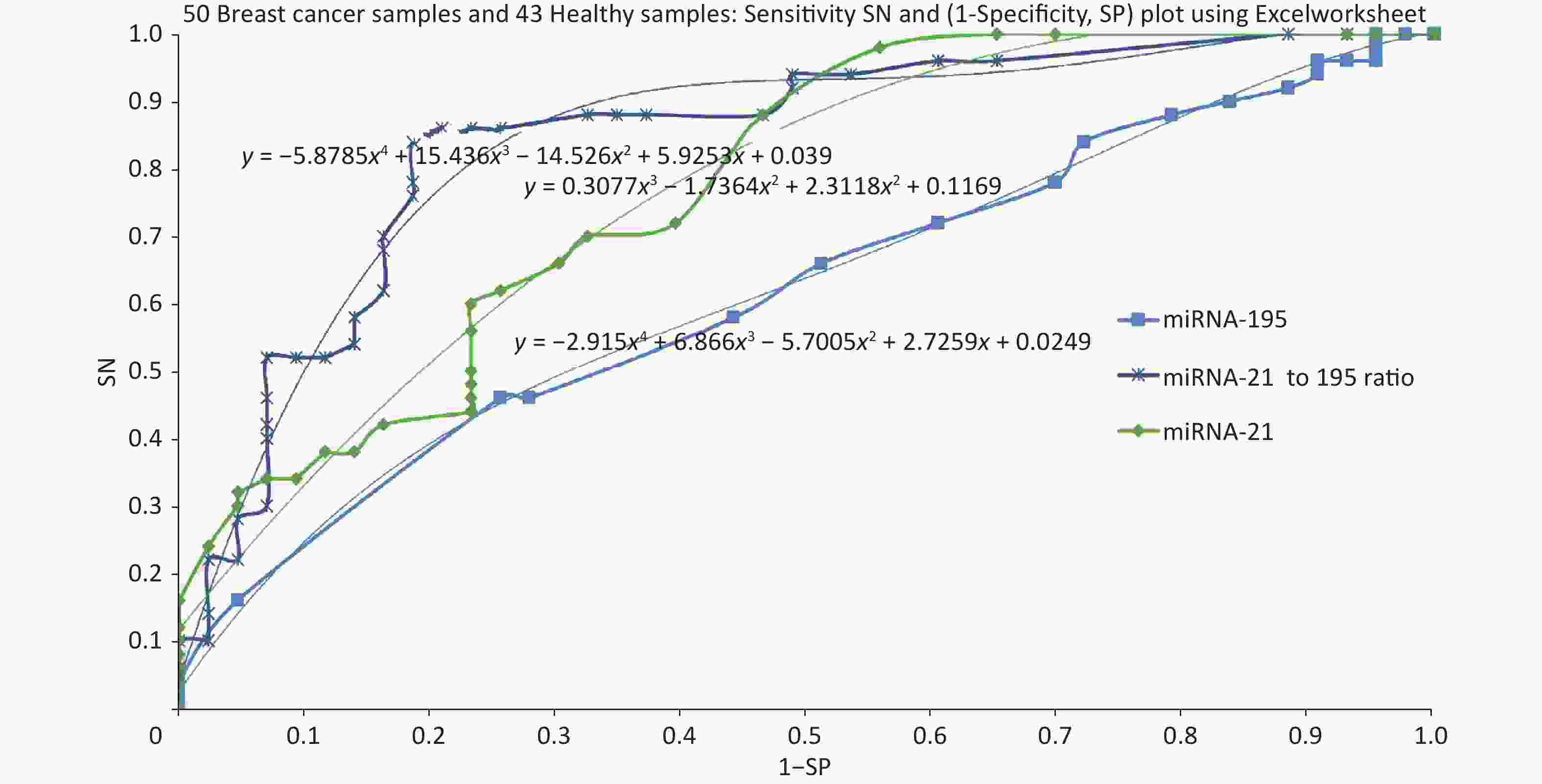
Specificity and sensitivity are estimated keeping various screening threshold levels and ROC was plotted for sensitivity Vs (1-Speocificity). Based on this data, ROC was plotted using miRNA-21, miRNA-195 followed by the ratio of miRNA-21 to miRNA-195. Figure 2 shows the ROC plots for 3 cases and the area under curve is 0.77 for miRNA-21, 0.62 for miRNA-195. Further we evaluated the ROC with a combination of miRNA-21 and miRNA-195 expression as a ratio of 21 to 195 which has resulted in an AUC of 0.87.
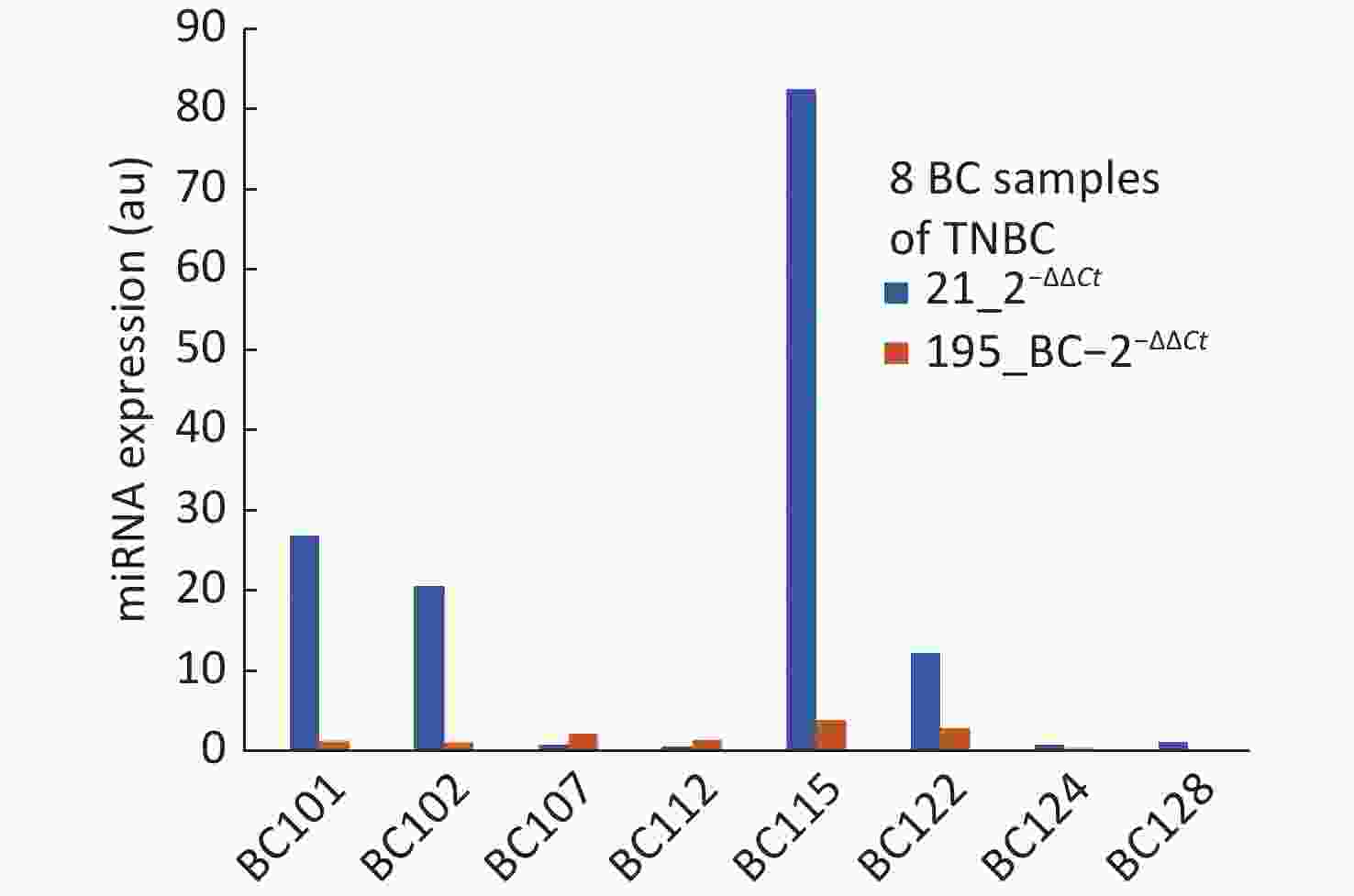
The objective of this study was to develop proof of concept towards realizing a screening system with a direct reading of miRNA expression using molecular beacon probe. An instrument was developed using miRNA-21 as the circulating biomarker [8]. An additional biomarker to improve the specificity was one of the major objectives for this study and mir195 was chosen. Meanwhile, in comparison with various reports about miRNA-21 and 195, our present study showed higher expression of miRNA-21 and lower expression of miRNA-195 in breast cancer participants than healthy controls at an earlier stage. The two biomarkers have proved to improve the specificity of screening compared to using miRNA-21 alone. However, the study should be conducted with more samples so that the miRNA population density gives a clear picture for their range of expression values. Some of the breast cancer samples were collected after surgery which has created more outliers in the distribution of miRNA expression. One participant (BC 108) who showed low expression of miRNA-21, had undergone traditional ayurvedic treatment, still miRNA-21 expressed higher than miRNA-195. Much information is not available for the healthy samples other than their age and apparently, they are healthy. Some of the healthy samples have shown abnormal values of miRNA-21 and 195. The ROC curves plotted for miRNA-21, MiRNA-195 and combination of miRNA-21 and 195 showed that the AUC were 0.77, 0.62 and 0.87 respectively. The analysis of samples which were reported as TNBC, revealed the outcome of miRNA-21 and 195 expressions as shown in Figure 3. TNBC being a challenge in curing breast cancer, the miRNA expressions are valid to detect TNBC, but for BC107 and BC112 which were post-surgery samples. This is indicative of prognosis possibility. To reduce referral in screening of breast cancer, we aimed to evaluate the potential of a combination of an oncogenic miRNA21 and a tumour suppressor miRNA195 for improved specificity. The results of this study demonstrate that miRNA-21 an oncogene have shown elevated levels in breast cancer which is highly linked with hormonal status of ER, PR and HER2 and grade I to grade III. The miRNA-195 tumour suppressor gene was found to be higher in healthy controls than that of miRNA-21. Hence, identifying the level of oncogene and a tumour suppressor gene in breast cancer serum has become a signature to distinguish early breast cancer participants from healthy controls. This outcome could be easily used for developing POC mode of screening by incorporating direct reading of miRNA using molecular beacon probe. The authors express their gratitude to Department of science and Technology/ Technology Development Program/Biomedical Device and Technology Development, Govt. Of India for the grant allocated for this project.
doi: 10.3967/bes2023.067
Improved Specificity for Breast Cancer Screening Using an Oncogenic (miRNA-21) and a Gene Suppressor (miRNA-195) miRNA in the Serum for a Point of Care (POC) Screening Solution
-
-
[1] Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2021; 71, 209−49. doi: 10.3322/caac.21660 [2] Bish A, Ramirez A, Burgess C, et al. Understanding why women delay in seeking help for breast cancer symptoms. J Psychosom Res, 2005; 58, 321−6. doi: 10.1016/j.jpsychores.2004.10.007 [3] McKinney SM, Sieniek M, Godbole V, et al. International evaluation of an AI system for breast cancer screening. Nature, 2020; 577, 89−94. doi: 10.1038/s41586-019-1799-6 [4] Maruvada P, Wang W, Wagner PD, et al. Biomarkers in molecular medicine: cancer detection and diagnosis. BioTechniques, 2005; 38, S9−15. doi: 10.2144/05384SU04 [5] Zou RY, Loke SY, Tang YC, et al. Development and validation of a circulating microRNA panel for the early detection of breast cancer. Br J Cancer, 2022; 126, 472−81. doi: 10.1038/s41416-021-01593-6 [6] Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer, 2006; 6, 259−69. doi: 10.1038/nrc1840 [7] Hajian-Tilaki K. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform, 2014; 48, 193−204. doi: 10.1016/j.jbi.2014.02.013 [8] Salim B, Athira MV, Kandaswamy A, et al. Microfluidic device for novel breast cancer screening by blood test using miRNA beacon probe. Biomed Microdevices, 2017; 19, 89. doi: 10.1007/s10544-017-0230-z -





 下载:
下载:






 Quick Links
Quick Links