-
Alzheimer’s disease (AD), characterized by progressive cognitive decline, is a neurodegenerative disease and the most common cause of dementia[1]. AD can be divided into preclinical stage, mild cognitive impairment (MCI), and AD dementia[2]. Typical pathological changes in AD include senile plaques deposition, neurofibrillary tangles, synaptic loss, and endolysosomal dysregulations[3,4]. Beta-amyloid (Aβ) is the main component of senile plaques, and its aggregation has been recognized as the cardinal pathological hallmark of AD[5]. The endolysosomal system is a critical site for the amyloidogenic processing of amyloid precursor protein (APP)[6]. Therefore, abnormal endolysosomal transport of APP could contribute partly to Aβ generation in AD progression.
The degradation of APP involves both the non-amyloid and amyloid degeneration pathways[3]. During non-amyloidogenic processing, APP is first cleaved by α-secretase (mainly metalloproteinase 10, ADAM10) to soluble APPα (sAPPα) and C-terminal fragments of 83 amino acids (C83). Then, C83 is cleaved by γ-secretase into the P3 peptide and APP intracellular domain (AICD)[6]. In the amyloidogenic pathway, APP undergoes first cleavage by BACE1 to sAPPβ and C-terminal fragment of 99 amino acids (C99), which is then further processed by γ-secretase to Aβ and AICD[7].
The transport of APP and its relative secretases in the endolysosomal system are essential for Aβ production[8]. After endoplasmic reticulum (ER) synthesis, APP is transported to the Golgi apparatus for various posttranslational modifications. Some APP exits the Golgi through the trans-Golgi network (TGN)[9]. Subsequently, some APP is transported to early endosomes, whereas others are secreted into the cell membrane for non-amyloid degradation or internalization. Endocytic APP is further sorted into early endosomes[4]. Early endosomes are sorting hubs, and their optimal acidic environment is essential for BACE1 activity[8,10]. From early endosomes, APP and its related products are transported to the TGN through the retromer-mediated pathway, sorted to the cell membrane by recycling endosomes, or targeted to late endosomes and lysosomes for further amyloid degradation[11]. Thus, factors regulating APP intracellular transport and endocytosis in the cell membrane may partially determine the rate of APP amyloid degradation.
Synaptic loss, an important early pathological change in AD, is associated with cognitive impairment[12]. Toxic Aβ and Tau could lead to synaptic loss or dysfunction[13]. Synaptic vesicle glycoprotein 2A (SV2A), an essential transporter-like synaptic vesicle protein, belongs to the major facilitator superfamily (MFS)[14,15]. SV2A is expressed in almost all synapses and serves as a biomarker of synaptic density[16]. Studies have shown that SV2A is essential for synaptic vesicular processes as it could modulate the size of the releasable pool and interact with proteins essential for neurotransmitter release[17]. SV2A could regulate the synaptic release of neurotransmitters by influencing the calcium sensitivity of synaptic vesicles[18]. [11C] UCB-J Positron Emission Tomography (PET) indicated that SV2A binding was reduced in the hippocampus of the early AD group compared to that in the cognitively normal (CN) group[19]. Moreover, SV2A is the target of the antiepileptic drug levetiracetam (LEV), and its absence or dysfunction is associated with epilepsy[20]. Previous studies have shown that LEV, an SV2A-targeted drug, significantly improved cognitive function in patients with Parkinson’s disease (PD)[17,21]. In this study, we investigated the effects and mechanisms of action of SV2A on APP intracellular trafficking and amyloid degradation.
-
AAV9 encoding mice mutant full-length SV2A and the relative control were purchased from SyngenTech (Beijing, China). APP/PS1 male mice (Beijing Viewsolid Biotech; Beijing, China) were anesthetized with pentobarbital sodium and randomly assigned to AAV-control (AAV-con) or AAV-SV2A overexpression (AAV-SV2A) groups for AAV injection (n = 7 for each group). The AAV-con (viral titer: 4.96 × 1012 vg/mL; 0.8 μL at each site) and AAV-SV2A (viral titer: 2.31 × 1013 vg/mL; 0.8 μL at each site) were injected bilaterally into the CA1 of the hippocampus (anterior-posterior: -2.7 mm, medial-lateral: 1.8 mm, dorsal-ventral: 2.0 mm). The needle was left in place for 3 min, slowly retracted, and the skin wounds were sutured.
All animal experiments were performed according to the National Institutes of Health guidelines and conformed to the guidelines of the Ethics Committee of Xuanwu Hospital of Capital University.
-
SH-SY5Y and N2a cells were purchased from American Type Culture Collection. SH-SY5Y and N2a cells were cultured in Dulbecco’s minimum Eagle’s medium (DMEM; BD Biosciences, USA) supplemented with 10% fetal bovine serum (FBS; BD Biosciences) and 1% Pen-Strep solution (BD Biosciences) at 37 °C in a humidified atmosphere of 5% CO2 in air.
Primary neurons were isolated from embryonic day 18 APP/PS1 mice (were purchased from Beijing Weixuan Technology, Beijing, China), separated from AAV-injected mice. Briefly, the brains were dissected in ice-cold DMEM (BD Biosciences) supplemented with 2% FBS (BD Biosciences), carefully removed the meninges and digested with 2 mg/mL papain (Sangon Biotech, Shanghai, China) for 10 min. FBS was used to terminate the digestion. The cells were plated in 24-well plates coated with poly-D-lysine (Sigma, Germany). Neurobasal medium (Thermo Fisher Scientific, USA) with 0.25% GlutaMAX™ (Gibco, USA), 0.5% Pen-Strep solution (BD Biosciences) and 2% B27 (Thermo Fisher Scientific) were used to cultured neurons. The culture medium was half-replaced every 3 days.
Mouse SV2A (NM_022030) overexpression (oeSV2A) lentivirus, mouse SV2A (NM_022030) oeSV2A plasmids, human oeSV2A (NM_014849.5) plasmids, mouse shSV2A (GCGTAAAGATCGGGAAGAATT) plasmids, and human shSV2A (ACCTGTTCGAGTACAAGTTTG) plasmid were purchased from SyngenTech (Beijing, China).
For plasmid transfection, SH-SY5Y and N2a cells were plated in six-well plates at a density of 2 × 105 cells/well and grown for 24 h. According to the manufacturer’ s protocol, 2 μg DNA, 200 μL jetOPTIMUS buffer and 2 μL jetOPTIMUS reagent (Polylus) were mixed and incubated for 10 min at room temperature. The final mixture was added to the inoculated cells and incubated for 48 h.
For lentivirus transfection, mouse primary neurons were seeded at a cell density of 1.0 × 105/well in 24-well plates. Then, neurons were infected with SV2A-overexpressed lentivirus at a multiplicity of infection (MOI) of 80 in the presence of 8 μg/mL Polybrene (Sigma). After 72–96 h, the neurons were harvested for further analysis.
-
Cells were lysed in radioimmunoprecipitation assay buffer (RIPA; Solarbio, Beijing, China) to extract total protein. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with primary antibodies overnight at 4 °C and secondary antibodies for 1 h at room temperature. Proteins were visualized using a Tanon 4600SF. The following primary antibodies were used: SV2A (1:2,000, ab32942, Abcam, USA), APP (1:5,000, ab32136, Abcam), BACE1 (1:1,000, ab183612, Abcam), early endosome antigen-1 (EEA1, 1:1,000, #610456, BD Biosciences), Ras-related protein Rab 7a (Rab7, 1:1,000, ab137029, Abcam), anti-Rab11 (1:1000, #5589, Cell Signaling Technology, USA), lysosomal associated membrane protein-1 (LAMP1, 1:1,000, #99437, Cell signaling Technology), TGN46 (1:1,000, 13573-1-AP, Proteintech, USA), ATPase Na(+)/K(+) transporting subunit α1(ATP1A1, 1:20,000, ab76020, Abcam), Arl5b (1:1,000, 11694-1-AP, Proteintech), and cathepsin D (CTSD, 1:1,000, ab75852, Abcam).
-
SH-SY5Y and N2a cells were washed three times with ice-cold phosphate-buffered saline (PBS) and extracted with lysis buffer containing 100× protease inhibitor cocktail, 0.1 mol/L EDTA (Beyotime, Shanghai, China), PMSF (Solarbio, Beijing, China), and 1% Triton X-100. The supernatants were incubated with 4 μg agarose-conjugated antibody at 4 °C for 4 h. The samples were incubated with protein A/G beads (Millipore) at 4 °C overnight. After extensive washing, the immunoprecipitated beads were eluted with 1× loading buffer, boiled at 100 °C for 10 min and analyzed by western blotting.
-
Total RNA was extracted from cells using Trizol® LS reagent (Invitrogen, USA) according to the manufacturer’ s protocol. Then, 1 μg RNA was reversed-transcribed into cDNA using 5 × All-In-One RT Master Mix (Abm). PCR was performed in duplicate with TB Green Premix Ex Taq™ II (TaKaRa) on the 7,500 real-time PCR system (ThermoFisher Scientific). The average threshold cycle (Ct) of the fluorescent units was used to analyze the mRNA levels. The relative quantification of mRNA expression was calculated using the 2−ΔΔCt method after adjusting the levels to the corresponding internal GAPDH and normalized against control samples. The primer sequences used in this study are listed in Table 1.
Table 1. Sequences used for RT-qPCR
Target gene Forward primer Reverse primer Human SV2A GCCCAACAGTATGAAGCCATCCTAC ACCTCCACACCGTCAGCCATC Mouse SV2A CTCACTCTCAGTCAACAGCGTCTTC CACTCCACCGATCATCCAGAACATAC Human APP TGATGATGAGGATGTGGAGGATGG TGTGGTGGTGGTGGCAGTG Mouse APP GACTGACCACTCGACCAGGTTCTG CTTGTAAGTTGGATTCTAATATCCG -
Cells were fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature and blocked with 10% donkey serum for 30 min at 37 °C. Primary antibodies were diluted with 5% donkey serum albumin and incubated at 4 °C overnight. On the second day, the sections were washed and incubated with secondary antibodies conjugated to Alex Flour 488 (1:1,000, ab150105, Abcam and 1:1,000, ab150073, Abcam) and Alex Flour 594 (1:1,000, ab150108, abcam and 1:1,000, ab150076, abcam) for 1 h at 37 °C. Nuclei were stained with DAPI (Merck).
The frozen tissue samples were fixed with 4% PFA for 15 min at room temperature and permeabilized with 0.1% Triton X-100 for 15 min. The sections were then blocked with 10% donkey serum albumin for 30 min and incubated with the primary antibodies at 4 °C overnight. Alexa Fluor 488 (1:400, ab150105, Abcam; 1:400, ab150073, Abcam) and Alexa Fluor 594 (1:400, ab150108, Abcam; and 1:400, ab150076, Abcam) were used as secondary antibodies for 1 h at room temperature. Sections were coated with an antifade mounting medium containing DAPI and observed under a Zeiss LSM 800 confocal microscope and a Leica TCS SP8 confocal microscope.
The region of interest (ROI) selection adhered to the following criteria: intact cellular structures, excluding obvious background areas, randomly selecting five fields per sample and a total of 30–45 cells/group from three biological replicates for analysis. Colocalization quantification was performed using the JACOP plugin in ImageJ to calculate Mander’s overlap coefficient. Statistical analyses were conducted using GraphPad Prism 9.0, employing two-tailed unpaired t-tests. All experiments were independently repeated in triplicate, and statistical significance was defined as P < 0.05[22].
The following primary antibodies were used: APP (1:100, ab32136, abcam and 1:100, 14-9749-82, Thermo Fisher Scientific), EEA1 (1:50, #6104560, BD Biosciences and 1:100, sc-137130, Santa Cruz Biotechnology), Rab7 (1:100, ab137029, abcam), Rab11 (1:100, #5589, Cell Signaling Technology), LAMP1 (1:200, sc-20011, Santa Cruz Biotechnology), cathepsin D (1:100, ab75852, abcam), TGN46 (1:100, 13573-1-AP, Proteintech), ATP1A1 (1:500, ab76020, abcam) and 6E10 (1:500, 803014, BioLegend).
-
Cells were starved in serum-free DMEM for 30 min at room temperature and incubated with the anti-APP antibody (1:100, ab32136, abcam) in a complete medium containing 10 nmol/L HEPES (Beyotime) at 4 °C for 45 min. After washing with the complete medium containing 10 nmol/L HEPES, cells were returned to 37 °C for 10 min. Then, cells were fixed with 4% PFA for 15 min and blocked with 10% donkey serum for 30 min. The remaining surface APP was labeled with a secondary antibody conjugated to Alexa Fluor 488 (1:1,000, ab150073, Abcam). Subsequently, cells were permeabilized with 0.1% Triton for 5 min, and internalized APP was labeled with a secondary antibody conjugated to Alexa Fluor 594 (1:1,000, ab150076, Abcam). Images were acquired using a Zeiss LSM 800 confocal microscope[23].
-
The LysoTracker Red DND-99 (Thermo Fisher Scientific, L7528) probe was used to visualize lysosomes, and lysosome staining was performed as described[24]. SH-SY5Y and N2a cells were cultured at a density of 0.6×105 into glass coverslips and incubated with LysoTracker (75 nmol/L) for 1 h at 37 °C. The medium was aspirated, and the cells were washed with PBS three times to remove the unbound probe. Then, cells were fixed with 4% PFA (Solarbio) for 15 min and incubated with DAPI (Merck) for 10 min at room temperature. A Zeiss LSM 800 microscope was used to capture confocal images.
-
Commercial ELISA kits were used to determine secreted Aβ1-40 (Mlbio, ml001859), Aβ1-42 (Mlbio, ml002201), sAPPα (Mlbio, ml625251V), and sAPPβ (Mlbio, ml057880V) levels in SH-SY5Y and N2a cell culture supernatants.
-
All data are presented as mean ± standard error of the mean (SEM; immunofluorescence: 30–45 cells total per group from 3 biologically independent experiments; primary neurons: 25–30 cells total from 3 batches; other assays: n = 3 biologically independent replicates) and analyzed using GraphPad Prism. Unpaired two-tailed t-tests or two-way ANOVA were used to analyze the data. P < 0.05 was considered statistically significant.
-
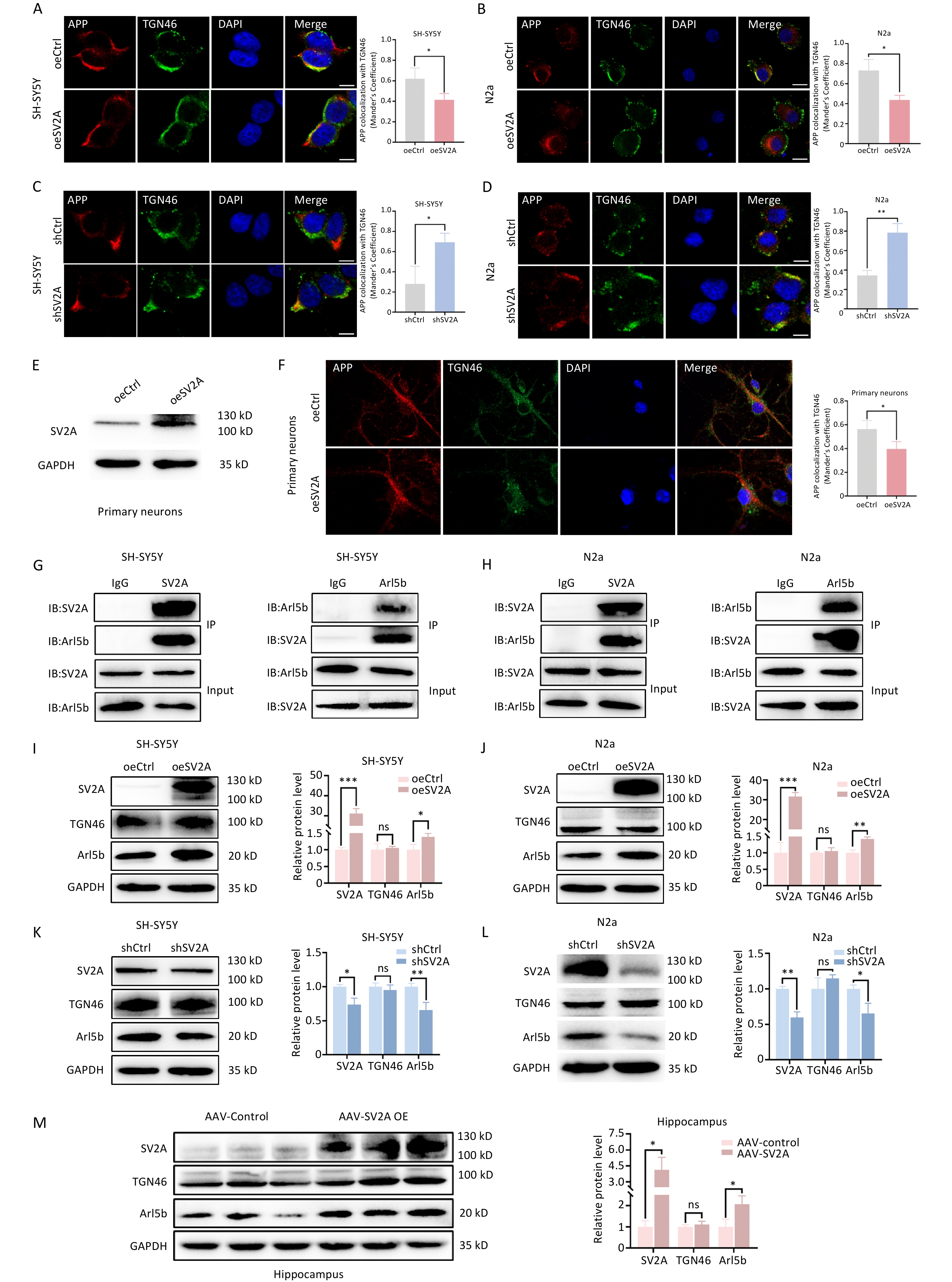
To investigate the effect of SV2A on APP distribution in the TGN, colocalization of APP and TGN46, a recognized biomarker of the TGN, was performed using an immunocytochemistry confocal assay. The results showed that the colocalization of APP with TGN46 decreased significantly in SH-SY5Y and N2a cells transfected with the oeSV2A plasmid (Figure 1A and 1B), whereas it increased significantly in cells transfected with the shSV2A plasmid (Figure 1C and 1D). Colocalization was also decreased in mouse primary neurons transfected with the SV2A-overexpressed lentivirus (Figure 1E and 1F). Moreover, the expression of TGN46 was not different between SV2A-overexpressed or knockdown cells and hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (Figure 1I-1L). These results demonstrate that SV2A reduced the distribution of APP in the TGN.
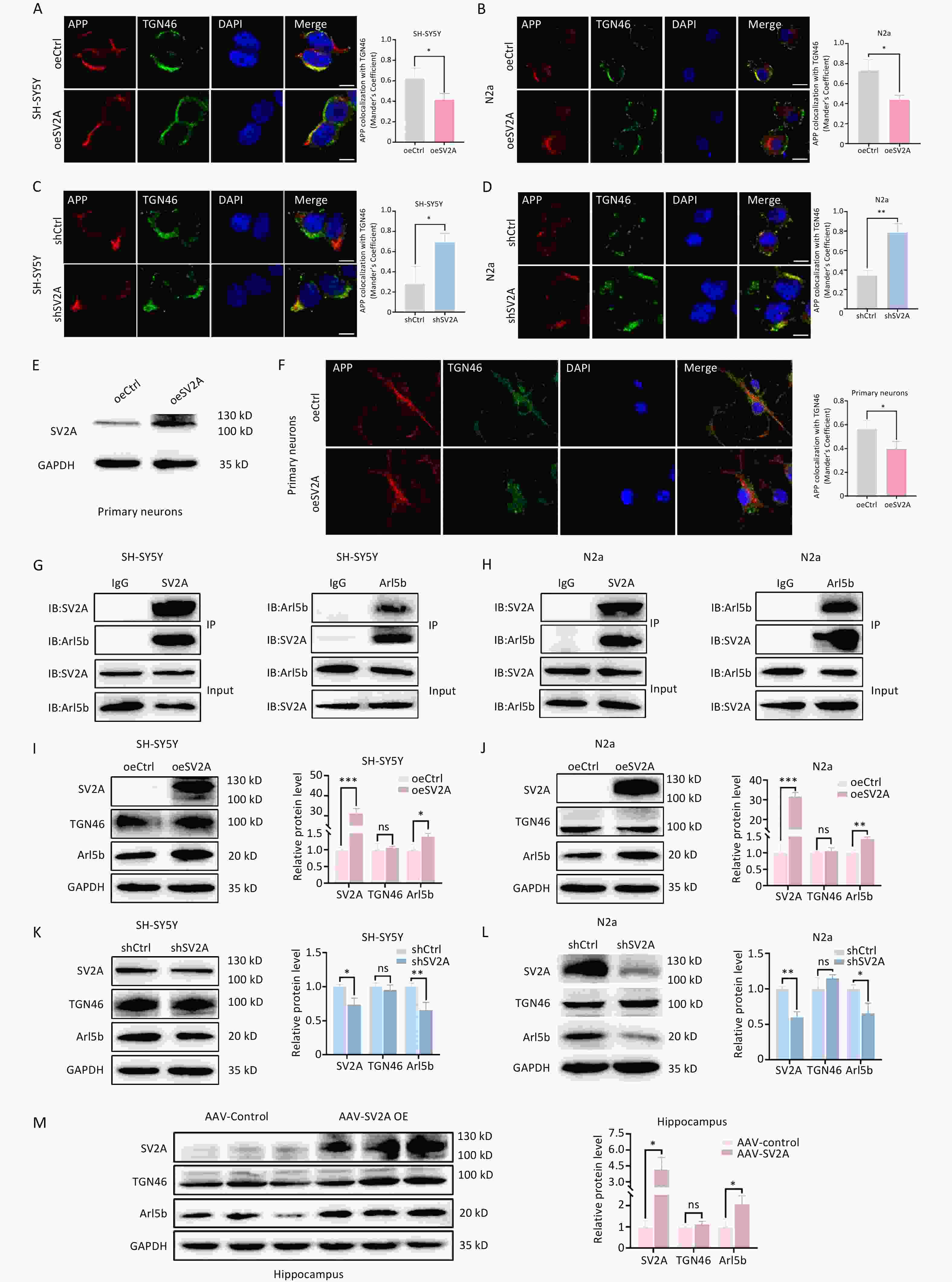
Figure 1. Effect of SV2A on APP distribution in the trans-Golgi network and Ar15b expression. (A–B) Colocalization of APP (red) with TGN46 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Colocalization of APP (red) with TGN46 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) Protein levels of SV2A in primary neurons transfected with SV2A-overexpressed lentivirus. (F) Colocalization of APP (red) with TGN46 (green) in mice primary neurons transfected with SV2A-overexpressed lentivirus. (G) Interaction of SV2A and Arl5b in SH-SY5Y cells by Co-IP assay. (H) Interaction between SV2A and Arl5b in N2a cells by Co-IP assay. I and J. Protein levels of SV2A, TGN46, and Arl5b in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (K–L) Protein levels of SV2A, TGN46, and Arl5b in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (M) Protein levels of SV2A, TGN46 and Arl5b in hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significantly different. Note: AAV, Adeno-associated virus; Arl5b, ADP-ribosylation factor 5b; APP, amyloid precursor protein; Co-IP, Co-immunoprecipitation; SV2A, synaptic vesicle glycoprotein 2A; TGN, the trans-Golgi network.
Arl5b, located mainly in the TGN, promotes the transport of APP from the TGN to early endosomes. Arl5b depletion induced APP accumulation, APP processing and Aβ generation in the TGN[9]. To investigate the mechanisms by which SV2A regulates APP distribution in the TGN, the effects of SV2A on Arl5b expression and the interaction between SV2A and Arl5b were observed in vitro and in vivo. A notable interaction was detected between SV2A and Arl5b in SH-SY5Y and N2a cells using a Co-IP assay (Figure 1G and 1H). Moreover, significant upregulation of Arl5b was observed in SH-SY5Y and N2a cells transfected with the oeSV2A plasmid (Figure 1I and 1J), as well as in the hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (Figure 1M). Conversely, the protein level of Arl5b was downregulated in cells transfected with the shSV2A plasmid (Figure 1K and 1L). These results indicate that SV2A reduces APP distribution in the TGN via the partial transport enhancement of APP from the TGN to early endosomes, which is mediated by Arl5b upregulation.
-
To further investigate the effect of SV2A on APP distribution in early endosomes, the protein levels of the early endosome biomarker EEA1 and the colocalization of APP and EEA1 were analyzed in vitro and in vivo. The results showed that the protein levels of EEA1 were markedly downregulated in SH-SY5Y and N2a cells overexpressing SV2A (Figure 2A and 2B), as well as in the hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (Figure 2E). In contrast, EEA1 was significantly upregulated in cells with SV2A knockdown (Figure 2C and 2D). Moreover, the colocalization of APP with EEA1 was reduced in SV2A-overexpressed cells and mouse primary neurons (Figure 2F, 2G, and 2J); however, it increased in SV2A-knockdown cells (Figure 2H and 2I). Notably, SV2A may decrease the number of early endosomes and distribution of APP in early endosomes.

Figure 2. Effect of SV2A on APP distribution in early endosomes of neurons. (A–B) Protein levels of SV2A and EEA1 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A and EEA1 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) Protein levels of SV2A and EEA1 in hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). (F–G) Colocalization of APP (red) with EEA1 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (H–I) Colocalization of APP (red) and EEA1 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with EEA1 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; EEA1, early endosome antigen-1; SV2A, synaptic vesicle glycoprotein 2A.
The reason SV2A reduced APP distribution in early endosomes, considering that SV2A-mediated upregulation of Arl5b promotes APP transport from the TGN to the early endosomes, is unclear. Early endosomes are the sorting hubs of the endocytic pathway[25]. APP in early endosomes can be sorted into late endosomes, recycling endosomes, or the TGN[4]. Moreover, APP in late endosomes is mainly transported to lysosomes, whereas APP in recycling endosomes is transported to the cell membrane. Thus, APP is thought to be transported mostly out of early endosomes in oeSV2A-transfected cells.
-
C99, the amyloid degradation product of APP, could be cleaved by γ-secretase to Aβ in late endosomes[26,27]. To investigate the effect of SV2A on APP distribution in late endosomes, the protein levels of the late endosome biomarker Rab7 and the colocalization of APP and Rab7 were examined in vitro and in vivo. The results showed that Rab7 levels were downregulated in SH-SY5Y and N2a cells transfected with oeSV2A plasmids (Figure 3A and 3B), as well as in the hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (Figure 3E). In contrast, Rab7 was significantly upregulated in cells transfected with the shSV2A plasmid (Figure 3C and 3D). Moreover, the colocalization of APP with Rab7 was reduced in SV2A-overexpressed cells and mouse primary neurons (Figure 3F, 3G, and 3J), whereas it was enhanced in SV2A-knockdown cells (Figure 3H and 3I). These results showed that SV2A decreased the number of late endosomes and APP distribution in late endosomes, indicating that late endosomes are not the main diversion direction for APP in early endosomes or that APP in late endosomes could mostly be transported to lysosomes.
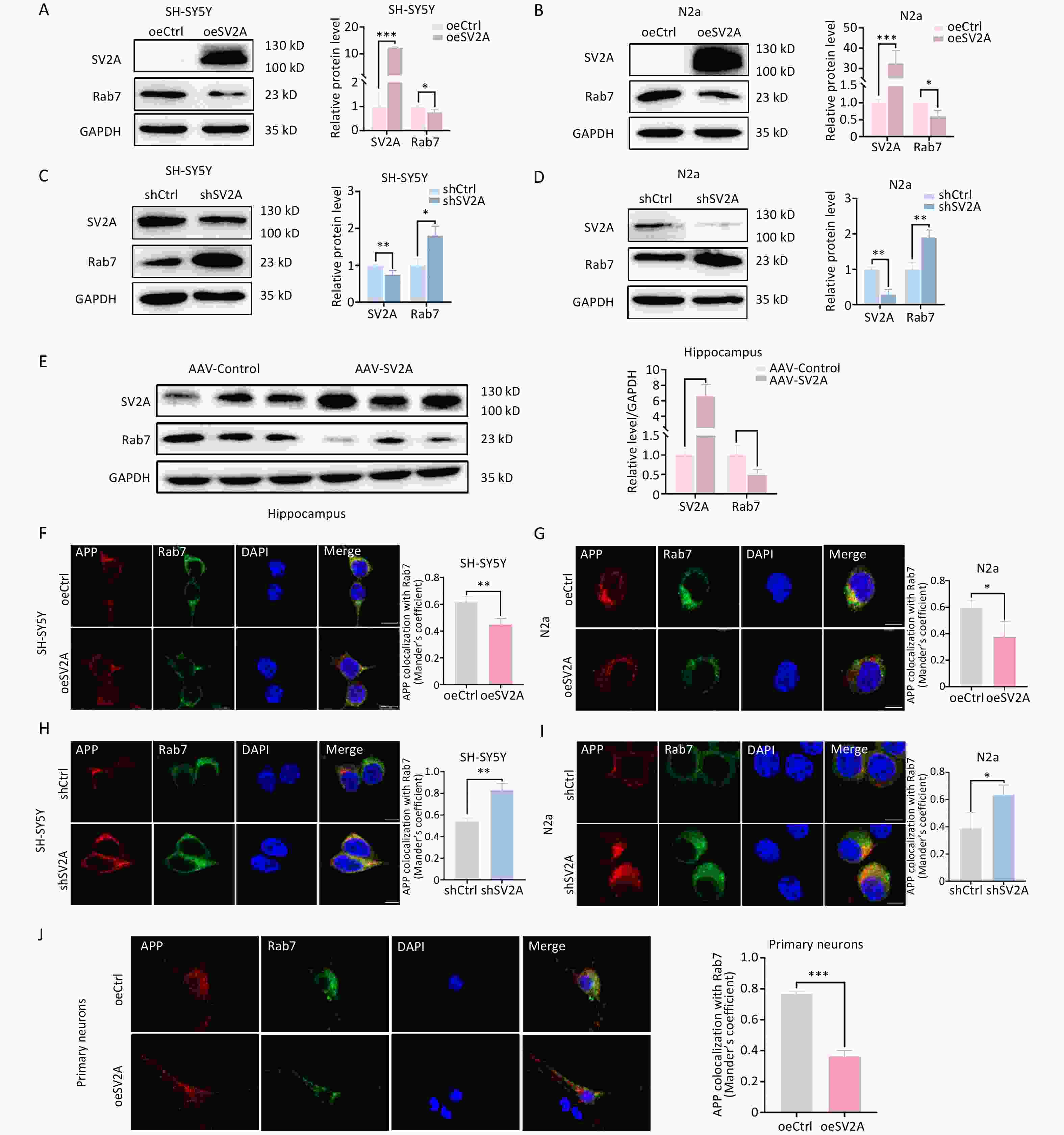
Figure 3. Effect of SV2A on APP distribution in the late endosomes of neurons. (A–B) Protein levels of SV2A and Rab7 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A and Rab7 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. E. Protein levels of SV2A and Rab7 in hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). (F–G) Colocalization of APP (red) and Rab7 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (H–I) Colocalization of APP (red) and Rab7 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with Rab7 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; Rab7, Ras-related protein Rab 7a; SV2A, synaptic vesicle glycoprotein 2A.
-
Lysosomes are essential for the clearance of unwanted cellular components, including damaged or misfolded proteins[26,28]. As mentioned earlier, APP in late endosomes is primarily transported to lysosomal organelles[29]. To investigate the effect of SV2A on APP transport in lysosomes, the protein levels of the lysosomal biomarker LAMP1 and the colocalization of APP with LAMP1 were examined in vitro and in vivo. The results showed that LAMP1 expression was downregulated in SH-SY5Y and N2a cells transfected with the oeSV2A plasmid (Figure 4A and 4B), as well as in the hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (Figure 4E). Contrastingly, LAMP1 was significantly upregulated in cells transfected with the shSV2A plasmid (Figure 4C and 4D). In addition, the colocalization of APP with LAMP1 decreased in SV2A-overexpressed cells and mouse primary neurons (Figure 4F, 4G, and 4J), whereas it increased in SV2A knockdown cells (Figure 4H and 4I). These results indicate that SV2A reduced the number of lysosomes and the distribution of APP in lysosomes.
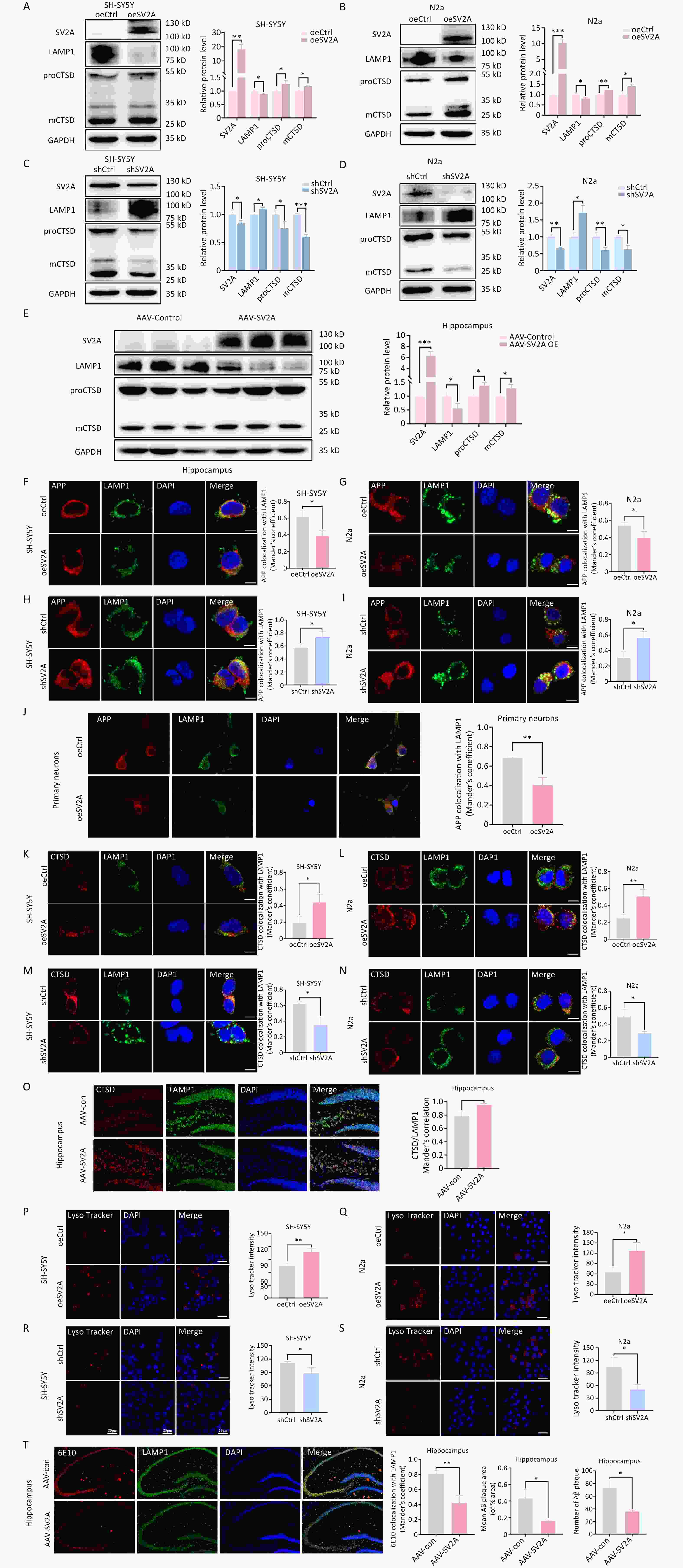
Figure 4. Effect of SV2A on APP distribution in lysosomes and cathepsin D expression of neurons. (A–B) Protein levels of SV2A, LAMP1, and cathepsin D in SH-SY5Y and N2a cells transfected with SV2A-overexpression plasmids. (C–D) Protein levels of SV2A, LAMP1, and cathepsin D in SH-SY5Y and N2a cells transfected with shSV2A plasmids. E. Protein levels of SV2A, LAMP1 and cathepsin D in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n =7/group). (F–G) Colocalization of APP (red) with LAMP1 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpression plasmids. (H–I) Colocalization of APP (red) with LAMP1 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with LAMP1 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. (K–L) Colocalization of cathepsin D (red) with LAMP1 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (M–N) Colocalization of cathepsin D (red) with LAMP1 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (O) Colocalization of cathepsin D (red) with LAMP1 (green) in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n = 4/group). (P–Q) LysoTracker staining in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (R–S) LysoTracker staining in SH-SY5Y and N2a cells transfected with shSV2A. (T) Colocalization of 6E10 (red) with LAMP1 (green) in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n = 4/group). Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; LAMP1, lysosomal associated membrane protein-1; SV2A, synaptic vesicle glycoprotein 2A.
Cathepsin-D (CTSD), the major lysosomal protease, is essential for lysosomal protein hydrolysis[24]. Mature CTSD (mCTSD) is synthesized by processing the precursor CTSD (proCTSD) in lysosomes, and mainly exerts its activity in a lysosomal acidic environment[30,31]. To investigate the effects of SV2A on lysosomal function, we examined CTSD expression and lysosomal staining in vitro and in vivo. The results showed that the protein levels of mCTSD and proCTSD were significantly increased in SH-SY5Y and N2a cells transfected with oeSV2A plasmids (Figure 4A and 4B), as well as in the hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (Figure 4E). However, mCTSD and proCTSD were markedly downregulated in cells transfected with shSV2A (Figure 4C and 4D). Moreover, immunofluorescence staining revealed that the colocalization of CTSD and LAMP1 increased in SV2A-overexpressed cells (Figure 4K and 4L), as well as in the hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (Figure 4O). Conversely, colocalization was reduced in SV2A-knockdown cells (Figure 4M and N). These experiments suggest that SV2A enhances the expression of CTSD, which possesses hydrolytic activity. Consistent with these observations, the fluorescence intensity of the LysoTracker Red dye, a probe sensitive to lysosomal pH, increased in SV2A-overexpressed cells (Figure 4P and 4Q) and decreased in SV2A-downregulation cells (Figure 4R and 4S). These results indicated that SV2A enhanced lysosomal function.
Lysosomes are the major site for Aβ generation[26,28]. To observe the effects of SV2A on Aβ localization in lysosomes, the colocalization of the classical Aβ antibody 6E10 and LAMP1 was experimented. The co-labeling experiments revealed that the colocalization of 6E10 and LAMP1 decreased in the hippocampal tissues of APP/PS1 mice injected with AAV-SV2A. Moreover, the number of Aβ plaques and mean Aβ plaque area, pathological hallmarks of AD, were reduced in hippocampal tissues with SV2A overexpression (Figure 4T).
These results showed that SV2A reduced the number of lysosomes, APP distribution and Aβ generation in lysosomes, although it could enhance lysosomal functions. These results also demonstrate that late endosome–lysosomes are not the main diversion direction for APP in early endosomes.
-
APP in early endosomes can be transported to recycling endosomes in addition to late endosomes[4]. To investigate the effect of SV2A on APP distribution in recycling endosomes, protein levels of the recycling biomarker Rab11 and the localization of APP with Rab11 were examined in vitro and in vivo. The results showed that Rab11 protein levels were upregulated in SH-SY5Y and N2a cells transfected with the oeSV2A plasmid (Figure 5A and 5B), as well as in the hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (Figure 5E). Contrastingly, Rab11 was significantly downregulated in cells transfected with the shSV2A plasmid (Figure 5C and 5D). Furthermore, the colocalization of APP with Rab11 was increased in SV2A-overexpressed cells and mouse primary neurons (Figure 5F, 5G, and 5J), whereas it was reduced in SV2A-knockdown cells (Figure 5H and 5I). These results demonstrate that SV2A increases the number of recycling endosomes and the distribution of APP in the recycling endosomes.
Figure 5. Effect of SV2A on APP distribution in neuronal recycling endosomes. (A–B) Protein levels of SV2A and Rab11 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A and Rab11 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) Protein levels of SV2A and Rab11 in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). (F–G) Colocalization of APP (red) and Rab11 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (H–I) Colocalization of APP (red) and Rab11 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with Rab11 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; Rab11, Ras-related protein Rab 11; SV2A, synaptic vesicle glycoprotein 2A.
-
The cell membrane is an important site for non-amyloid degradation of APP[3]. It is also the main site of APP transport from recycling endosomes. To explore the effect of SV2A on APP distribution in the cell membrane, the protein expression of the plasma membrane biomarker ATP1A1 and the localization of APP with ATP1A1 were explored in vitro and in vivo. The results showed that the expression of ATP1A1 was not altered in SH-SY5Y and N2a cells transfected with oeSV2A or shSV2A plasmids (Figure 6A to 6D) or in hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (Figure 6E). However, the colocalization of APP and ATP1A1 was increased in SV2A-overexpressed cells and mouse primary neurons (Figure 6F, 6G, and 6J), whereas it was reduced in cells transfected with shSV2A plasmids (Figure 6H and 6I).
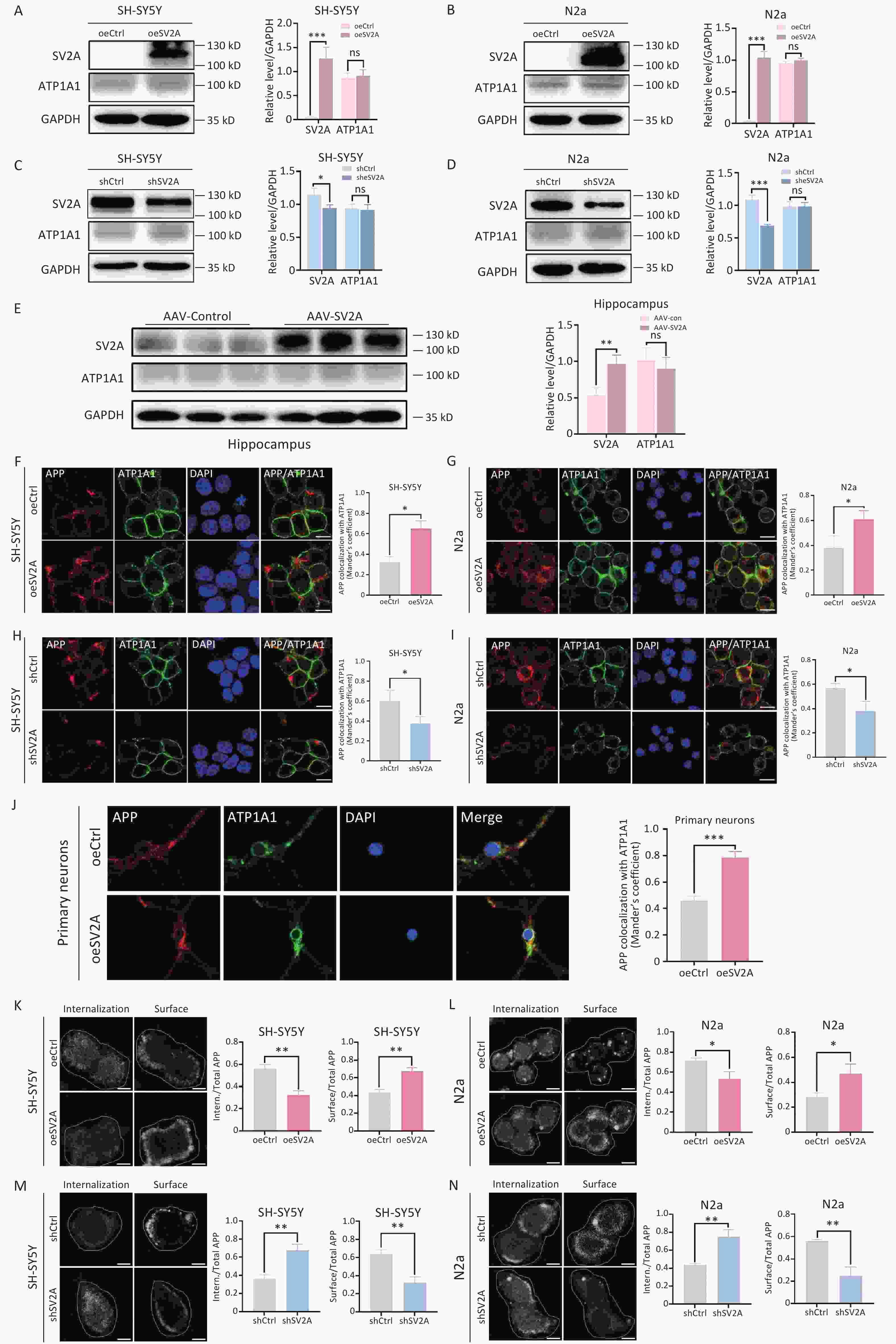
Figure 6. of SV2A on APP distribution in the cell surface of neurons. (A–B) Protein levels of SV2A and ATP1A1 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A and ATP1A1 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) Protein levels of SV2A and ATP1A1 in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). (F–G) Colocalization of APP (red) and ATP1A1 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (H–I) Colocalization of APP (red) and ATP1A1 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with ATP1A1 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. (K–L) Internalization of APP in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (M–N) Internalization of APP in SH-SY5Y and N2a cells transfected with shSV2A plasmids. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). Scale bar = 25 μm. *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; ATP1A1, ATPase Na (+)/K (+) transporting subunit α1; SV2A, synaptic vesicle glycoprotein 2A.
In the present study, antibody internalization assay was performed to investigate the effect of SV2A on APP endocytosis. The results showed that the internalization of APP decreased significantly in SH-SY5Y and N2a cells transfected with oeSV2A plasmids (Figure 6K and 6L), whereas it increased obviously in cells transfected with shSV2A plasmids (Figure 6M and 6N), indicating that SV2A reduced APP endocytosis.
These results demonstrate that SV2A promotes APP distribution in the cell surface and inhibits APP internalization. This indicates that the recycling endosomes-cell surface route is the main direction of diversion for APP in early endosomes.
-
To further investigate the effects of SV2A on APP amyloid degradation, the levels of APP, BACE1, and APP cleavage products were examined in vitro. The results showed that the protein levels of BACE1 were significantly reduced in SH-SY5Y and N2a cells overexpressing SV2A (Figure 7A and 7B), whereas the levels were elevated in SV2A-knockdown cells (Figure 7C and 7D). Protein levels of APP did not show differential expression. The mRNA levels of APP were downregulated obviously in SV2A-overexpressed cells (Figure 7E), but were significantly upregulated in SV2A-knockdown cells (Figure 7F). These results indicate that SV2A did not affect the protein levels of APP; however, it inhibited APP synthesis at the transcriptional level.

Figure 7. Effect of SV2A on APP amyloid degradation in vitro. (A–B) Protein levels of SV2A, APP, and BACE1 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A, APP, and BACE1 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) mRNA levels of SV2A and APP in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (F) mRNA levels of SV2A and APP in SH-SY5Y and N2a cells transfected with shSV2A plasmids. G and H. Levels of sAPPα and sAPPβ in the medium of SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (I–J) Levels of Aβ1-40 and Aβ1-42 in the medium of SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. K and L. Levels of sAPPα and sAPPβ in the medium of SH-SY5Y and N2a cells transfected with shSV2A plasmids. M and N. Levels of Aβ1-40 and Aβ1-42 in the medium of SH-SY5Y and N2a cells transfected with shSV2A plasmids. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significantly different. AAV, Adeno-associated virus; APP, amyloid precursor protein; BACE1, SV2A, synaptic vesicle glycoprotein 2A.
Subsequently, the levels of sAPPβ, Aβ1-40 and Aβ1-42 (products of APP amyloidogenic cleavage) significantly decreased in SV2A-overexpressed cells (Figure 7H-7J), whereas the levels were enhanced in SV2A-knockdown cells (Figure 7L-7N). The levels of sAPPα (APP non-amyloidogenic cleavage products) were not different in the SV2A-overexpressed or knockdown cells (Figure 7G and 7K). Thus, although SV2A slowed the amyloid degradation of APP, it did not affect the nonamyloid degradation of APP.
-
Senile plaques, formed by Aβ aggregation, are one of the major neuropathological hallmarks of AD[32]. Aβ binds to the cellular prion protein (PrPC) and metabotropic glutamate receptor 5 (mGluR5), and inositol triphosphate (IP3) is generated, increasing calcium release. Increased eukaryotic elongation factor 2 (eEF2) phosphorylation leads to the disruption of synaptic protein translation. Moreover, Aβ-PrPC binding activates Fyn to disrupt synapses[13]. Synaptic dysfunction and loss are other early pathological features of AD that correlate with cognitive decline[33]. SV2A, a vesicle protein specifically expressed in synapses, is a synaptic density biomarker[34]. SV2A also plays an important role in vesicle transport, exocytosis, and neurotransmitter release[35]. Previous studies have shown that SV2A gene and protein expression decreases in patients with epilepsy and that SV2A is the molecular target of the antiepileptic drug LEV[36]. [11C] UCB-J PET observed that the expression of SV2A significantly reduced in the nigrostriatal system of patients with PD[37]. Our previous studies showed that SV2A levels are significantly decreased in the serum and cerebrospinal fluid (CSF) of patients with AD and are recognized as ideal biomarkers for the early diagnosis of AD[38]. Currently, the mechanisms by which SV2A regulates APP amyloid and non-amyloid cleavage products remain unclear.
Impaired membrane transport can lead to aberrant Aβ generation and AD[39]. The TGN plays an important role in the processing and transport of newly synthesized proteins. As BACE1 and γ-secretase components are located in these subcellular compartments, the TGN is also a significant site of APP processing and Aβ generation[40,41]. BACE1 is the rate-limiting enzyme in the APP amyloid degradation pathway[42]. The acidic environment of the TGN contributes to BACE1 activity[43]. Previous studies have shown that BACE1 is transported from the TGN to the cell membrane via the activating protein-1 (AP-1)/ ADP-ribosylation factor 1 (Arf1)-/Arf4-pathway. APP is mainly transported from the TGN to early endosomes via the AP4-/Arl5b-dependent pathway[6,40]. Arl5b, a member of the small GTPase family, is responsible for APP transport from the TGN to early endosomes. Arl5b downregulation results in APP accumulation and increases Aβ production in the TGN[9]. In this study, the interaction between SV2A and Arl5b was observed in SH-SY5Y and N2a cells, and SV2A overexpression increased the protein levels of Arl5b in vitro and in vivo. Moreover, immunostaining showed that SV2A overexpression decreased the colocalization of APP and TGN46 (Figure 1). These results proposed that SV2A may decrease APP distribution in the TGN through APP transport from the TGN to early endosomes by Arl5b, which reduces APP cleavage by BACE1 and γ-secretase, resulting in the lower generation of Aβ in TGN.
The endolysosomal network consists of dynamic intracellular membranous organelles, including early endosomes, late endosomes, recycling endosomes, autophagosomes, and lysosomes[4,44]. The endolysosomal system regulates the recycling and degradation of proteins[45]. The amyloid degradation of APP occurs primarily in the endolysosomal system[6]. Early endosomes are critical sorting centers for endocytosis and provide an ideal acidic environment for BACE1 activity. BACE1 could cleave APP to sAPPβ and C99 in early endosomes. APP and its products are then transported to lysosomes via late endosomes. C99 was further cleaved to Aβ and AICD by γ-secretase in late endosomes or lysosomes[46,47]. Impaired endolysosomal system is a significant early pathological hallmark of AD and can be recognized as a potential therapeutic target[48]. The dysfunction of the endolysosomal system results in the aggregation of Aβ and misfolded proteins[6,49]. However, the mechanisms by which SV2A regulates cellular trafficking of APP in the endolysosomal system remain unclear.
In this study, the expression of endolysosomal biomarkers and the colocalization of APP with related biomarkers were explored to investigate the effect of SV2A on APP intracellular transport. SV2A downregulated the expression of EEA1, Rab7 and LAMP1, but upregulated the expression of Rab11 in vitro and in vivo, indicating that SV2A may influence the number of early endosomes, late endosomes, lysosomes, and recycling endosomes. Moreover, SV2A reduced the localization of APP in early endosomes, late endosomes, and lysosomes, but increased its localization in recycling endosomes (Figure 2-5). Therefore, SV2A could promote APP transport in the TGN-early endosome-recycling pathway, but reduce APP transport in the early endosome-late endosome-lysosome pathway. However, CTSD, the key lysosomal hydrolase, was significantly upregulated by SV2A.
The cell membrane is a major site of APP non-amyloid degradation[50]. APP, located in the early endosomes, is transported to the cell membrane via recycling endosomes[51]. Additionally, some APP in the cell membrane can be internalized into early endosomes for further processing[52]. In this study, SV2A increased APP distribution in the cell membrane (Figure 6). Furthermore, the antibody internalization assay showed that SV2A reduced APP endocytosis (Figure 6). These results indicate that SV2A promotes APP distribution in the cell membrane via the recycling endosome-cell membrane route and decreases APP internalization in the cell membrane.
APP could be processed by BACE1 and γ-secretase sequentially to generate sAPPβ and Aβ[41]. sAPPα played an important role in neuroprotection and was generated from the nonamyloid cleavage of APP by α- (ADAM10)and γ-secretase[53]. In this study, SV2A reduced the levels of sAPPβ, Aβ1-40 and Aβ1-42, but had no effect on sAPPα, indicating that SV2A slows down the amyloid degradation of APP, but does not affect the non-amyloid degradation of APP (Figure 7).
This study demonstrates that SV2A promotes APP transport from the TGN to early endosomes by upregulating Arl5b and APP transport from early endosomes to recycling endosomes-cell membranes, which slows APP amyloid degradation.
AD is a multifactorial neurodegenerative disorder[54]. Currently, AD drugs mainly focus on a single target and cause adverse effects[5,55]. AD drugs sanctioned by the US Food and Drug Administration (FDA) could improve symptoms but do not slow AD progression or cure AD[56]. Recent studies have suggested that the endolysosomal system is essential for neuronal homeostasis[57]. The present study indicates that the SV2A-mediated regulation of APP intracellular transport may be a potential target for AD therapeutic intervention.
-
Our study is the first to indicate that SV2A can slow down the amyloid degradation of APP by promoting APP transport from early endosomes to the recycling endosomes-cell membrane pathway rather than the late endosomes-lysosomes pathway. Consequently, the underlying mechanisms by which SV2A regulates the intracellular transport of APP in AD will provide novel targets for AD therapy and drug development.
doi: 10.3967/bes2025.049
Synaptic Vesicle Glycoprotein 2A Slows down Amyloidogenic Processing of Amyloid Precursor Protein via Regulating Its Intracellular Trafficking
-
Abstract:
Objective To reveal the effects and potential mechanisms by which synaptic vesicle glycoprotein 2A (SV2A) influences the distribution of amyloid precursor protein (APP) in the trans-Golgi network (TGN), endolysosomal system, and cell membranes and to reveal the effects of SV2A on APP amyloid degradation. Methods Colocalization analysis of APP with specific tagged proteins in the TGN, ensolysosomal system, and cell membrane was performed to explore the effects of SV2A on the intracellular transport of APP. APP, β-site amyloid precursor protein cleaving enzyme 1 (BACE1) expressions, and APP cleavage products levels were investigated to observe the effects of SV2A on APP amyloidogenic processing. Results APP localization was reduced in the TGN, early endosomes, late endosomes, and lysosomes, whereas it was increased in the recycling endosomes and cell membrane of SV2A-overexpressed neurons. Moreover, Arl5b (ADP-ribosylation factor 5b), a protein responsible for transporting APP from the TGN to early endosomes, was upregulated by SV2A. SV2A overexpression also decreased APP transport from the cell membrane to early endosomes by downregulating APP endocytosis. In addition, products of APP amyloid degradation, including sAPPβ, Aβ1-42, and Aβ1-40, were decreased in SV2A-overexpressed cells. Conclusion These results demonstrated that SV2A promotes APP transport from the TGN to early endosomes by upregulating Arl5b and promoting APP transport from early endosomes to recycling endosomes-cell membrane pathway, which slows APP amyloid degradation. -
Key words:
- Alzheimer’s disease /
- Amyloid precursor protein /
- Amyloid degradation /
- Synaptic vesicle glycoprotein 2A /
- Endolysosomal system
The authors declare no competing interests.
All animal experiments were performed according to the National Institutes of Health guidelines and conformed to the guidelines of the Ethics Committee of Xuanwu Hospital of Capital University ([2021]225).
注释:1) Authors’ contributions: 2) Competing interests: 3) Ethics: -
Figure 1. Effect of SV2A on APP distribution in the trans-Golgi network and Ar15b expression. (A–B) Colocalization of APP (red) with TGN46 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Colocalization of APP (red) with TGN46 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) Protein levels of SV2A in primary neurons transfected with SV2A-overexpressed lentivirus. (F) Colocalization of APP (red) with TGN46 (green) in mice primary neurons transfected with SV2A-overexpressed lentivirus. (G) Interaction of SV2A and Arl5b in SH-SY5Y cells by Co-IP assay. (H) Interaction between SV2A and Arl5b in N2a cells by Co-IP assay. I and J. Protein levels of SV2A, TGN46, and Arl5b in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (K–L) Protein levels of SV2A, TGN46, and Arl5b in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (M) Protein levels of SV2A, TGN46 and Arl5b in hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significantly different. Note: AAV, Adeno-associated virus; Arl5b, ADP-ribosylation factor 5b; APP, amyloid precursor protein; Co-IP, Co-immunoprecipitation; SV2A, synaptic vesicle glycoprotein 2A; TGN, the trans-Golgi network.
Figure 2. Effect of SV2A on APP distribution in early endosomes of neurons. (A–B) Protein levels of SV2A and EEA1 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A and EEA1 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) Protein levels of SV2A and EEA1 in hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). (F–G) Colocalization of APP (red) with EEA1 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (H–I) Colocalization of APP (red) and EEA1 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with EEA1 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; EEA1, early endosome antigen-1; SV2A, synaptic vesicle glycoprotein 2A.
Figure 3. Effect of SV2A on APP distribution in the late endosomes of neurons. (A–B) Protein levels of SV2A and Rab7 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A and Rab7 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. E. Protein levels of SV2A and Rab7 in hippocampal tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). (F–G) Colocalization of APP (red) and Rab7 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (H–I) Colocalization of APP (red) and Rab7 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with Rab7 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; Rab7, Ras-related protein Rab 7a; SV2A, synaptic vesicle glycoprotein 2A.
Figure 4. Effect of SV2A on APP distribution in lysosomes and cathepsin D expression of neurons. (A–B) Protein levels of SV2A, LAMP1, and cathepsin D in SH-SY5Y and N2a cells transfected with SV2A-overexpression plasmids. (C–D) Protein levels of SV2A, LAMP1, and cathepsin D in SH-SY5Y and N2a cells transfected with shSV2A plasmids. E. Protein levels of SV2A, LAMP1 and cathepsin D in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n =7/group). (F–G) Colocalization of APP (red) with LAMP1 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpression plasmids. (H–I) Colocalization of APP (red) with LAMP1 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with LAMP1 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. (K–L) Colocalization of cathepsin D (red) with LAMP1 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (M–N) Colocalization of cathepsin D (red) with LAMP1 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (O) Colocalization of cathepsin D (red) with LAMP1 (green) in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n = 4/group). (P–Q) LysoTracker staining in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (R–S) LysoTracker staining in SH-SY5Y and N2a cells transfected with shSV2A. (T) Colocalization of 6E10 (red) with LAMP1 (green) in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n = 4/group). Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; LAMP1, lysosomal associated membrane protein-1; SV2A, synaptic vesicle glycoprotein 2A.
Figure 5. Effect of SV2A on APP distribution in neuronal recycling endosomes. (A–B) Protein levels of SV2A and Rab11 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A and Rab11 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) Protein levels of SV2A and Rab11 in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). (F–G) Colocalization of APP (red) and Rab11 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (H–I) Colocalization of APP (red) and Rab11 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with Rab11 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. Scale bar = 25 μm. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; Rab11, Ras-related protein Rab 11; SV2A, synaptic vesicle glycoprotein 2A.
Figure 6. of SV2A on APP distribution in the cell surface of neurons. (A–B) Protein levels of SV2A and ATP1A1 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A and ATP1A1 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) Protein levels of SV2A and ATP1A1 in hippocampus tissues of APP/PS1 mice injected with AAV-SV2A (n = 7/group). (F–G) Colocalization of APP (red) and ATP1A1 (green) in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (H–I) Colocalization of APP (red) and ATP1A1 (green) in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (J) Colocalization of APP (red) with ATP1A1 (green) in mouse primary neurons transfected with SV2A-overexpressed lentivirus. (K–L) Internalization of APP in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (M–N) Internalization of APP in SH-SY5Y and N2a cells transfected with shSV2A plasmids. Data are presented as mean ± SEM (immunofluorescence: total 30–45 cells/group from 3 biologically independent experiments; other assays: n = 3 biologically independent replicates). Scale bar = 25 μm. *P < 0.05, **P < 0.01, ***P < 0.001. AAV, Adeno-associated virus; APP, amyloid precursor protein; ATP1A1, ATPase Na (+)/K (+) transporting subunit α1; SV2A, synaptic vesicle glycoprotein 2A.
Figure 7. Effect of SV2A on APP amyloid degradation in vitro. (A–B) Protein levels of SV2A, APP, and BACE1 in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (C–D) Protein levels of SV2A, APP, and BACE1 in SH-SY5Y and N2a cells transfected with shSV2A plasmids. (E) mRNA levels of SV2A and APP in SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (F) mRNA levels of SV2A and APP in SH-SY5Y and N2a cells transfected with shSV2A plasmids. G and H. Levels of sAPPα and sAPPβ in the medium of SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. (I–J) Levels of Aβ1-40 and Aβ1-42 in the medium of SH-SY5Y and N2a cells transfected with SV2A-overexpressed plasmids. K and L. Levels of sAPPα and sAPPβ in the medium of SH-SY5Y and N2a cells transfected with shSV2A plasmids. M and N. Levels of Aβ1-40 and Aβ1-42 in the medium of SH-SY5Y and N2a cells transfected with shSV2A plasmids. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significantly different. AAV, Adeno-associated virus; APP, amyloid precursor protein; BACE1, SV2A, synaptic vesicle glycoprotein 2A.
Table 1. Sequences used for RT-qPCR
Target gene Forward primer Reverse primer Human SV2A GCCCAACAGTATGAAGCCATCCTAC ACCTCCACACCGTCAGCCATC Mouse SV2A CTCACTCTCAGTCAACAGCGTCTTC CACTCCACCGATCATCCAGAACATAC Human APP TGATGATGAGGATGTGGAGGATGG TGTGGTGGTGGTGGCAGTG Mouse APP GACTGACCACTCGACCAGGTTCTG CTTGTAAGTTGGATTCTAATATCCG -
[1] 2024 Alzheimer's disease facts and figures. Alzheimers Dement, 2024; 20, 3708-821. [2] Chen ZY, Zhang Y. Animal models of Alzheimer's disease: applications, evaluation, and perspectives. Zool Res, 2022; 43, 1026−40. doi: 10.24272/j.issn.2095-8137.2022.289 [3] Orobets KS, Karamyshev AL. Amyloid precursor protein and Alzheimer's disease. Int J Mol Sci, 2023; 24, 14794. doi: 10.3390/ijms241914794 [4] Szabo MP, Mishra S, Knupp A, et al. The role of Alzheimer's disease risk genes in endolysosomal pathways. Neurobiol Dis, 2022; 162, 105576. doi: 10.1016/j.nbd.2021.105576 [5] Rajendran K, Krishnan UM. Mechanistic insights and emerging therapeutic stratagems for Alzheimer's disease. Ageing Res Rev, 2024; 97, 102309. doi: 10.1016/j.arr.2024.102309 [6] Van Acker ZP, Bretou M, Annaert W. Endo-lysosomal dysregulations and late-onset Alzheimer's disease: impact of genetic risk factors. Mol Neurodegener, 2019; 14, 20. doi: 10.1186/s13024-019-0323-7 [7] Takasugi N, Komai M, Kaneshiro N, et al. The pursuit of the "inside" of the amyloid hypothesis-is C99 a promising therapeutic target for Alzheimer's disease? Cells, 2023; 12, 454. [8] Zhuravleva V, Vaz-Silva J, Zhu M, et al. Rab35 and glucocorticoids regulate APP and BACE1 trafficking to modulate Aβ production. Cell Death Dis, 2021; 12, 1137. doi: 10.1038/s41419-021-04433-w [9] Toh WH, Tan JZA, Zulkefli KL, et al. Amyloid precursor protein traffics from the Golgi directly to early endosomes in an Arl5b- and AP4-dependent pathway. Traffic, 2017; 18, 159−75. doi: 10.1111/tra.12465 [10] Mishra S, Knupp A, Szabo MP, et al. The Alzheimer's gene SORL1 is a regulator of endosomal traffic and recycling in human neurons. Cell Mol Life Sci, 2022; 79, 162. doi: 10.1007/s00018-022-04182-9 [11] Eggert S, Thomas C, Kins S, et al. Trafficking in Alzheimer's disease: modulation of APP transport and processing by the transmembrane proteins LRP1, SorLA, SorCS1c, sortilin, and calsyntenin. Mol Neurobiol, 2018; 55, 5809−29. doi: 10.1007/s12035-017-0806-x [12] Perdigão C, Barata MA, Araújo MN, et al. Intracellular trafficking mechanisms of synaptic dysfunction in Alzheimer's disease. Front Cell Neurosci, 2020; 14, 72. [13] Tzioras M, McGeachan RI, Durrant CS, et al. Synaptic degeneration in Alzheimer disease. Nat Rev Neurol, 2023; 19, 19−38. doi: 10.1038/s41582-022-00749-z [14] Löscher W, Gillard M, Sands ZA, et al. Synaptic vesicle glycoprotein 2A ligands in the treatment of epilepsy and beyond. CNS Drugs, 2016; 30, 1055−77. doi: 10.1007/s40263-016-0384-x [15] Bradberry MM, Chapman ER. All-optical monitoring of excitation-secretion coupling demonstrates that SV2A functions downstream of evoked Ca2+ entry. J Physiol, 2022; 600, 645−54. doi: 10.1113/JP282601 [16] Mecca AP, O'Dell RS, Sharp ES, et al. Synaptic density and cognitive performance in Alzheimer's disease: a PET imaging study with [11C]UCB-J. Alzheimers Dement, 2022; 18, 2527−36. doi: 10.1002/alz.12582 [17] Wu PP, Cao BR, Tian FY, et al. Development of SV2A ligands for epilepsy treatment: a review of levetiracetam, brivaracetam, and padsevonil. Neurosci Bull, 2024; 40, 594−608. doi: 10.1007/s12264-023-01138-2 [18] Tokudome K, Okumura T, Shimizu S, et al. Synaptic vesicle glycoprotein 2A (SV2A) regulates kindling epileptogenesis via GABAergic neurotransmission. Sci Rep, 2016; 6, 27420. doi: 10.1038/srep27420 [19] Mecca AP, Chen MK, O'Dell RS, et al. In vivo measurement of widespread synaptic loss in Alzheimer's disease with SV2A PET. Alzheimers Dement, 2020; 16, 974−82. doi: 10.1002/alz.12097 [20] Harper CB, Small C, Davenport EC, et al. An epilepsy-associated SV2A mutation disrupts synaptotagmin-1 expression and activity-dependent trafficking. J Neurosci, 2020; 40, 4586−95. doi: 10.1523/JNEUROSCI.0210-20.2020 [21] Lin CY, Chang MC and Jhou HJ. Effect of levetiracetam on cognition: a systematic review and meta-analysis of double-blind randomized placebo-controlled trials. CNS Drugs, 2024; 38, 1−14. doi: 10.1007/s40263-023-01058-9 [22] Knupp A, Mishra S, Martinez R, et al. Depletion of the AD risk gene SORL1 selectively impairs neuronal endosomal traffic independent of Amyloidogenic APP processing. Cell Rep, 2020; 31, 107719. doi: 10.1016/j.celrep.2020.107719 [23] Musardo S, Therin S, Pelucchi S, et al. The development of ADAM10 endocytosis inhibitors for the treatment of Alzheimer's disease. Mol Ther, 2022; 30, 2474−90. doi: 10.1016/j.ymthe.2022.03.024 [24] Hossain MI, Marcus JM, Lee JH, et al. Restoration of CTSD (cathepsin D) and lysosomal function in stroke is neuroprotective. Autophagy, 2021; 17, 1330−48. doi: 10.1080/15548627.2020.1761219 [25] Tian YL, Kang QJ, Shi XM, et al. SNX-3 mediates retromer-independent tubular endosomal recycling by opposing EEA-1-facilitated trafficking. PLoS Genet, 2021; 17, e1009607. doi: 10.1371/journal.pgen.1009607 [26] Sannerud R, Esselens C, Ejsmont P, et al. Restricted location of PSEN2/γ-secretase determines substrate specificity and generates an intracellular Aβ pool. Cell, 2016; 166, 193−208. doi: 10.1016/j.cell.2016.05.020 [27] McKendell AK, Houser MCQ, Mitchell SPC, et al. In-depth characterization of endo-lysosomal Aβ in intact neurons. Biosensors (Basel), 2022; 12, 663. [28] Gallwitz L, Schmidt L, Marques ARA, et al. Cathepsin D: analysis of its potential role as an amyloid beta degrading protease. Neurobiol Dis, 2022; 175, 105919. doi: 10.1016/j.nbd.2022.105919 [29] Meyer H, Kravic B. The endo-lysosomal damage response. Annu Rev Biochem, 2024; 93, 367−87. doi: 10.1146/annurev-biochem-030222-102505 [30] Di YQ, Han XL, Kang XL, et al. Autophagy triggers CTSD (cathepsin D) maturation and localization inside cells to promote apoptosis. Autophagy, 2021; 17, 1170−92. doi: 10.1080/15548627.2020.1752497 [31] Prieto Huarcaya S, Drobny A, Marques ARA, et al. Recombinant pro-CTSD (cathepsin D) enhances SNCA/α-Synuclein degradation in α-Synucleinopathy models. Autophagy, 2022; 18, 1127−51. doi: 10.1080/15548627.2022.2045534 [32] Al-Kuraishy HM, Jabir MS, Al-Gareeb AI, et al. Evaluation and targeting of amyloid precursor protein (APP)/amyloid beta (Aβ) axis in amyloidogenic and non-amyloidogenic pathways: a time outside the tunnel. Ageing Res Rev, 2023; 92, 102119. doi: 10.1016/j.arr.2023.102119 [33] Eckman EA, Clausen DM, Solé-Domėnech S, et al. Nascent Aβ42 fibrillization in synaptic endosomes precedes plaque formation in a mouse model of Alzheimer's-like β-amyloidosis. J Neurosci, 2023; 43, 8812−24. doi: 10.1523/JNEUROSCI.1318-23.2023 [34] Mecca AP, Chen MK, O'Dell RS, et al. Association of entorhinal cortical tau deposition and hippocampal synaptic density in older individuals with normal cognition and early Alzheimer's disease. Neurobiol Aging, 2022; 111, 44−53. doi: 10.1016/j.neurobiolaging.2021.11.004 [35] Rossi R, Arjmand S, Bærentzen SL, et al. Synaptic Vesicle Glycoprotein 2A: features and functions. Front Neurosci, 2022; 16, 864514. doi: 10.3389/fnins.2022.864514 [36] Stout KA, Dunn AR, Hoffman C, et al. The synaptic vesicle glycoprotein 2: structure, function, and disease relevance. ACS Chem Neurosci, 2019; 10, 3927−38. doi: 10.1021/acschemneuro.9b00351 [37] Wang WY, Wang YR, Xu LM, et al. Presynaptic terminal integrity is associated with glucose metabolism in Parkinson's disease. Eur J Nucl Med Mol Imaging, 2025; 52, 1510−19. doi: 10.1007/s00259-024-06993-3 [38] Wang XL, Zhang XM, Liu J, et al. Synaptic vesicle glycoprotein 2 A in serum is an ideal biomarker for early diagnosis of Alzheimer's disease. Alzheimers Res Ther, 2024; 16, 82. doi: 10.1186/s13195-024-01440-9 [39] Fourriere L, Gleeson PA. Amyloid β production along the neuronal secretory pathway: dangerous liaisons in the Golgi? Traffic, 2021; 22, 319-27. [40] Tan JZA, Fourriere L, Wang JQ, et al. Distinct anterograde trafficking pathways of BACE1 and amyloid precursor protein from the TGN and the regulation of amyloid-β production. Mol Biol Cell, 2020; 31, 27−44. doi: 10.1091/mbc.E19-09-0487 [41] Hur JY. γ-secretase in Alzheimer's disease. Exp Mol Med, 2022; 54, 433−46. doi: 10.1038/s12276-022-00754-8 [42] Cui YT, Zhang XM, Liu J, et al. Myeloid ectopic viral integration site 2 accelerates the progression of Alzheimer's disease. Aging Cell, 2024; 23, e14260. doi: 10.1111/acel.14260 [43] Fourriere L, Cho EHJ, Gleeson PA. Segregation of the membrane cargoes, BACE1 and amyloid precursor protein (APP) throughout the Golgi apparatus. Traffic, 2022; 23, 158−73. doi: 10.1111/tra.12831 [44] Herman M, Randall GW, Spiegel JL, et al. Endo-lysosomal dysfunction in neurodegenerative diseases: opinion on current progress and future direction in the use of exosomes as biomarkers. Philos Trans R Soc Lond B Biol Sci, 2024; 379, 20220387. doi: 10.1098/rstb.2022.0387 [45] Lai SSM, Ng KY, Koh RY, et al. Endosomal-lysosomal dysfunctions in Alzheimer's disease: pathogenesis and therapeutic interventions. Metab Brain Dis, 2021; 36, 1087−100. doi: 10.1007/s11011-021-00737-0 [46] Behl T, Kaur D, Sehgal A, et al. Exploring the potential role of rab5 protein in endo-lysosomal impairment in Alzheimer's disease. Biomed Pharmacother, 2022; 148, 112773. doi: 10.1016/j.biopha.2022.112773 [47] Houser MCQ, Mitchell SPC, Sinha P, et al. Endosome and lysosome membrane properties functionally link to γ-secretase in live/intact cells. Sensors (Basel), 2023; 23, 2651. doi: 10.3390/s23052651 [48] Mishra S, Jayadev S, Young JE. Differential effects of SORL1 deficiency on the endo-lysosomal network in human neurons and microglia. Philos Trans R Soc Lond B Biol Sci, 2024; 379, 20220389. doi: 10.1098/rstb.2022.0389 [49] Afghah Z, Khan N, Datta G, et al. Involvement of endolysosomes and aurora kinase A in the regulation of amyloid β protein levels in neurons. Int J Mol Sci, 2024; 25, 6200. doi: 10.3390/ijms25116200 [50] Shen Q, Wu XL, Zhang Z, et al. Gamma frequency light flicker regulates amyloid precursor protein trafficking for reducing β-amyloid load in Alzheimer's disease model. Aging Cell, 2022; 21, e13573. doi: 10.1111/acel.13573 [51] Filippone A, Praticò D. Endosome dysregulation in down syndrome: a potential contributor to Alzheimer disease pathology. Ann Neurol, 2021; 90, 4−14. doi: 10.1002/ana.26042 [52] Burrinha T, Martinsson I, Gomes R, et al. Upregulation of APP endocytosis by neuronal aging drives amyloid-dependent synapse loss. J Cell Sci, 2021; 134, jcs255752. doi: 10.1242/jcs.255752 [53] Song C, Li SF, Mai Y, et al. Dysregulated expression of miR-140 and miR-122 compromised microglial chemotaxis and led to reduced restriction of AD pathology. J Neuroinflammation, 2024; 21, 167. doi: 10.1186/s12974-024-03162-z [54] Rao RV, Subramaniam KG, Gregory J, et al. Rationale for a multi-factorial approach for the reversal of cognitive decline in Alzheimer's disease and MCI: a review. Int J Mol Sci, 2023; 24, 1659. doi: 10.3390/ijms24021659 [55] Adepoju VA, Onyezue OI, Jamil S, et al. Lecanemab unveiled: exploring Alzheimer's treatment advancements, assessing strengths, limitations, and its therapeutic landscape position. Biomed Environ Sci, 2024; 37, 428−31. [56] Shastri D, Raj V, Lee S. Revolutionizing Alzheimer's treatment: harnessing human serum albumin for targeted drug delivery and therapy advancements. Ageing Res Rev, 2024; 99, 102379. doi: 10.1016/j.arr.2024.102379 [57] Asiamah EA, Feng BF, Guo RY, et al. The contributions of the endolysosomal compartment and autophagy to APOEɛ4 allele-mediated increase in Alzheimer's disease risk. J Alzheimers Dis, 2024; 97, 1007−31. doi: 10.3233/JAD-230658 -




 下载:
下载:



 Quick Links
Quick Links