-
Brucellosis is a global public health issue that severely affects human health, Brucella melitensis is currently the predominant species in China. Brucella spondylitis is the primary cause of the debilitating and disabling complications[1]. The lumbar vertebra was the most commonly affected site, followed by the thoracic, cervical, thoracolumbar, and lumbosacral segments, and back pain, fever, sweating, and fatigue were the most common symptoms[2]. However, the diagnosis of Brucella spondylitis is challenging owing to its wide spectrum of clinical presentations, cross-reactions with other bacteria, and low strain isolation rate. Therefore, a timely and accurate diagnosis of spinal brucellosis is crucial for implementing an effective therapeutic plan and improving treatment outcomes. Droplet digital polymerase chain reaction (ddPCR) is widely used for low-abundance nucleic acid detection and is useful for diagnosing infectious diseases[3]. Therefore, this study aimed to evaluate the ddPCR approach for the diagnosis of brucellosis with spondylitis to improve its clinical diagnostic capacity.
The diagnosis of brucellosis was based on the Diagnostic Criteria for Human Brucellosis (WS269-2019) in China[4]. Serological tests/isolate strains, clinical manifestations, and contact history were used to identify the cases. This study included 96 surgical patients with Brucella spondylitis, all of whom had osteoarticular complications and a history of contact with livestock. They were admitted to Ningxia Fourth People’s Hospital between December 2021 and September 2022. Fasting peripheral venous blood (5–10 mL) was collected for brucellosis serological testing, and 96 connective tissue and blood samples from around the bone tissue were collected to assess the diagnostic value of ddPCR. Thirty-two serum samples were collected from healthy individuals and used as controls. DNA was extracted from all samples using a QIAamp DNA Mini Kit (Qiagen 51304, Hilden, Germany).
Primers and probes were designed using Primer 5.0 software based on two highly conserved genes, bcsp31 gene of Brucella spp. (GenBank accession number M20404) and IS711 gene (GenBank accession number NC_003317), of Brucella melitensis 16M (Supplementary Table S1). The ddPCR amplification system and parameters were based on a previous study[5], and the ddPCR amplification was performed with a volume of 20 μL.
Brucella melitensis vaccine M5 (Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences) DNA was serially diluted ten-fold (initial concentration of 2,875,000 copies/μL) to assess the sensitivity (limit of detection) and detection efficiency of the ddPCR. The initial concentration of M5 DNA was determined using the developed ddPCR. Whole blood, and connective tissue, and blood from the area surrounding bone tissue samples were collected from 96 spondylitis brucellosis cases to assess the diagnostic efficiency of the ddPCR assay.
The course for 21/96 patients lasted over 20 months (chronic), and their serology results were negative in the Rose Bengal plate test (RBPT) and Serum Agglutination Test (SAT). The course for the remaining 75/96 patients (positive in RBPT and SAT), was less than six months, and they were classified into the acute group. Additionally, whole blood samples were categorized into two subgroups: ddPCR-I (direct ddPCR analysis of whole blood samples) and ddPCR-II (ddPCR analysis conducted five days after the whole blood was cultured in a dual-phase culture flask (bioMérieux, France)). The connective tissue and blood around the bone tissue were collected, ground,and used for DNA extraction, which was then directly detected by ddPCR [ddPCR(bt)]. All whole-blood samples were subjected to Brucella culture following standard bacteriological procedures. Moreover, 32 healthy participants (SAT negative) were included in a negative control group. Data were analyzed using Microsoft Excel (Microsoft Corp., Redmond, WA, USA). Receiver operating characteristic (ROC) curve analysis was performed using SPSS statistics software (version 21.0; IBM Corp., Armonk, NY, USA) to evaluate and compare the diagnostic efficiencies of the different methods.
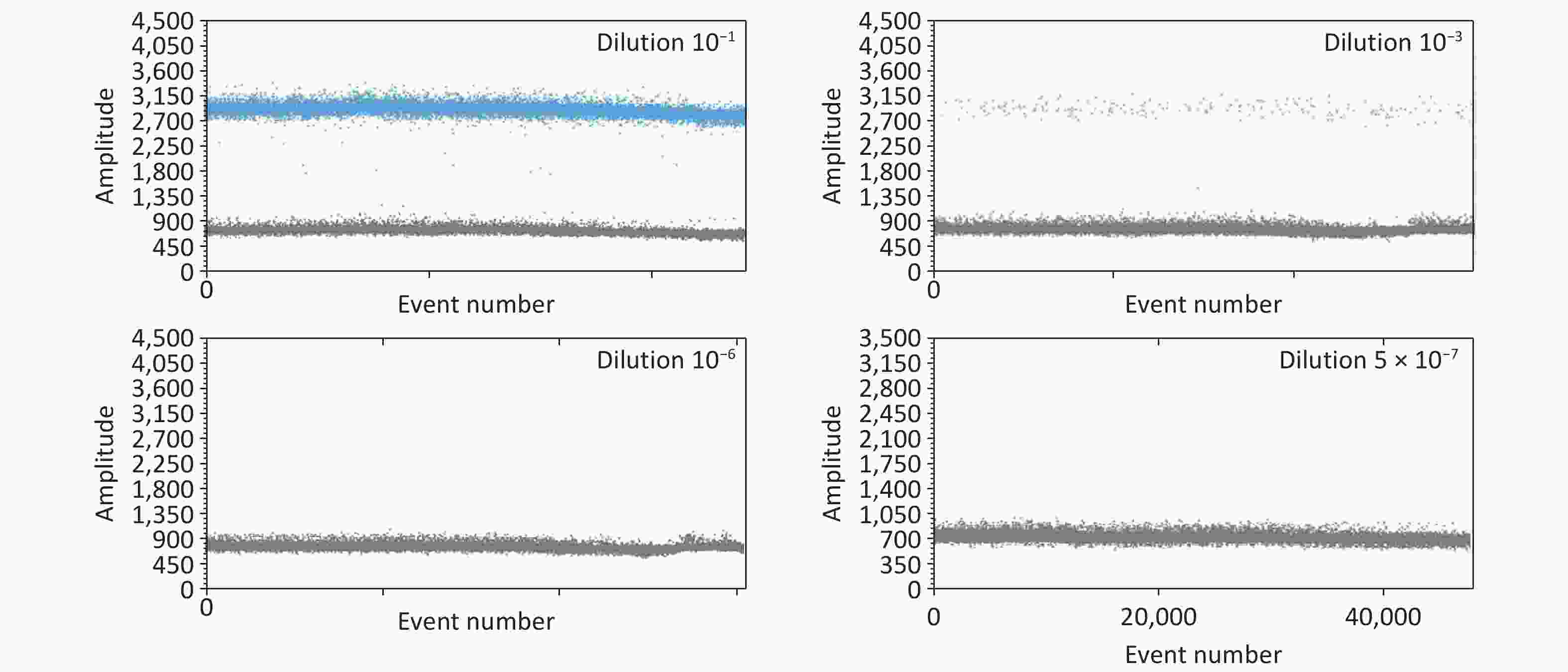
Blood culture results showed that 18 patients were positive and 67 were negative in the acute stage group; in the chronic stage group, only three were positive and 18 were negative. Although Brucella isolation is the gold standard for the diagnosis of human brucellosis, the Brucella isolation rate is low, the assays are time-consuming, and laboratory-acquired infection when isolating Brucella poses a risk that requires highly skilled personnel. The ddPCR method is more sensitive than the conventional PCR method, because it is based on Poisson statistics and is highly precise for nucleic acid quantitation in clinical samples[6]. Furthermore, the analytical stability of the ddPCR assay showed that all four primer sets had high amplification efficiencies at high concentrate titers (Supplementary Table S2). When the DNA copy number was low (i.e., four copies), primer P3 had the highest amplification efficiency, with the closest copies, and P3 was the most suitable primer for constructing the ddPCR approach (Supplementary Table S2). The limit of detection (LOD) for the P3 primer was 1.4 copies per reaction; concentrations below this limit were not detectable (Supplementary Table S3 and Figure 1). A previous study reported that the LOD for ddPCR was 1.87 copies per reaction with high repeatability[5], while another reported 5.9 copies per reaction, both of which are much lower than that of the quantitative PCR (qPCR; 54.9 copies per reaction)[7]. Another study reported positivity rates of 88.5% and 75.4% in whole blood samples from 61 SAT-positive patients detected by ddPCR and qPCR, respectively[5], confirming that ddPCR is more sensitive than qPCR. These data suggest that ddPCR is more sensitive than conventional or real-time (RT)-PCR methods and could be a useful diagnostic supplement to reduce misdiagnosis.
In the acute group, 75 confirmed seropositive SAT samples were analyzed using ddPCR; 35 (46.67%), 59 (78.67%), and 65 (86.67%) samples were positive in the ddPCR-I, ddPCR-II, and ddPCR(bt) groups, respectively (Supplementary Table S4). In the chronic group, 3 (14.29%), 3 (14.29%), and 13 (61.90%) samples were positive in the ddPCR-I, ddPCR-II, and ddPCR(bt) groups, respectively. Our study showed that the positive detection rates of connective tissue and blood around the bone tissue samples were higher than those of the other samples in the acute and chronic groups. Moreover, the diagnostic performance of ddPCR for chronic spinal brucellosis was excellent; 61.90% (13/21) of patients tested positive using ddPCR. Since the disease course of these patients was over 20 months, the antibody titer disappeared or was below the detection limit; therefore, these patients were RBPT- and SAT-negative. In the acute group, the detection rate in the connective tissue and blood around the bone tissue samples was also higher than that in Brucella culture samples and whole blood samples, which was almost equal to that of SAT. These results demonstrate that ddPCR is a reliable diagnostic tool for detecting spinal brucellosis and can provide important evidence to ensure proper treatment and minimize health damage. Li and colleagues demonstrated that ddPCR used for the detection of Brucella suis, is able to differentiate wild strains of Brucella from the S2 vaccine strain, with a LOD of 10 copies/µL[8]. Furthermore, receiver operating characteristic (ROC) curve analysis demonstrated that ddPCR(bt) had superior detection performance compared with ddPCR-I and ddPCR-II (Figure 2), with an area under the curve of 0.906 (Table 1). Brucella spp. are facultative intracellular pathogenic agents, and the bacterial concentration in the blood of patients with brucellosis is usually low and difficult to detect using conventional PCR and other methods[9]. Since ddPCR can detect low quantities of circulating DNA, and Brucella strains utilize the endoplasmic reticulum for intracellular replication during osteoarticular localization of the disease[10], connective tissue and blood around bone tissue samples should be a priority sample source for diagnosing spinal brucellosis. Despite these promising results, caution should be exercised when interpreting them because the results are prone to being affected by multiple factors such as DNA quality, reagents, experimental procedures, and experimental environment. Further studies with larger sample sizes and additional analyses are required to confirm the value of this technology in clinical settings.
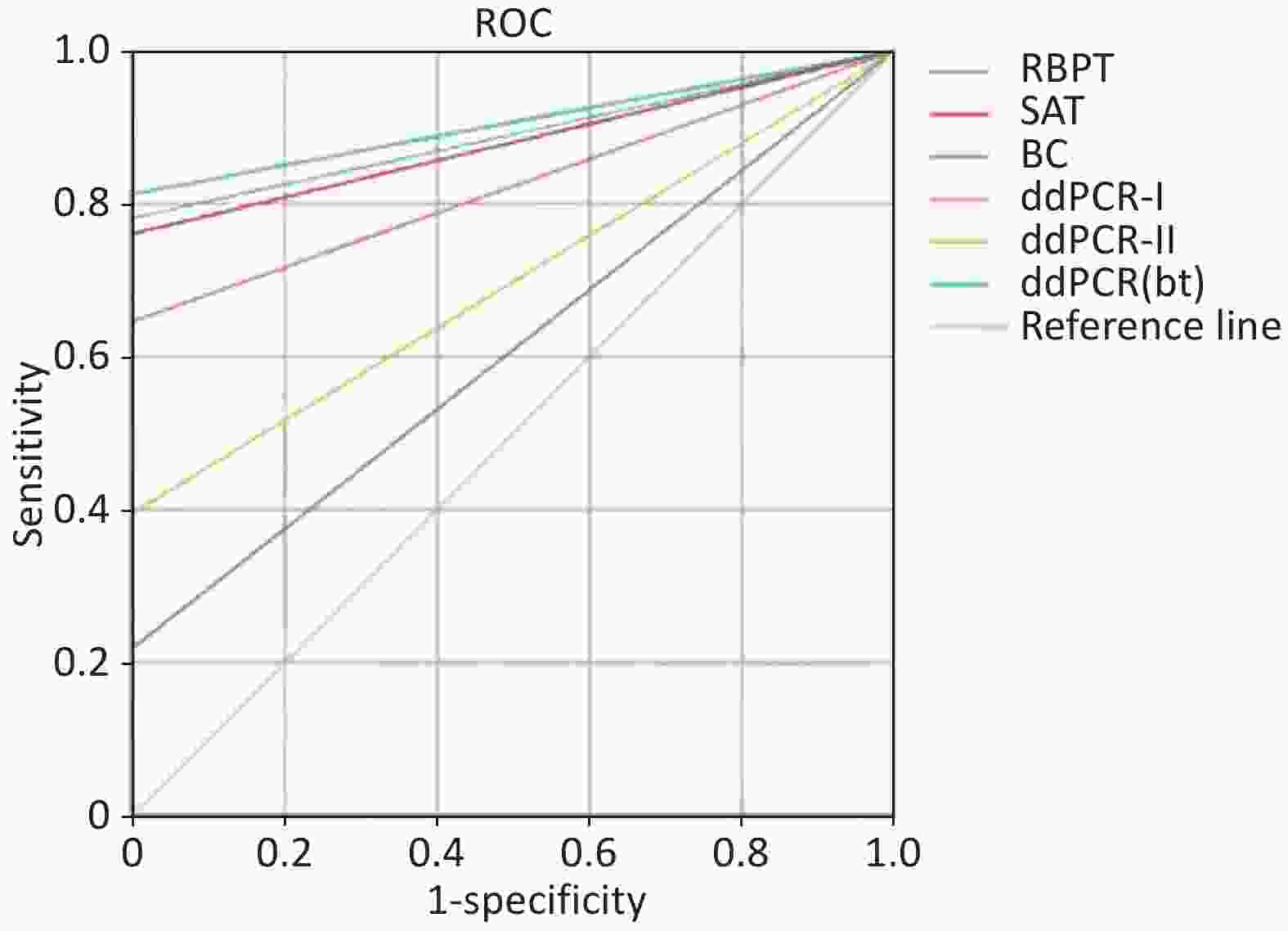
Figure 2. Receiver operating characteristic (ROC) curves and area under the curve (AUC) values of six detection methods used in this study.
Table 1. Receiver operating characteristic curve AUCs of ddPCR
Methods AUC Std. Error P-value 95% confidence interval Sensitivity Specificity Youden index RBPT 0.891 0.028 0.000 0.836 0.945 0.781 1.000 0.781 SAT 0.880 0.029 0.000 0.823 0.937 0.760 1.000 0.760 BC 0.609 0.053 0.071 0.505 0.714 0.219 1.000 0.219 ddPCR-Ⅰ 0.823 0.036 0.000 0.753 0.893 0.646 1.000 0.646 ddPCR-Ⅱ 0.698 0.047 0.001 0.606 0.790 0.396 1.000 0.396 ddPCR(bt) 0.906 0.026 0.000 0.856 0.957 0.813 1.000 0.813 Note. AUC, area under the curve; BC, bacterial culture; ddPCR, droplet digital polymerase chain reaction; ddPCR-I, direct ddPCR analysis of whole blood samples; ddPCR-II, ddPCR analysis after five days of whole blood enrichment; ddPCR(bt), ddPCR analysis of bone tissue; RBPT, Rose Bengal plate test; SAT, standard agglutination test. In this study, we developed a ddPCR method for detecting Brucella with excellent sensitivity and specificity. The limit of detection was 1.4 copies. These findings suggest that this assay could be a useful tool for diagnosing and managing spinal brucellosis, ensuring proper treatment, and minimizing damage to patient health.
doi: 10.3967/bes2025.085
Droplet Digital PCR for Diagnosing Brucellosis Spondylitis: Method Development and Evaluation
-
CWH and LGT performed targeted gene selection, designed primers and probes, and developed and evaluated ddPCR; LB and HM collected samples and epidemiological data of patients; LZG and CWH performed the data analysis, created the figures, and drafted the manuscript; JXF and LZG participated in the study design and critically reviewed the manuscript. All the authors approved the final version of the manuscript.
The authors declare no conflict of interest.
This study was conducted in accordance with the principles of the Declaration of Helsinki. The Ethics Committee of Ningxia Fourth People’s Hospital reviewed and approved this study (Grant No. 202201). Blood and tissue samples were collected for diagnostic purposes. All patients and their guardians provided informed consent, if applicable. All patient data were anonymized, and identifiable information, including names, identification numbers, home addresses, and telephone numbers, was discarded.
&These authors contributed equally to this work.
注释:1) Authors’ Contributions: 2) Competing Interests: 3) Ethics: -
Table 1. Receiver operating characteristic curve AUCs of ddPCR
Methods AUC Std. Error P-value 95% confidence interval Sensitivity Specificity Youden index RBPT 0.891 0.028 0.000 0.836 0.945 0.781 1.000 0.781 SAT 0.880 0.029 0.000 0.823 0.937 0.760 1.000 0.760 BC 0.609 0.053 0.071 0.505 0.714 0.219 1.000 0.219 ddPCR-Ⅰ 0.823 0.036 0.000 0.753 0.893 0.646 1.000 0.646 ddPCR-Ⅱ 0.698 0.047 0.001 0.606 0.790 0.396 1.000 0.396 ddPCR(bt) 0.906 0.026 0.000 0.856 0.957 0.813 1.000 0.813 Note. AUC, area under the curve; BC, bacterial culture; ddPCR, droplet digital polymerase chain reaction; ddPCR-I, direct ddPCR analysis of whole blood samples; ddPCR-II, ddPCR analysis after five days of whole blood enrichment; ddPCR(bt), ddPCR analysis of bone tissue; RBPT, Rose Bengal plate test; SAT, standard agglutination test. -
[1] Yang Z, Wu WG, Ou PC, et al. Discussion on treatment courses of brucellosis with spondylitis - a report of two cases. IDCases, 2023; 31, e01650. doi: 10.1016/j.idcr.2022.e01650 [2] Liang C, Wei W, Liang XW, et al. Spinal brucellosis in Hulunbuir, China, 2011-2016. Infect Drug Resist, 2019; 12, 1565−71. doi: 10.2147/IDR.S202440 [3] Li HY, Bai RL, Zhao ZY, et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci Rep, 2018; 38, BSR20181170. doi: 10.1042/BSR20181170 [4] Jiang H, Feng L, Lu JX. Updated guidelines for the diagnosis of human brucellosis - China, 2019. China CDC Wkly, 2020; 2, 487−9. doi: 10.46234/ccdcw2020.129 [5] Liu JY, Song ZC, Ta N, et al. Development and evaluation of a droplet digital PCR assay to detect Brucella in human whole blood. PLoS Negl Trop Dis, 2023; 17, e0011367. doi: 10.1371/journal.pntd.0011367 [6] Wang J, Zhao YY, Li JF, et al. IDH1 mutation detection by droplet digital PCR in glioma. Oncotarget, 2015; 6, 39651−60. doi: 10.18632/oncotarget.5630 [7] Wang Z, Chen YM, Deng HJ, et al. Quantification of intrahepatic cccDNA in HBV associated hepatocellular carcinoma by improved ddPCR method. J Virol Methods, 2022; 299, 114334. doi: 10.1016/j.jviromet.2021.114334 [8] Li WY, Zhang S, Dang S, et al. Establishment of an A/T-rich specifically MGB probe digital droplet PCR assays based on SNP for Brucella wild strains and vaccine strains. Diagn Microbiol Infect Dis, 2024; 110, 116432. doi: 10.1016/j.diagmicrobio.2024.116432 [9] Yagupsky P, Morata P, Colmenero JD. Laboratory diagnosis of human brucellosis. Clin Microbiol Rev, 2019; 33, e00073−19. [10] Giambartolomei GH, Arriola Benitez PC, Delpino MV. Brucella and osteoarticular cell activation: partners in crime. Front Microbiol, 2017; 8, 256. -




 下载:
下载:



 Quick Links
Quick Links