-
In the present study, we used a proteomics approach based on a two-dimensional electrophoresis (2-DE) reference map to investigate protein expression in the ovarian tissues of pubertal Swiss-Webster mice subjected to carbon ion radiation (CIR). Among the identified proteins, ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) is associated with the cell cycle[1] and that it influences proliferation in ovarian tissues. We analyzed the expression of UCH-L1 and the proliferation marker proliferation cell nuclear antigen (PCNA) following CIR using immunoblotting and immunofluorescence. The proteomics and biochemical results provide insight into the underlying mechanisms of CIR toxicity in ovarian tissues.
High dose radiotherapy has radically increased long-term survival in young cancer patients but it can cause ovarian failure and infertility in women of child-bearing age. Novel radiotherapies may overcome these problems in the future, but for the moment it is important to understand the risks involved with high linear energy transfer (LET) radiation exposure and to characterize the types of genetic alterations that can occur in ovarian tissues following pelvic radiotherapy.
A total of 12 female Swiss webster mice (Lanzhou University School of Medicine, China) aged 4 weeks and weighing 14-20 g were used. All animals were kept in 22 ± 2 ℃, 60% ± 10% humidity and light : dark cycle 12 h : 12 h. All animals had free access to deionized water in glass bottles with a rubber stopper and rodent feed (Keaoxieli Laboratory Animal Feed, Beijing, China). All fed procedures were approved Lanzhou University School of Medicine. Mice were randomly divided into two groups: the control (CK) and 4 Gy carbon ion radiation group (CIR). Mice were placed in a chamber and received whole-body carbon-ion irradiation by irradiation dose during a single exposure. Irradiations were conducted using a 6 cm-spread-out Bragg peak beam (270 MeV/μ) of accelerated 12C6+ ions at a nominal dose rate of 0.5 Gy min−1 at the Heavy Ion Research Facility (Institute of Modern Physics, Chinese Academy of Sciences, Lanzhou, China). The whole body of each mouse was traversed by carbon ions with a dose-averaged LET estimated at 10 Kev/μm[2]. The carbon ion is equipped with a passive beam delivery system. Dosimetric data were controlled automatically by a microcomputer during irradiation. Particle fluence was determined from an air-ionization chamber signal according to the calibration of the detector (PTW-UNIDOS, PTW-Freiburg Co., Wiesbaden, Germany)[3].
Mice were killed by cervical dislocation at 2 days after irradiation. The fat and connective tissues adhering to ovaries were removed and then were prepared for extraction proteins, paraffin sections and extraction total RNA. Protein samples (400 μg in 350 μL rehydration buffer) were loaded respectively onto 17 cm immobilized pH gradient (IPG) strips (pH 3 to 10, Bio-Rad Laboratories, Hercules, USA) on an IPG phoresis (IPGphor) isoelectric focusing system. Differential protein spots were manually cut from 2-DE gels, mass spectrometry (MS) and tandem MS data for protein identification were obtained by using a matrix-assisted laser desorption/ionization tandem mass spectrometry (MALDI-TOF-TOF) instrument (4800 proteomics analyzer; Applied Biosystems).
The polyvinylidene difluoride membranes (PVDF) were blocked with 5% skimmed milk in Tris buffer saline, then immunoblotted with the rabbit polyclonal immunoglobulin G (IgG) anti-UCH-L1 (cat. #SC-25800), anti-PCNA (Cat. no. SC-7907) (Santa Cruz Biotechnology, California, USA) and anti-β-actin (Cat. no. AP0060) (Bioworld technology, Inc, Ltd, Nanjing, China), and a horseradish peroxidase-labelled secondary antibody (cat. #ZB2301) (Beijing Zhongshanjinqiao Biotechnology Co., Beijing, China). Ovary sections (5 μm) were immersed in 3% H2O2 (30% H2O2diluted in methanol) for 30 min and incubated in 1% Triton X-100 (1 mLTriton X-100 diluted with 99 mLPBS) for 30 min. Sections were then blocked for 25 min at room temperature in 5% bovine serum albumin (BSA) (5 g BSA in 100 mLPBS) and incubated with the primary antibody (1:500) at 4 ℃ overnight. The donkey anti-rabbit IgG (1:100 dilution in Tris-buffered saline) secondary antibody Alexa Fluor 555 (cat. #A-21432; Invitrogen, Carlsbad, CA, USA) and Alexa Fluor 647 (cat. #A-31573; Invitrogen) were added dropwise and incubated at 37 ℃ for 1 h. Samples were washed with 0.01 mol/L (pH 7.2-7.4) phosphate-buffered saline (PBS) twice for 5 min each and then counterstained with 4, 6-diamidino-2-phenylindole (DAPI) (5 μg/mL; Sigma-Aldrich, St. Louis, MO, USA) for 10 min. Samples were then washed with PBS twice for 5 min each, followed by mounting with a glycerin-sodium bicarbonate wet sealant. Confocal laser microscope (LSM, Zeiss, Germany) and camera section on a Now red (PCNA) and green (UCH-L1) fluorescence was positive.
Ovary total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Primers of two genes were designed with Primer 3.0 plus, which were synthesized by Takara (Takara, Dalian, China). The actin used as a reference gene to quantify the amount of UCH-L1 mRNA. The length of the β-actin product was 171 bp. The sequences of the forward and reverse primers were 5'-CATCCGTAAAGACCTCTATGCCAAC-3' and 5'-ATGGAGCCACCGATCCACA-3', respectively. The length of the UCH-L1 product was 131 bp. The sequences of the forward and reverse primers were 5'-CGAAGATAGAGCCAAGTGTTTCGAG-3' and 5'-CGTCCACGTTGTTGAACAGAATA-3', respectively. Polymerase chain reaction (PCR) was proceeded on the Funglyn Biotech 3000 (FTC) quantitative real time PCR (qPCR) system (Funglyn Biotech Inc., Scarborough, ON, Canada) using two-step SYBR®quantitative reverse transcription polymerase chain reaction (QRT-PCR) kit (Takara, Dalian, China). β-actin was applied as an internal control and the expression levels of the target gene in different groups were carried out with the Delta-Delta-Ct (DDCt) method[4].
The data were analyzed with Statistical Product and Service Solutions (SPSS) statistical software (SPSS 19.0, Inc., Chicago, IL).
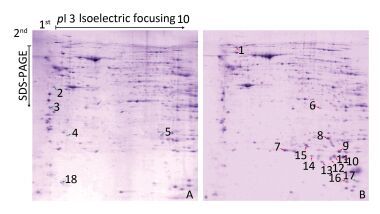
Radiation-induced apoptotic and inflammatory degeneration of mouse ovarian follicles had therefore reached serious levels at 2 days after irradiation, as previously reported[5]. Therefore, 2 days after radiation is the critical time to explore the radiation-induced infertility factors. Figure 1 illustrates representative protein profiles of CK and CIR. The 2-DE gels showed that a total of 18 differentially expressed proteins were found between the CK and CIR groups, the name and function of these proteins are described in Table 1.
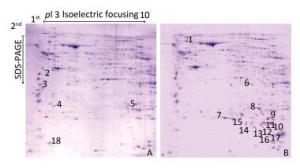
Figure 1. The protein profiles and differentially expressed spots of ovaries in mice after CIR. 400µg proteins were separated using isoelectric focusing (IEF) of pH 3-10, followed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie Brilliant Blue (CCB) R250 staining. (A) control group; (B) 4 Gy group. Green and red arrows represent decreased and increased expression, respectively. Spot numbers of representative proteins are in accordance with Table 1.
Table 1. Differential Expression of Proteins in Mice Ovaries
Protein Spot Protein Name Abbreviation Accession (NCBI) pI/Mw PeptidesMatcheda ProteinScore Average FoldChangeb Spot 1 Heat shock protein 90, beta (Grp94), member 1[Mus musculus] Hsp90b1 gi|14714615 4.74/92717.4 16 184 12.300 ± 2.060 Spot 2 Laminin receptor[Mus musculus] Rpsa gi|171948782 4.8/32943.5 13 666 0.293 ± 0.028 Spot 3 Keratin, type Ⅰ cytoskeletal 13[Mus musculus] Krt13 gi|37994713 4.88/33925.4 5 67 0.340 ± 0.017 Spot 4 Ubiquitin carboxyl-terminal hydrolase isozyme L1, partial[Mus musculus] Uchl1 gi|399106762 5.02/16462.3 1 107 0.016 ± 0.005 Spot 5 Annexin[Mus musculus] Anxa1 gi|70912321 7.59/38995.1 9 46 0 Spot 6 Alpha-fetoprotein, partial[Mus musculus] Afp gi|191765 5.47/48791.6 7 99 22.370 ± 1.822 Spot 7 Aldo-keto reductase family 1 member C18 [Mus Musculus] Akr1c18 gi|19527284 6.02/37666 16 640 5.280 ± 0.490 Spot 8 Beta-crystallin B2[Mus musculus] Crybb2 gi|6681035 6.5/23480.4 2 87 29.440 ± 3.815 Spot 9 Mitogen-activated protein kinase p38 beta[Mus musculus] Mapk1 gi|6273387 6.53/25861.3 8 56 51.620 ± 5.218 Spot 10 Unknown[Mus musculus] gi|40556944 9.24/10359.2 5 49 140.800 ± 17.950 Spot 11 Psip1 protein[Mus musculus] Psip1 gi|116284033 9.66/29411.4 7 45 97.240 ± 8.310 Spot 12 Unnamed protein product[Mus musculus] gi|74149487 9.37/25567.1 7 45 19.190 ± 1.396 Spot 13 Lysozyme C-1 precursor[Mus musculus] Lyz1 gi|7305247 9.55/17239.6 4 49 17.100 ± 1.400 Spot 14 Chimerin mKIAA0716 protein[Mus musculus] mKIAA0716 gi|28972363 7.97/234544.2 22 64 1.7460 ± 0.128 Spot 15 Ig H-chain V-D-region, partial[Mus musculus] IgHvd gi|194583 7.68/ 10120.9 4 35 119.370 ± 11.040 Spot 16 Synaptonemal complex protein 1[Mus musculus] Sycp1 gi|1661003 5.29/82651.7 17 56 25.190 ± 2.580 Spot 17 KRAB-zinc finger protein[Mus musculus] Tce8 gi|40796098 9.27/76185.8 13 63 0.323 ± 0.023 Spot 18 Glutathione S-transferase Mu 2[Mus musculus] Gstm2 gi|6680121 6.9/25871 7 245 0 Note. aPeptides matched refers to the matched peptides to the amino acids of the full-length protein. bMatch between control and 4 Gy radiation group (gels of 3 × carbon ion radiation vs. control; n = 3 in each group). Values shown are mean ± SEM (P < 0.05). pI, Isoelectric point. Mw, molecular weight. NCBI, National Center of Biotechnology Information. The present study aimed to use a comparative proteomics approach to investigate the molecular mechanisms underlying injury in pubertal mice ovaries following exposure to high CIR as a model to understand the damage that may occur in children receiving environmental radiation or radiotherapy during childhood and adolescence. The results revealed that high dose CIR caused proteomic changes in the ovaries of pubertal mice, and the differentially expressed proteins may be associated with severe acute injury in ovarian tissues. Furthermore, 18 differentially expressed proteins were identified in the ovaries using 2-DE methods. These results suggest that the ovarian impairment caused by high dose CIR is multifactorial and involves multiple pathways. The result of Gene Ontology (GO) annotations of the 16 differentially expressed proteins (two proteins were unknown), including Hsp90b1, Rpsa, Krt13, Uchl1, Afp, Anxa1, Akr1c18, Crybb2, Psip1, Mapk1, Lyz1, mKIAA0716, IgHvd, Sycp1, Tce8, and Gstm2, are shown in Figure 2. Regarding the biological processes, the majority of proteins were involved in cellular process (13%), developmental process (7%), metabolic process (11%), reproductive process (10%), multicellular organismal process (7%), biological regulation (9%). Regarding the cellular components, most of the proteins were located in the organelles (21%), cells (25%), membrane-enclosed lumen (11%), extracellular region (14%), macromolecular complex (12%) and membrane (9%). Regarding molecular function, 49% of the proteins participated in binding, 12% in structural molecule activity, 18% in catalytic activity, 6% in protein binding transcription factor activity and 3% in molecular transducer activity.
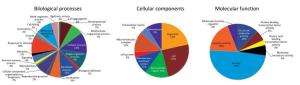
Figure 2. The annotation of biological process, cellular component and molecular function of differentially expressed proteins in mice ovaries after CIR. The original GO annotations were downloaded from the NCBI Entrez Gene database and analyzed further. The percentages are the total hits divided by the number of annotated proteins for each GO category.
The protein-protein interaction network was analyzed using a publicly available program (http://string-db.org), with the interaction network (Figure 3) including ten proteins (Hsp90b1, Rpsa, Krt13, Uchl1, Anxa1, Akr1c18, Crybb2, Psip1, Mapk1, and Lyz1) involved in ten biological processes, including complement and coagulation cascades, staphylococcus aureus infection, legionellosis, leishmaniasis, pertussis, american trypanosomiasis, platelet activation, systemic lupus erythematosus, the pahagosome and tuberculosis. Among the differential proteins identified, UCH-L1 was involved in the ubiquitin-proteasome system and it has also been implicated in cellular pathways such as proliferation, apoptosis and cell migration[1].
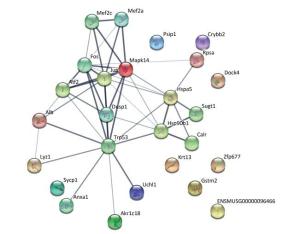
Figure 3. The interaction network of differentially expressed proteins, their interacting proteins and upstream kinases. The interaction network as analyzed by STRING software (http://string-db.org). In this network, nodes are proteins; lines represent functional associations between proteins. The blue lines between bodes represent functional associations between proteins and the thickness of the lines represents the level of confidence in association reported.
Ovaries are very sensitive to low LET radiation such as X-rays and gamma-rays, which destroy follicles, and damage the ovaries by triggering follicular cell apoptosis, chromosomal damage and oxidative stress. However, it is unknown whether high LET radiation has a damaging effect on follicles, and there are no reference values[6]. Other researchers have found that low doses of high LET ions can cause premature ovarian failure, and this has raised concerns regarding ovarian damage due to the clinical use of high-LET particles for cancer treatment[7]. In testicular germ cells, UCH-L1 expression is responsible for an early apoptotic wave during spermatogenesis and tight regulation of UCH-L1 is important because high levels cause excessive apoptosis in the ovaries and testes of transgenic mice[8]. In addition to germ cells, oocytes also express UCH-L1 and expression is significantly higher in the single oocyte of Gad mice compared with normal mice, suggesting UCH-L1 affects the ability of sperm and oocytes to combine, thereby preventing multiple fertilized oocytes[9]. UCH-L1 also plays a role in oocyte formation and inhibition of UCH-L1 in porcine oocytes caused them to stall at the first meiosis-post-transition point. This was accompanied by the formation of the cell cycle dependent kinase 1 (CDK1)-cyclinB1 complex, an increase in histone H1 kinase activity and a decrease in the levels of monomeric ubiquitin[10]. Apoptosis controls the number of oocytes and eliminates defective oocytes during oogenesis, UCH-L1 is closely correlated with apoptosis and gives that UCH-L1-dependent apoptosis is important for normal reproductive function during prepubertal development. UCH-L1 is proposed to play a role in the selective elimination of abnormal oocytes during prepubertal development of ovaries[11]. In the present study was investigated the function of UCH-L1 in ovaries following CIR. We measured UCH-L1 expression using immunoblotting (Figure 4A, B) and Real-time PCR (Figure 4C), and determined its cellular localization by immunofluorescence (Figure 5). The expression of UCH-L1 was significantly decreased in the irradiated groups (Figure 4B) and the mRNA levels of UCH-L1 (Figure 4C) were consistent with the immunoblotting results (Figure 4B). The result of immunofluorescence showed the UCH-L1 and PCNA were clearly visible and markedly stronger in control group (Figure 5). However, the immunofluorescent signal of UCH-L1 and PCNA were weakened in the CIR group (Figure 5). The observed decrease in UCH-L1 expression following CIR suggests that radiation may impact the ubiquitin-proteasome pathway, which may subsequently affect apoptosis in ovarian cells.

Figure 4. (A) Western blotting analysis of UCH-L1 and PCNA expression in mice ovaries; (B) Relative protein expression compared with control group; Values represent the average ± SEM from four gels per group. Pound sign denote values that are significantly different from control. #P < 0.001 with Student's t-test analysis. (C) Relative UCH-L1 mRNA expression compared with control group; Values represent the average ± SEM from four gels per group. Pound sign denote values that are significantly different from control. #P < 0.001 with Student's t-test analysis.
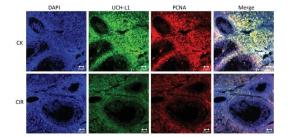
Figure 5. Immunofluorescence localization analysis of UCH-L1 and PCNA in mice ovaries. Staining was performed at 400 × magnification. DAPI (blue), UCH-L1 (green) and PCNA (red). Bar = 20 μm. CK, control; CIR, carbon ion radiation.
In summary, our results showed that high dose CIR causes acute injury to ovaries and induces degeneration of ovarian follicles in pubertal mice. The combined comparative proteomics and bioinformatics analysis indicated that the observed toxicological effects are multifactorial and involve multiple pathways. Among the identified differentially expressed proteins, UCH-L1 directly affects the function and location of numerous proteins, and may therefore be associated with follicular deg-eneration and induction of apoptosis in ovarian cells.
doi: 10.3967/bes2017.040
Differential Proteomics Reveals the Potential Injury Mechanism Induced by Heavy Ion Radiation in Mice Ovaries
-
-
Figure 1. The protein profiles and differentially expressed spots of ovaries in mice after CIR. 400µg proteins were separated using isoelectric focusing (IEF) of pH 3-10, followed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie Brilliant Blue (CCB) R250 staining. (A) control group; (B) 4 Gy group. Green and red arrows represent decreased and increased expression, respectively. Spot numbers of representative proteins are in accordance with Table 1.
Figure 2. The annotation of biological process, cellular component and molecular function of differentially expressed proteins in mice ovaries after CIR. The original GO annotations were downloaded from the NCBI Entrez Gene database and analyzed further. The percentages are the total hits divided by the number of annotated proteins for each GO category.
Figure 3. The interaction network of differentially expressed proteins, their interacting proteins and upstream kinases. The interaction network as analyzed by STRING software (http://string-db.org). In this network, nodes are proteins; lines represent functional associations between proteins. The blue lines between bodes represent functional associations between proteins and the thickness of the lines represents the level of confidence in association reported.
Figure 4. (A) Western blotting analysis of UCH-L1 and PCNA expression in mice ovaries; (B) Relative protein expression compared with control group; Values represent the average ± SEM from four gels per group. Pound sign denote values that are significantly different from control. #P < 0.001 with Student's t-test analysis. (C) Relative UCH-L1 mRNA expression compared with control group; Values represent the average ± SEM from four gels per group. Pound sign denote values that are significantly different from control. #P < 0.001 with Student's t-test analysis.
Table 1. Differential Expression of Proteins in Mice Ovaries
Protein Spot Protein Name Abbreviation Accession (NCBI) pI/Mw PeptidesMatcheda ProteinScore Average FoldChangeb Spot 1 Heat shock protein 90, beta (Grp94), member 1[Mus musculus] Hsp90b1 gi|14714615 4.74/92717.4 16 184 12.300 ± 2.060 Spot 2 Laminin receptor[Mus musculus] Rpsa gi|171948782 4.8/32943.5 13 666 0.293 ± 0.028 Spot 3 Keratin, type Ⅰ cytoskeletal 13[Mus musculus] Krt13 gi|37994713 4.88/33925.4 5 67 0.340 ± 0.017 Spot 4 Ubiquitin carboxyl-terminal hydrolase isozyme L1, partial[Mus musculus] Uchl1 gi|399106762 5.02/16462.3 1 107 0.016 ± 0.005 Spot 5 Annexin[Mus musculus] Anxa1 gi|70912321 7.59/38995.1 9 46 0 Spot 6 Alpha-fetoprotein, partial[Mus musculus] Afp gi|191765 5.47/48791.6 7 99 22.370 ± 1.822 Spot 7 Aldo-keto reductase family 1 member C18 [Mus Musculus] Akr1c18 gi|19527284 6.02/37666 16 640 5.280 ± 0.490 Spot 8 Beta-crystallin B2[Mus musculus] Crybb2 gi|6681035 6.5/23480.4 2 87 29.440 ± 3.815 Spot 9 Mitogen-activated protein kinase p38 beta[Mus musculus] Mapk1 gi|6273387 6.53/25861.3 8 56 51.620 ± 5.218 Spot 10 Unknown[Mus musculus] gi|40556944 9.24/10359.2 5 49 140.800 ± 17.950 Spot 11 Psip1 protein[Mus musculus] Psip1 gi|116284033 9.66/29411.4 7 45 97.240 ± 8.310 Spot 12 Unnamed protein product[Mus musculus] gi|74149487 9.37/25567.1 7 45 19.190 ± 1.396 Spot 13 Lysozyme C-1 precursor[Mus musculus] Lyz1 gi|7305247 9.55/17239.6 4 49 17.100 ± 1.400 Spot 14 Chimerin mKIAA0716 protein[Mus musculus] mKIAA0716 gi|28972363 7.97/234544.2 22 64 1.7460 ± 0.128 Spot 15 Ig H-chain V-D-region, partial[Mus musculus] IgHvd gi|194583 7.68/ 10120.9 4 35 119.370 ± 11.040 Spot 16 Synaptonemal complex protein 1[Mus musculus] Sycp1 gi|1661003 5.29/82651.7 17 56 25.190 ± 2.580 Spot 17 KRAB-zinc finger protein[Mus musculus] Tce8 gi|40796098 9.27/76185.8 13 63 0.323 ± 0.023 Spot 18 Glutathione S-transferase Mu 2[Mus musculus] Gstm2 gi|6680121 6.9/25871 7 245 0 Note. aPeptides matched refers to the matched peptides to the amino acids of the full-length protein. bMatch between control and 4 Gy radiation group (gels of 3 × carbon ion radiation vs. control; n = 3 in each group). Values shown are mean ± SEM (P < 0.05). pI, Isoelectric point. Mw, molecular weight. NCBI, National Center of Biotechnology Information. -
[1] Brinkmann K, Schell M, Hoppe T, et al. Regulation of the DNA damage response by ubiquitin conjugation. Front Genet, 2015; 10, 98. https://www.researchgate.net/profile/Kerstin_Brinkmann/publication/273756063_Regulation_of_the_DNA_damage_response_by_ubiquitin_conjugation/links/550a55a40cf26198a63a7f61.pdf?disableCoverPage=true [2] Di CX, Sun C, Li HY, et al. Diallyl disulfide enhances carbon ion beams-induced apoptotic cell death in cervical cancer cells through regulating Tap73/ΔNp73. Cell Cycle, 2015; 14, 3725-33. doi: 10.1080/15384101.2015.1104438 [3] Li HY, Zhang H. Proteome analysis for profiling infertility markers in male mouse sperm after carbon ion radiation. Toxicology, 2013; 5, 306, 85-92. http://www.ncbi.nlm.nih.gov/pubmed/23435181 [4] Li HY, He YX, Zhang H, et al. Differential proteome and gene expression reveal response to carbon ion irradiation in pubertal mice testes. Toxicol Lett, 2014; 21, 433-44. https://www.researchgate.net/publication/259807325_Differential_proteome_and_gene_expression_reveal_response_to_carbon_ion_irradiation_in_pubertal_mice_testes [5] Lee JC, Yoon YD. Gamma-radiation-induced follicular degeneration in the prepubertal mouse ovary. Mutat Res, 2005; 578, 247-55. doi: 10.1016/j.mrfmmm.2005.05.019 [6] Huang S, Dang Y, Li F, et al. Biological intensity-modulated radiotherapy plus neoadjuvant chemotherapy for multiple peritoneal metastases of ovarian cancer: A case report. Oncol Lett, 2015; 9, 1239-43. https://www.researchgate.net/profile/Shigao_Huang/publication/272097469_Biological_intensity-modulated_radiotherapy_plus_neoadjuvant_chemotherapy_for_multiple_peritoneal_metastases_of_ovarian_cancer_A_case_report/links/5639545d08aed5314d221efa.pdf?inViewer=0&pdfJsDownload=0&origin=publication_detail [7] Mishra B, Ortiz L, Luderer U, et al. Charged iron particles, components of space radiation, destroy ovarian follicles. Hum Reprod, 2016; 31, 1816-26. doi: 10.1093/humrep/dew126 [8] Susor A, Liskova L, Toralova T, et al. Role of ubiquitin C-terminal hydrolase-L1 in antipolyspermy defense of mammalian oocytes. Biol Reprod, 2010; 82, 1151-61. doi: 10.1095/biolreprod.109.081547 [9] Sekiguchi S, Kwon J, Yoshida E, et al. Localization of ubiquitin C-terminal hydrolase L1 in mouse ova and its function in the plasma membrane to blockpolyspermy. Am J Pathol, 2006; 169, 1722-9. doi: 10.2353/ajpath.2006.060301 [10] Susor A, Ellederova Z, Jelinkova L, et al. Proteomic analysis of porcine oocytes during in vitro maturation reveals essential role for the ubiquitin C-terminal hydrolase-L1. Reproduction, 2007; 134, 559-68. doi: 10.1530/REP-07-0079 [11] Gu YQ, Che QJ, Gu Z, et al. Ubiquitin carboxyl-terminal hydrolase L1 contributes to the oocyte selective elimination in prepubertal mouse ovaries. Acta Physiologica Sinica, 2009; 61, 175-84. https://www.researchgate.net/publication/24308735_Ubiquitin_carboxyl-terminal_hydrolase_L1_contributes_to_the_oocyte_selective_elimination_in_prepubertal_mouse_ovaries -




 下载:
下载:



 Quick Links
Quick Links