-
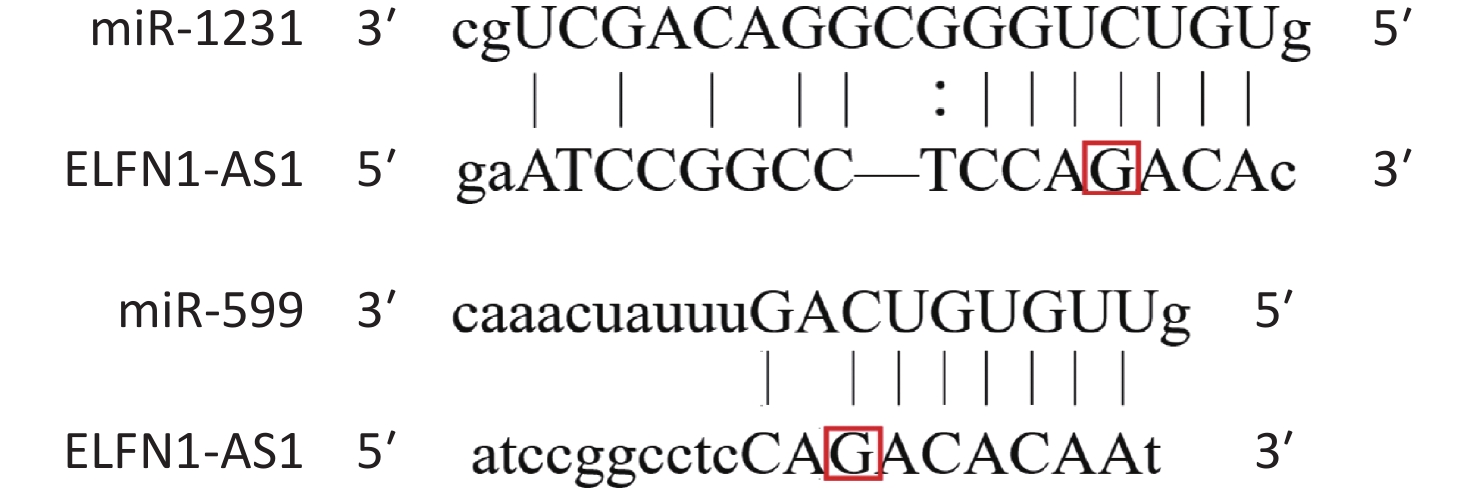
Colorectal cancer (CRC) is a common and deadly disease, with over two million new cases and one million deaths in 2020[1]. Risk factors include lack of exercise, obesity, high red meat consumption, low intake of fiber, smoking, and alcohol consumption, as well as genetic factors[2,3]. ELFN1-AS1, located on chromosome 7p22.3, expresses a pro-oncogenic lncRNA[4-7]. Wang et al. reported that exosomal ELFN1-AS1 from osteosarcoma cells could mediate macrophage M2 polarization by sponging miR-138-5p and miR-1291 to promote osteosarcoma tumorigenesis[4]. Lei et al. found that ELFN1-AS1 could promote CRC proliferation and migration by modulating the miR-4644/TRIM44 axis[5]. Zhang et al. confirmed that ELFN1-AS1 could facilitate the progression of esophageal cancer by promoting GFPT1 expression via sponging miR-183-3p[6]. Jie et al. found that ELFN1-AS1 could accelerate the proliferation, invasion, and migration of ovarian cancer cells by directly interacting with miR-497-3p and then regulating CLDN4 expression[7]. These results suggest that ELFN1-AS1 promotes the progression of various cancers, including CRC, osteosarcoma, esophageal cancer, and ovarian cancer, by acting as a competing endogenous RNA (ceRNA) and modulating miRNA expression. Bioinformatics analysis shows that a single nucleotide polymorphism (rs3735664 G>A) is located at the binding site of ELFN1-AS1 to miR-599 and miR-1231 (Supplementary Figure S1 available in www.besjournal.com)[8]. MiR-599 and miR-1231 could be adsorbed by some sponges, which in turn promotes the progression of CRC[9,10]. Therefore, the rs3735664 polymorphism may be involved in the development and progression of CRC by affecting the ceRNA function of ELFN1-AS1.
In this study, 1,000 individuals of Chinese Han ethnicity were recruited, 500 of whom were diagnosed with sporadic CRC, confirmed by pathology. The other 500 were cancer-free individuals who had no family history of cancer, had no intestinal disease, and were enrolled in hospital physical examinations (Supplementary Table S1 available in www.besjournal.com). Patients with other digestive diseases were excluded. The average age of patients and cancer-free individuals was 59.18 ± 7.46 and 59.70 ± 7.63, respectively. Male participants made up the majority, with 274 (54.8%) of the CRC patients and 283 (56.6%) of the cancer-free individuals being male. Of the patients, 248 (49.6%) had I/II tumors, and 252 (50.4%) had III/IV tumors.
Table S1. Demographic and clinicopathological characteristics of CRC patients and cancer-free individuals
Characteristics CRC patients (n = 500) Cancer-free individuals (n = 500) P Age (mean ± SD) 59.18 ± 7.46 59.70 ± 7.63 0.28 Gender, n (%) Female 226 (45.2) 217 (43.4) 0.57 Male 274 (54.8) 283 (56.6) TMN stages, n (%) I + II 248 (49.6) III + IV 252 (50.4) Oral mucosal cells were collected from each individual, and genomic DNA was extracted using the Chelex-100 method. A single nucleotide polymorphism (rs3735664 G>A) was amplified by polymerase chain reaction (PCR) and sequenced using an ABI 3730XL sequencer. The PCR conditions were pre-denaturation at 95 ℃ for 5 min; denaturation at 94 ℃ for 30 s; annealing at 58 ℃ for 30 s; extension at 72 ℃ for 30 s, 35 cycles; extension at 72 ℃ for 10 min. The PCR primer sequences were as follows: 5’-CGTCTCGGAGTGAATGACAG-3’, 5’-AGGTACCACCTGGTCTCCTG-3’. The sequencing results were analyzed using Chromas software.
Tumor and adjacent non-tumor tissues were collected from 26 CRC patients who had not received preoperative radio- and chemotherapies. Human embryonic kidney cell line (HEK-293T) and CRC cell lines (HCT116 and HT29) were purchased from Shanghai EK-Bioscience Biotechnology Co., Ltd. ELFN1-AS1 full-length sequence with the rs3735664 G (WT) and A (MUT) allele was inserted into the pcDNA3.1 vector to create ELFN1-AS1 overexpression plasmids. An empty pcDNA3.1 vector was used as a negative control (NC). As per protocols, plasmids were transfected into HCT116 and HT29 cells using Lipofectamine 3000 (Invitrogen, USA). All cells were collected 48 hours later for further research. Total RNA was extracted from CRC tissues and cell lines using TRIzol reagent (Invitrogen, USA) according to the product manual, and cDNA was synthesized using a reverse transcription kit (Takara, Japan) according to the supplier's instructions. Thereafter, qRT-PCR was performed using an ABI Real-Time PCR system (Applied Biosystems, USA) and SYBR Green qPCR Master Mix (Takara, Japan). U6 and GAPDH were used as internal controls for miRNA and lncRNA, respectively. The relative expression levels of all genes were calculated using the 2−ΔΔCt method. The sequences of the related primers are presented in Supplementary Table S2 available in www.besjournal.com. All participants provided written informed consent, and the study protocol was approved by the Ethical Committee of Yancheng First People's Hospital (2021-K- 117).
Table S2. The primer sequences for qRT-PCR
Gene Forward (5’ to 3’ ) Reverse (5’ to 3’) ELFN1-AS1 ACTCTCAGCCCCCACCTAGT ATTCAACGGAAGAGGAAGCA miR-599 GCCGAGGTTGTGTCAGTTT CTCAACTGGTGTCGTGGAGT miR-1231 CCTCAACTGAATTGCCGACTC CTCAACTGGTGTCGTGGAGTC GAPDH CTCTGATTTGGTCGTATTGGGC CCTGGAAGATGGTGATGGGATT U6 CTCGCTTCGGCAGCACA AACGCTTCACGAATTTGCGT The case-control study found a significant association between ELFN1-AS1 rs3735664 polymorphism and CRC susceptibility (Table 1). Individuals with rs3735664 AA genotype or carrying the A allele had a significantly lower risk of developing CRC compared to those with the GG genotype or carrying the G allele (OR = 0.53, 95% CI = 0.34−0.84, P = 0.006; OR = 0.75, 95% CI = 0.62−0.91, P = 0.003, respectively).
Table 1. Association of ELFN1-AS1 rs3735664 polymorphism with CRC susceptibility, n (%)
Genotype/allele CRC patients (n = 500) Cancer-free individuals (n = 500) OR (95% CI)a Pa Power GG 268 (53.6) 230 (46.0) 1 GA 197 (39.4) 213 (42.6) 0.79 (0.61–1.03) 0.083 0.514 AA 35 (7.0) 57 (11.4) 0.53 (0.34–0.84) 0.006 0.506 PHWEb 0.472 G 733 (73.3) 673 (67.3) 1 A 267 (26.7) 327 (32.7) 0.75 (0.62–0.91) 0.003 0.488 Note. a: Adjusted for age and gender; b: PHWE for the controls. Furthermore, the study found a correlation between ELFN1-AS1 rs3735664 polymorphism and the TNM stage in CRC patients (Table 2). Patients with the rs3735664 GA or AA genotype were less likely to develop stage III + IV tumors compared to those with the GG genotype (GA vs. GG: OR = 0.61, 95% CI = 0.42−0.89, P = 0.009; AA vs. GG: OR = 0.25, 95% CI = 0.11−0.56, P = 0.001). CRC patients carrying the rs3735664 A allele were less likely to develop stage III + IV tumors compared to CRC patients carrying the G allele (A vs. G: OR = 0.58, 95% CI = 0.44−0.77, P < 0.001).
Table 2. Association of ELFN1-AS1 rs3735664 polymorphism with the TNM stage in colorectal cancer, n (%)
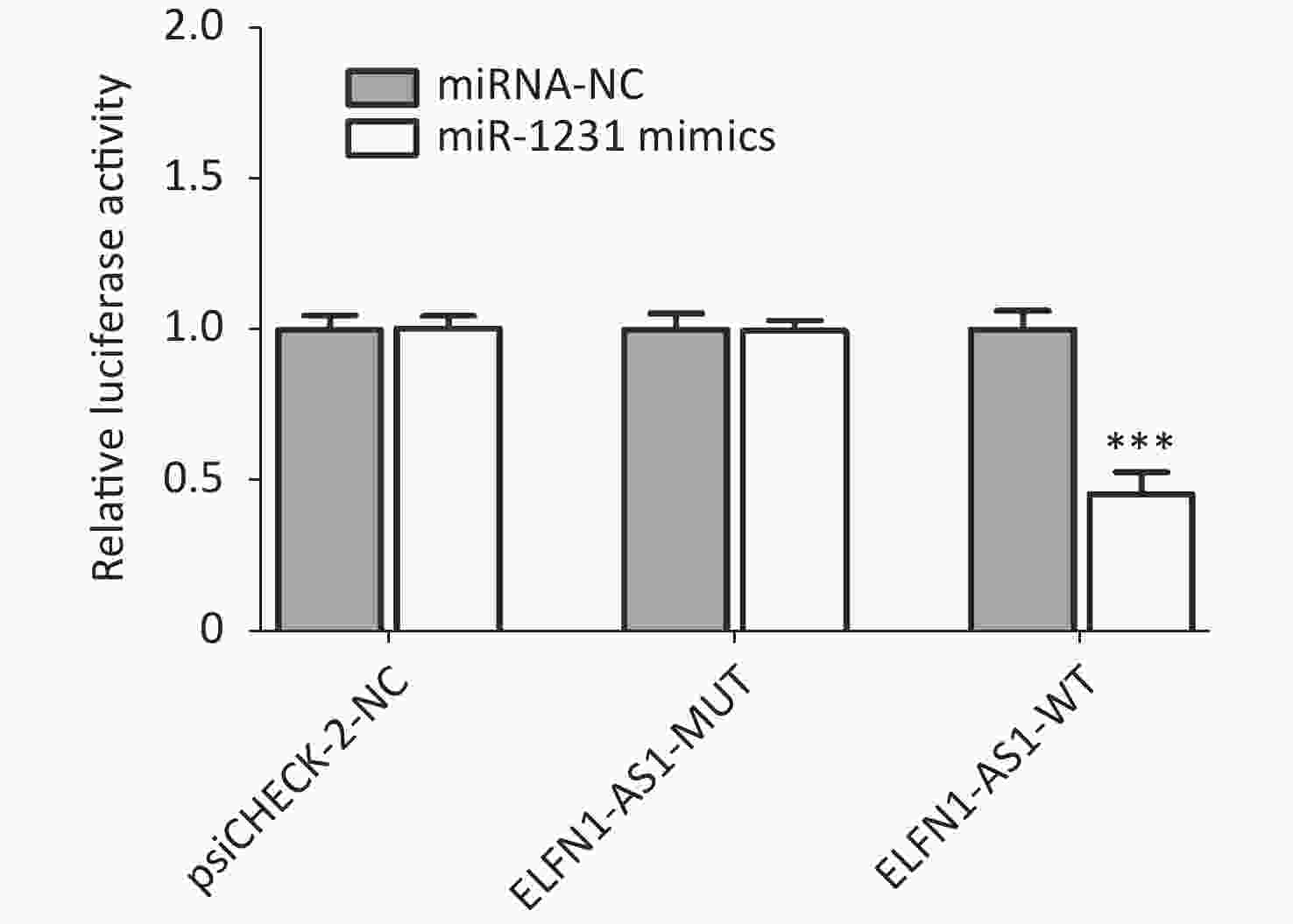
Genotype/allele III + IV (n = 252) I + II (n = 248) OR (95% CI)a Pa Power GG 153 (60.7) 115 (46.4) 1 GA 90 (35.7) 107 (43.1) 0.61 (0.42–0.89) 0.009 0.508 AA 9 (3.6) 26 (10.5) 0.25 (0.11–0.56) 0.001 0.591 G 396 (78.6) 337 (67.9) 1 A 108 (21.4) 159 (32.1) 0.58 (0.44–0.77) < 0.001 0.695 Note. a: Adjusted for age and gender. The study also found a significant negative correlation between ELFN1-AS1 expression and miR-1231 expression in colorectal tissues of rs3735664 GG and GA genotype (tumor tissues with GG genotype: r = −0.801, P = 0.001; adjacent non-tumor tissues with GG genotype: r = −0.844, P = 0.005; tumor tissues with GA genotype: r = −0.783, P = 0.012) (Table 3). The overexpression study showed that ELFN1-AS1 with the rs3735664 G allele (WT) could reduce miR-1231 expression in CRC cells, while ELFN1-AS1 with A allele (MUT) was unable to do so (Supplementary Figure S2 available in www.besjournal.com). ELFN1-AS1 sequence containing rs3735664 G (WT) and A (MUT) alleles was subcloned into the psiCHECK-2 vector. The recombinant dual-luciferase vectors (ELFN1-AS1-WT or ELFN1-AS1-MUT) were cotransfected into HEK-293T cells with miR-1231 mimic and miRNA NC, respectively. After 48 hours of transfection, the luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega, USA). The results of the luciferase assay showed that the rs3735664 A allele (MUT) was able to block the binding of ELFN1-AS1 to miR-1231 (Supplementary Figure S3 available in www.besjournal.com). This suggests that ELFN1-AS1 carrying the G allele may downregulate miR-1231 expression in CRC cells by adsorbing miR-1231. MiR-1231 has been linked to a negative association with tumor size, TNM stage, lymph node invasion, and poor prognosis in CRC patients[10]. The circTDRD3 promoted HIF1α expression by sponging miR-1231, which in turn contributes to the growth and metastasis of CRC[10]. Thus, the hypothesis is that the rs3735664 A allele may reduce the CRC-promoting role of ELFN1-AS1 by blocking its adsorption to miR-1231, leading to a decrease in CRC risk and progression.
Table 3. Correlation between ELFN1-AS1 and miRNA expression in colorectal tissues with different rs3735664 genotypes
Tissue types miRNAs Genotype Number
of tissueCorrelation
testP r Adjacent non-
tumor tissuesmiR-599 GG 13 0.353 −0.192 GA 9 0.512 −0.276 AA 4 0.535 −0.463 miR-1231 GG 13 0.005 −0.844 GA 9 0.217 −0.451 AA 4 0.193 −0.813 Tumor tissues miR-599 GG 13 0.474 −0.374 GA 9 0.612 −0.197 AA 4 0.416 −0.592 miR-1231 GG 13 0.001 −0.801 GA 9 0.012 −0.783 AA 4 0.135 −0.764 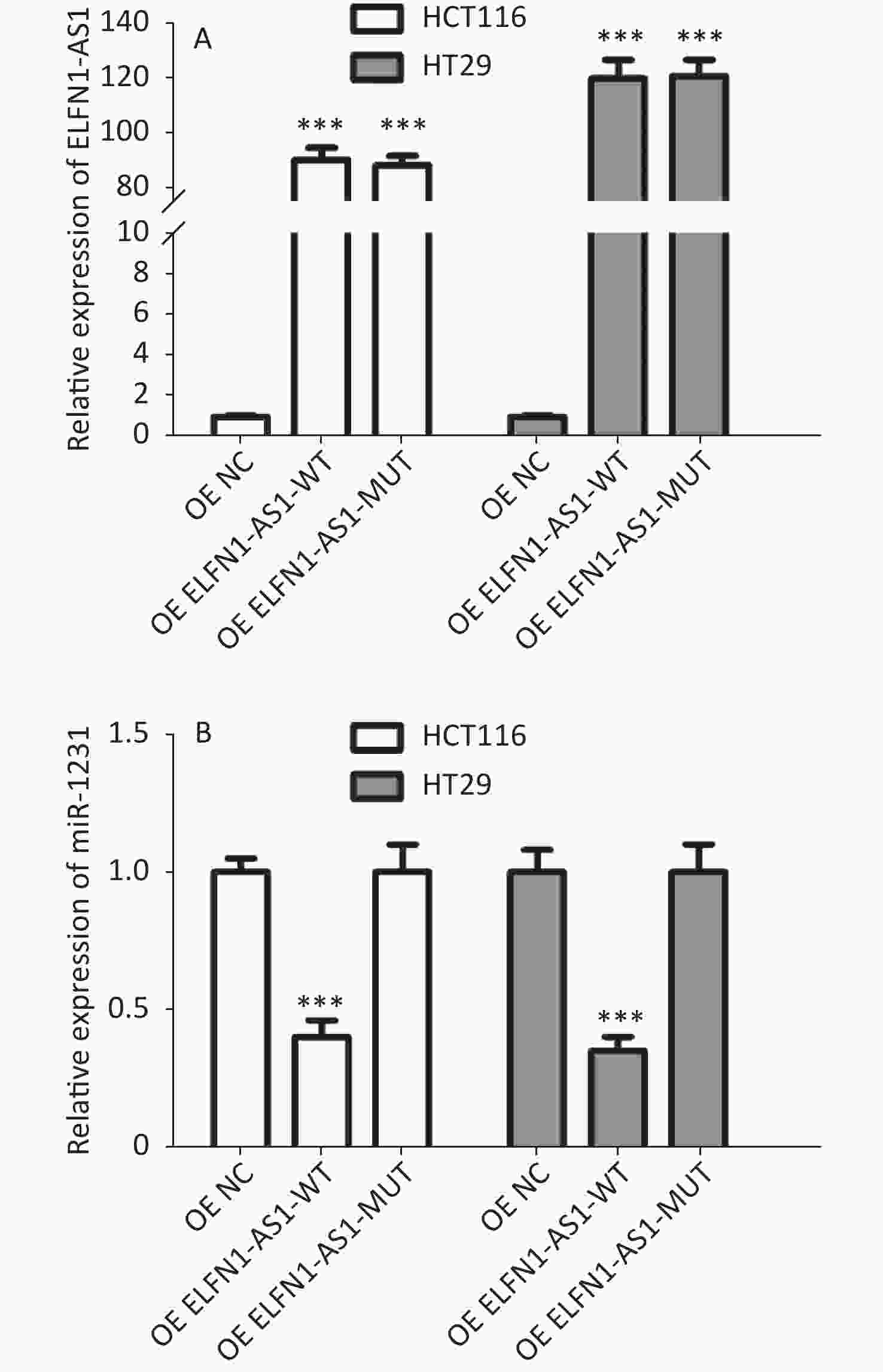
Figure S2. Effect of overexpression of ELFN1-AS1 carrying different rs3735664 alleles on miR-1231 expression in CRC cells (A: Relative expression of ELFN1-AS1 in CRC cells; B: Relative expression of miR-1231 in CRC cells)
The current study provides insights into the role of lncRNA polymorphisms in CRC and reveals a new genetic marker for the disease. However, there are some limitations to be noted. Further research is needed to examine the interaction between the rs3735664 polymorphism and environmental risk factors and its effect on CRC susceptibility, and the relationship between the rs3735664 polymorphism and the survival prognosis of CRC patients is yet to be explored.
Despite these limitations, the findings suggest that the ELFN1-AS1 rs3735664 polymorphism is associated with both CRC susceptibility and tumor stage in Chinese Han populations. The polymorphism holds promise as a biomarker for predicting CRC risk and progression.
doi: 10.3967/bes2023.045
rs3735664 Polymorphism Affecting ELFN1-AS1 Adsorption on miR-1231 is Associated with Colorectal Cancer Susceptibility and Tumor Stage
-
-
S1. Demographic and clinicopathological characteristics of CRC patients and cancer-free individuals
Characteristics CRC patients (n = 500) Cancer-free individuals (n = 500) P Age (mean ± SD) 59.18 ± 7.46 59.70 ± 7.63 0.28 Gender, n (%) Female 226 (45.2) 217 (43.4) 0.57 Male 274 (54.8) 283 (56.6) TMN stages, n (%) I + II 248 (49.6) III + IV 252 (50.4) S2. The primer sequences for qRT-PCR
Gene Forward (5’ to 3’ ) Reverse (5’ to 3’) ELFN1-AS1 ACTCTCAGCCCCCACCTAGT ATTCAACGGAAGAGGAAGCA miR-599 GCCGAGGTTGTGTCAGTTT CTCAACTGGTGTCGTGGAGT miR-1231 CCTCAACTGAATTGCCGACTC CTCAACTGGTGTCGTGGAGTC GAPDH CTCTGATTTGGTCGTATTGGGC CCTGGAAGATGGTGATGGGATT U6 CTCGCTTCGGCAGCACA AACGCTTCACGAATTTGCGT Table 1. Association of ELFN1-AS1 rs3735664 polymorphism with CRC susceptibility, n (%)
Genotype/allele CRC patients (n = 500) Cancer-free individuals (n = 500) OR (95% CI)a Pa Power GG 268 (53.6) 230 (46.0) 1 GA 197 (39.4) 213 (42.6) 0.79 (0.61–1.03) 0.083 0.514 AA 35 (7.0) 57 (11.4) 0.53 (0.34–0.84) 0.006 0.506 PHWEb 0.472 G 733 (73.3) 673 (67.3) 1 A 267 (26.7) 327 (32.7) 0.75 (0.62–0.91) 0.003 0.488 Note. a: Adjusted for age and gender; b: PHWE for the controls. Table 2. Association of ELFN1-AS1 rs3735664 polymorphism with the TNM stage in colorectal cancer, n (%)
Genotype/allele III + IV (n = 252) I + II (n = 248) OR (95% CI)a Pa Power GG 153 (60.7) 115 (46.4) 1 GA 90 (35.7) 107 (43.1) 0.61 (0.42–0.89) 0.009 0.508 AA 9 (3.6) 26 (10.5) 0.25 (0.11–0.56) 0.001 0.591 G 396 (78.6) 337 (67.9) 1 A 108 (21.4) 159 (32.1) 0.58 (0.44–0.77) < 0.001 0.695 Note. a: Adjusted for age and gender. Table 3. Correlation between ELFN1-AS1 and miRNA expression in colorectal tissues with different rs3735664 genotypes
Tissue types miRNAs Genotype Number
of tissueCorrelation
testP r Adjacent non-
tumor tissuesmiR-599 GG 13 0.353 −0.192 GA 9 0.512 −0.276 AA 4 0.535 −0.463 miR-1231 GG 13 0.005 −0.844 GA 9 0.217 −0.451 AA 4 0.193 −0.813 Tumor tissues miR-599 GG 13 0.474 −0.374 GA 9 0.612 −0.197 AA 4 0.416 −0.592 miR-1231 GG 13 0.001 −0.801 GA 9 0.012 −0.783 AA 4 0.135 −0.764 -
[1] Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2021; 71, 209−49. doi: 10.3322/caac.21660 [2] Ramzi NH, Chahil JK, Lye SH, et al. Role of genetic & environment risk factors in the aetiology of colorectal cancer in Malaysia. Indian J Med Res, 2014; 139, 873−82. [3] Sargazi S, Abghari AZ, Sarani H, et al. Relationship between CASP9 and CASP10 gene polymorphisms and cancer susceptibility: evidence from an updated meta-analysis. Appl Biochem Biotechnol, 2021; 193, 4172−96. doi: 10.1007/s12010-021-03613-w [4] Wang BM, Wang X, Li P, et al. Osteosarcoma cell-derived exosomal ELFN1-AS1 mediates macrophage M2 polarization via sponging miR-138-5p and miR-1291 to promote the tumorgenesis of osteosarcoma. Front Oncol, 2022; 12, 881022. doi: 10.3389/fonc.2022.881022 [5] Lei R, Feng LC, Hong D. ELFN1-AS1 accelerates the proliferation and migration of colorectal cancer via regulation of miR-4644/TRIM44 axis. Cancer Biomark, 2020; 27, 433−43. doi: 10.3233/CBM-190559 [6] Zhang CY, Lian HK, Xie LS, et al. LncRNA ELFN1-AS1 promotes esophageal cancer progression by up-regulating GFPT1 via sponging miR-183-3p. Biol Chem, 2020; 401, 1053−61. doi: 10.1515/hsz-2019-0430 [7] Jie YK, Ye L, Chen H, et al. ELFN1-AS1 accelerates cell proliferation, invasion and migration via regulating miR-497-3p/CLDN4 axis in ovarian cancer. Bioengineered, 2020; 11, 872−82. doi: 10.1080/21655979.2020.1797281 [8] Miao YR, Liu W, Zhang Q, et al. lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res, 2018; 46, D276−80. doi: 10.1093/nar/gkx1004 [9] Jiang ZP, Tai QW, Xie XJ, et al. EIF4A3-induced circ_0084615 contributes to the progression of colorectal cancer via miR-599/ONECUT2 pathway. J Exp Clin Cancer Res, 2021; 40, 227. doi: 10.1186/s13046-021-02029-y [10] Fu ZM, Zhang PS, Zhang RC, et al. Novel hypoxia-induced HIF1α-circTDRD3-positive feedback loop promotes the growth and metastasis of colorectal cancer. Oncogene, 2023; 42, 238−52. doi: 10.1038/s41388-022-02548-8 -
 22436Supplementary Materials.pdf
22436Supplementary Materials.pdf

-





 下载:
下载:





 Quick Links
Quick Links