-
Vibrio parahaemolyticus, a gram-negative halophilic bacterium that naturally inhabits coastal waters, causes gastroenteritis, skin infections, and septicemia in human beings[1]. This bacterium produces multiple virulence factors, including thermostable direct hemolysin (TDH), TDH-related hemolysin (TRH), type III secretion system 1 (T3SS1), T3SS2, type VI secretion system 1 (T6SS1), and T6SS2[1]. Furthermore, V. parahaemolyticus forms biofilms on the surface, which help it in adapting to unfavorable conditions[2]. Mature biofilm formation requires special structures, including lateral and polar flagella, exopolysaccharide (EPS), and type IV pili[2].
CpsQ, a c-di-GMP-binding regulatory protein, is encoded by the cpsQ-mfpABC operon[3]. Although CpsQ plays important roles in biofilm formation and the expression of RTX matrix proteins and capsules[4], whether it can regulate other genes remains obscure. In this study, nucleotides 35 to 616 of cpsQ were deleted from the genome of V. parahaemolyticus RIMD2210633 (wild-type, WT) using the suicide plasmid pDS132 to generate the cpsQ deletion mutant (ΔcpsQ)[5], and then the RNA sequencing (RNA-seq) and several phenotypic and molecular experimental assays were employed to explore the genes regulated by CpsQ.
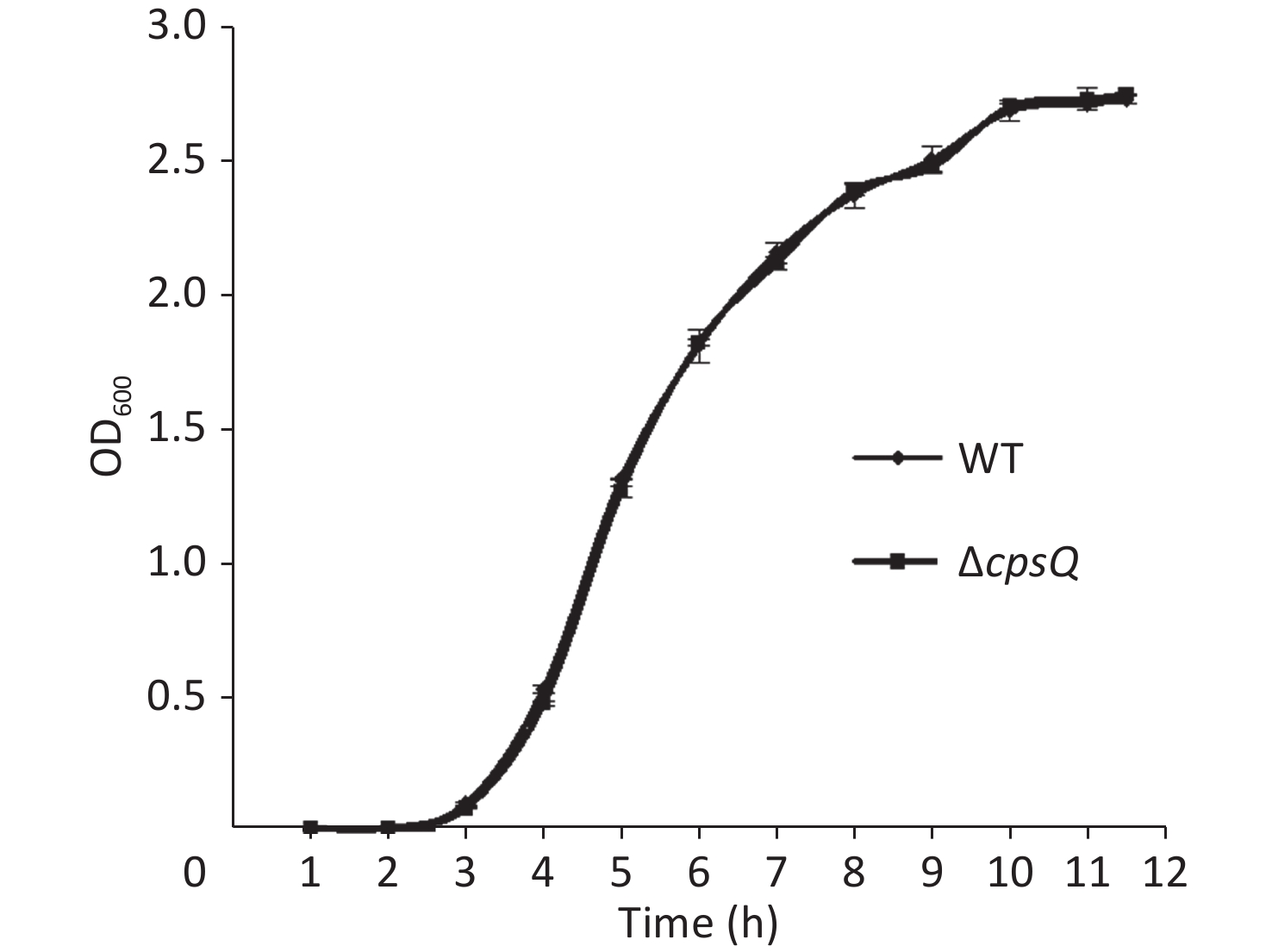
Overnight cell cultures of WT and ΔcpsQ in 2.5% (w/v) Bacto heart infusion (HI) broth (BD Biosciences, Franklin Lakes, NJ, USA) at 37 °C were serially diluted 10-fold using phosphate-buffered saline (PBS) buffer (pH 7.2); then, 200 μL of the diluted cells was spread onto an HI plate, followed by statical incubation at 37 °C for 48 h. Colonies were randomly selected in triplicate for each strain, resuspended in PBS, and adjusted to obtain an OD600 value of 1.4 for each bacterial suspension (defined here as bacterial seeds); they were then diluted 50-fold using 10 mL HI broth and allowed to continuously grow at 37 °C with shaking at 200 rpm. The OD600 values of all cultures were measured at 1 h intervals to plot growth curves. As shown in Supplementary Figure S1, available in www.besjournal.com, the two strains showed similar growth rates under the studied growth conditions, which suggested that CpsQ did not regulate the growth of V. parahaemolyticus.
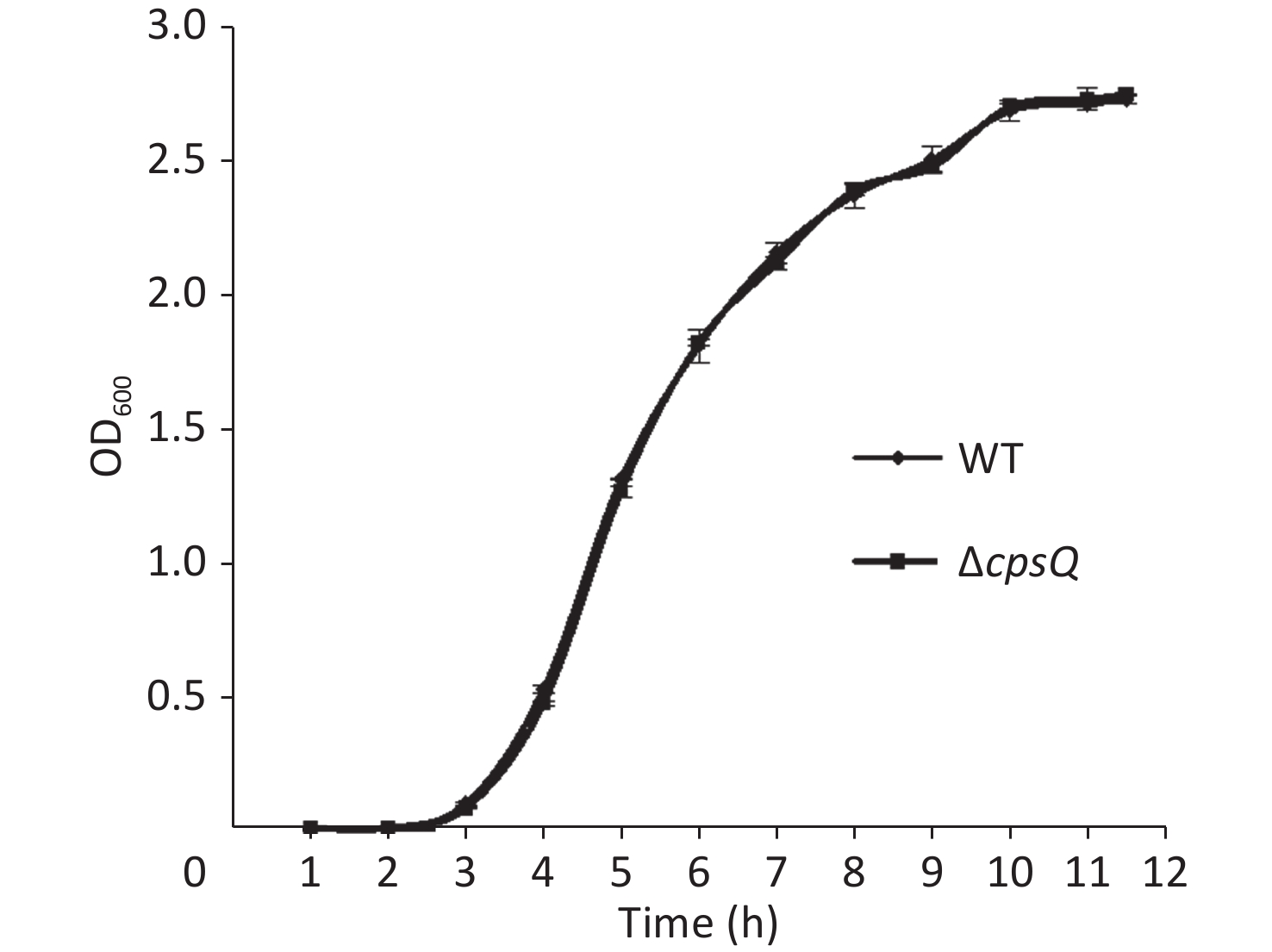
Figure S1. Growth curves of WT and ΔcpsQ. V. parahaemolyticus strains were grown in HI broth at 37 °C with shaking at 200 rpm, and the OD600 values of each culture were monitored at 1 h intervals. Experiments were performed thrice with three different colonies per experiment.
Two colonies of each strain were randomly harvested from the HI plate and placed in TRIzol reagent (Invitrogen, California, USA) for RNA extraction. RNA-related operations were performed at GENEWIZ Biotechnology Co., Ltd. (Suzhou, China). The mRNA profiles in ΔcpsQ (test group) were compared with those of WT (reference group) via RNA-seq to determine the genes that were regulated by CpsQ in V. parahaemolyticus. Significantly differentially expressed genes (DEGs) were analyzed using the DESeq2 (V1.6.3) software with at least two-fold changes in the ratio of mRNA levels (test/reference) and a P-value of < 0.05. As shown in Figure 1, CpsQ regulated the transcription of 567 genes, including 311 upregulated and 441 downregulated genes. Of these, 28 polar flagellar genes and 23 T6SS2 genes were down- and upregulated, respectively, in ΔcpsQ relative to those in WT (Supplementary Table S1, available in www.besjournal.com). A total of six genes encoding GGDEF- or EAL-domain proteins were significantly differentially expressed in ΔcpsQ relative to those in WT (Supplementary Table S1). Additionally, at least 31 putative regulatory genes, including 12 putative two-component system genes (highlighted with gray shadows in Supplementary Table S1), were significantly differentially expressed in ΔcpsQ relative to those in WT. Other genes such as antioxidative genes, T3SS2 genes, and putative outer membrane protein genes were also differentially expressed in ΔcpsQ and WT (Supplementary Table S1). The reliability of the transcriptome data was validated using quantitative PCR (qPCR) (Figure 2), which was performed as described previously[6]. The primers used in this study are listed in Supplementary Table S2, available in www.besjournal.com.
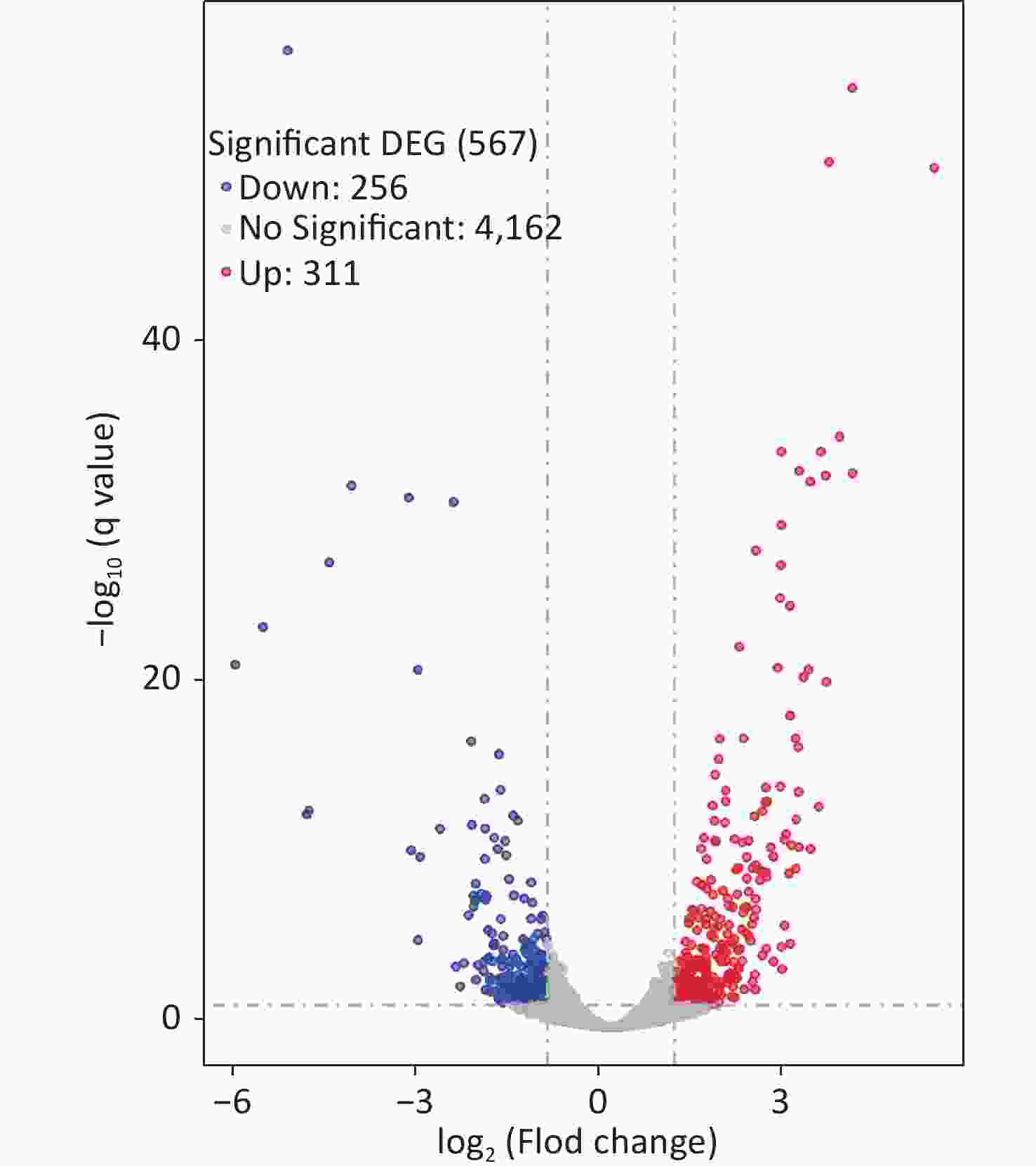
Figure 1. RNA-seq revealed the genes regulated by CpsQ on the HI agar plate. Volcano plot. Red, blue, and grey points indicate the upregulated, downregulated, and non-significant genes, respectively.
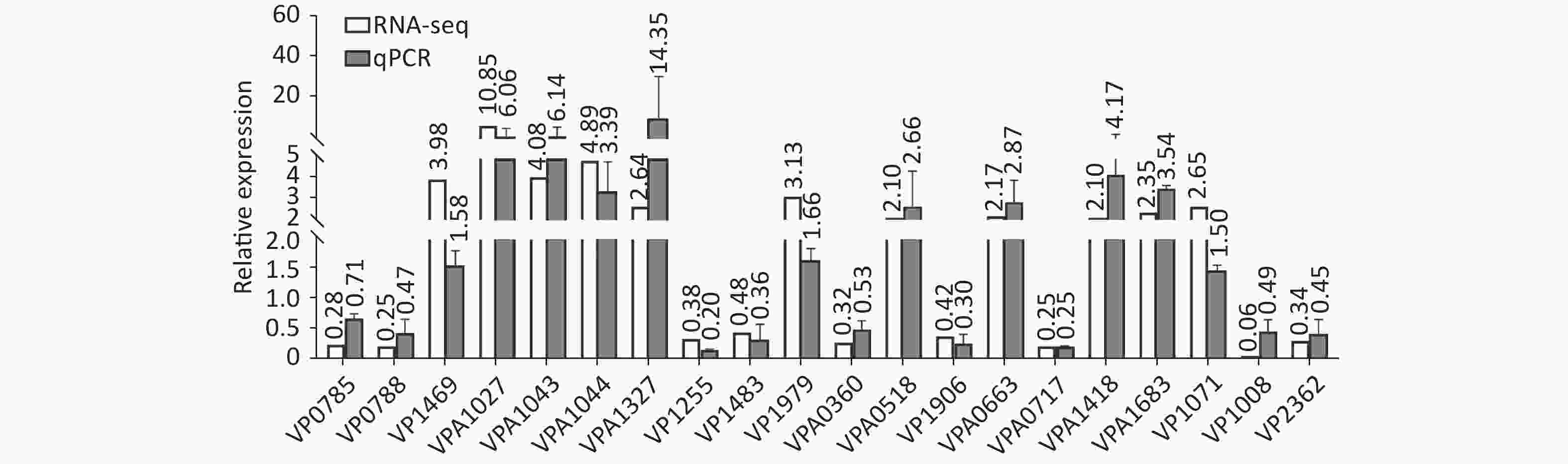
Figure 2. The transcriptional variation between ΔcpsQ (test) and WT (reference) was calculated for each target gene. A mean ratio of 2 was designated as the cutoff for determining statistical significance. The 16S rRNA gene was used as the internal control. WT, wild type.
Approximately 50% of the polar flagellar genes were activated by CpsQ (Supplementary Table S1). Therefore, we investigated whether CpsQ regulates polar flagellum-mediated swimming motility. Two microliters of bacterial seeds were inoculated onto a semi-solid swimming plate containing 1% Oxoid tryptone (Oxoid, Basingstoke, England), 2% NaCl (Merck, Darmstadt, Germany), and 0.5% Difco Noble agar (BD Biosciences). The diameters of swimming areas were measured per hour after incubation at 37 °C. Results showed that the swimming capacities of WT and ΔcpsQ were not significantly different under the studied growth conditions at all time points tested (Supplementary Figure S2A, available in www.besjournal.com), which contradicted the results of RNA-seq and qPCR (Supplementary Table S1 and Figure 2). To verify whether the contradiction is attributed to the different growth conditions, V. parahaemolyticus strains were grown in the same medium as the swimming plate but with no agar at 37 °C and harvested at the mid-log phase. qPCR assays were employed to detect the regulation of polar flagellar genes (VP0785 and VP0788) by CpsQ. As shown in Supplementary Figure S2B, the mRNA levels of VP0785 and VP0788 were similar in ΔcpsQ compared to those in WT, which suggested that CpsQ had no regulatory effect on the transcription of polar flagellar genes. Therefore, CpsQ did not regulate the swimming motility of V. parahaemolyticus.
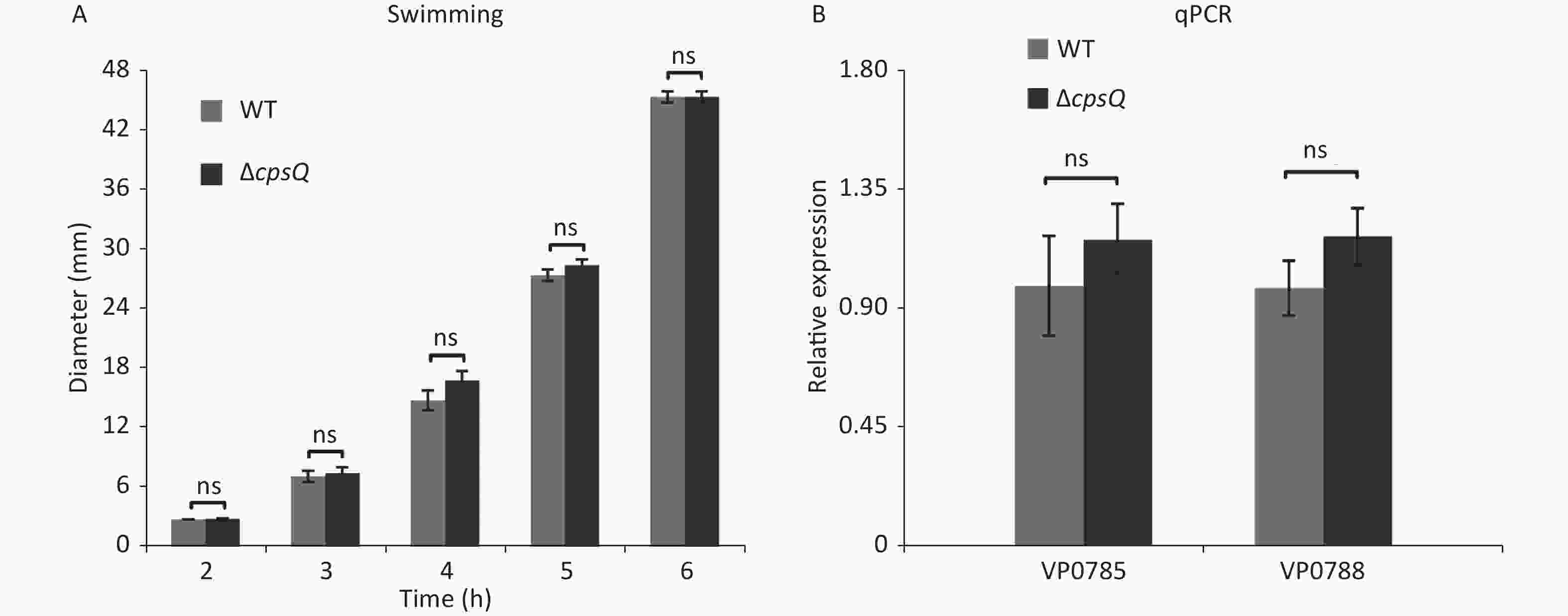
Figure S2. CpsQ did not affect the swimming motility of V. parahaemolyticus. (A) Swimming motility of WT and ΔcpsQ was evaluated by measuring the diameter of swimming areas in a semi-solid agar. (B) qPCR. Relative mRNA levels of each target gene were compared between WT and ΔcpsQ. Results were analyzed using paired Student’s t-test. ns, P > 0.05.
Six putative c-di-GMP metabolism-associated genes, including tpdA, are regulated by CpsQ (Supplementary Table S1). TpdA is a phosphodiesterase that plays a key role in the modulation of intracellular c-di-GMP levels, motility, and biofilm formation[7]. This prompted us to determine whether CpsQ regulates c-di-GMP production. The WT and ΔcpsQ colonies were randomly collected in triplicate from the HI plate, resuspended in 2 mL ice-cold PBS (pH 7.2), incubated at 100 °C for 5 min, and sonicated for 15 min (power, 100%; frequency, 37 kHz) in an ice-water bath. The intracellular c-di-GMP levels and total protein in the supernatants were measured using a c-di-GMP enzyme-linked immunosorbent assay (ELISA) kit (Mskbio, Wuhan, China) and Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Massachusetts, USA), respectively. Intracellular c-di-GMP levels are expressed as picomoles per gram of protein. As shown in Supplementary Figure S3 (available in www.besjournal.com), there were no significant differences in the intracellular c-di-GMP levels between WT and ΔcpsQ, suggesting that CpsQ did not affect the synthesis of c-di-GMP in V. parahaemolyticus.
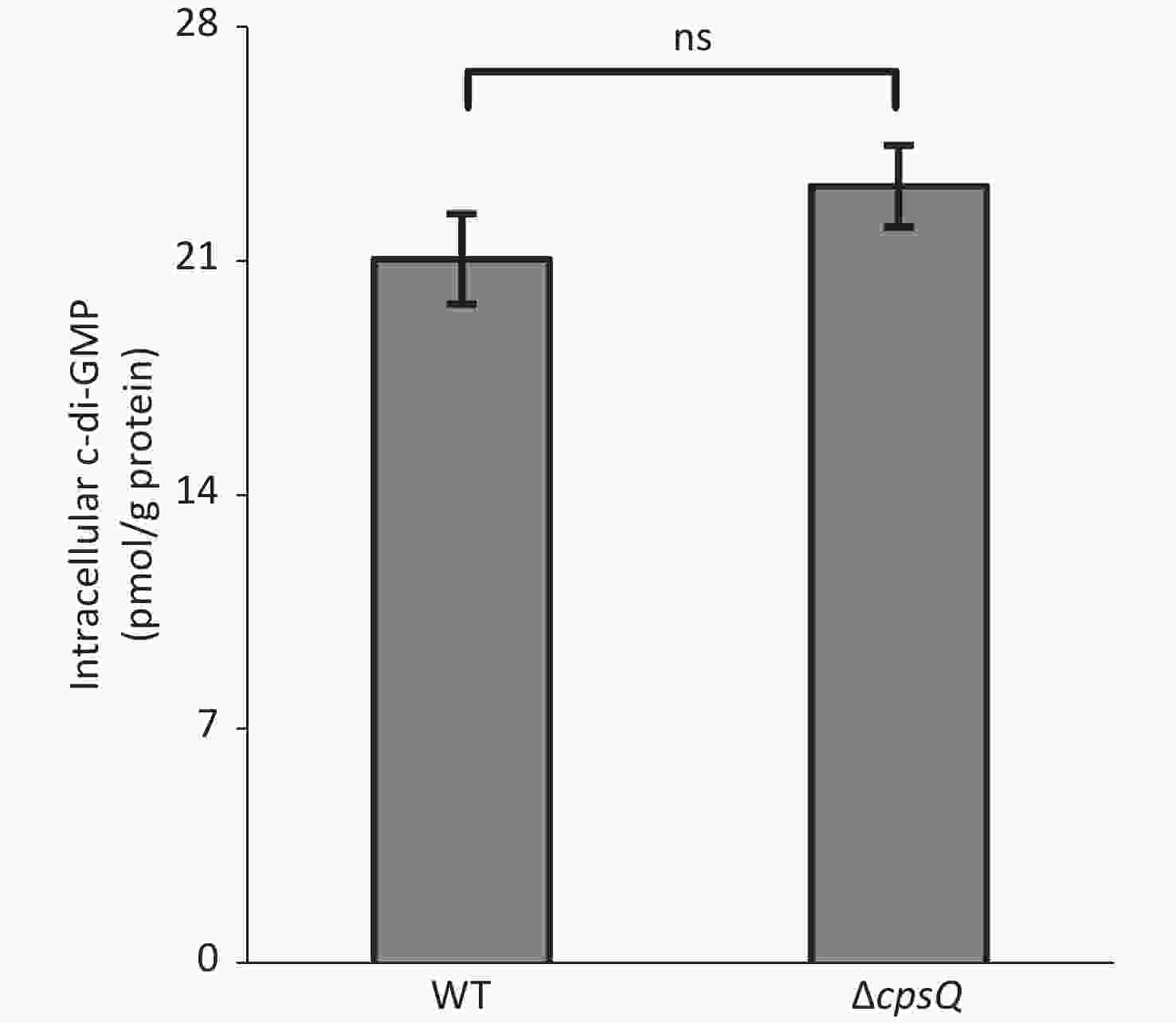
Figure S3. CpsQ did not affect the c-di-GMP pool in V. parahaemolyticus. Data are expressed as the mean ± SD of values from at least three independent experiments. Results were analyzed using paired Student’s t-test. ns, P > 0.05.
RNA-seq and qPCR data revealed that CpsQ repressed the transcription of T6SS2 (Supplementary Table S1 and Figure 2) genes. The T6SS2 gene cluster (VPA1024-1046) contains three operons—VPA1027-1024, VPA1043-1028, and VPA1044-1046[8]. VPA1043-1028 and VPA1044-1046 are transcribed in opposite directions and are adjacent to each other[8]. Thus, the first genes, VPA1027-1024 and VPA1043-1028, were selected as target genes to further investigate CpsQ-mediated gene expression. The regulatory DNA region of each target gene was cloned into the pHRP309 vector harboring the promoter-less lacZ reporter gene. Each recombinant plasmid was transferred into WT and ΔcpsQ. Colonies of the resulting transformants were lysed to measure β-galactosidase activity in the cellular extracts using a β-Galactosidase Enzyme Assay System (Promega, Madison, USA)[9]. As shown in Figure 3A, the promoter activity (Miller units) of VPA1027 or VPA1043 in the ΔcpsQ colony was significantly enhanced relative to that in the WT colony, suggesting that CpsQ repressed the promoter activities of T6SS2 genes in V. parahaemolyticus. In addition, E. coli 100 λpir (Epicenter) bearing pBAD33-cpsQ or pBAD33 and a recombinant lacZ plasmid were cultured in Luria-Bertani broth containing 0.1% L-arabinose and 20 μg/mL chloramphenicol at 37 °C and harvested when an OD600 value of 1.2 was reached, to test whether CpsQ regulates the T6SS2 gene expression in a heterologous host[10]. As shown in Figure 3B, the expression of CpsQ from pBAD33-cpsQ led to much lower promoter activities of VPA1027 and VPA1043 compared to those in strains bearing pBAD33. These findings indicated that CpsQ directly regulated the transcription of VPA1027-1024 and VPA1043-1028. Taken together, these results suggest that CpsQ may directly repress T6SS2 expression in V. parahaemolyticus.
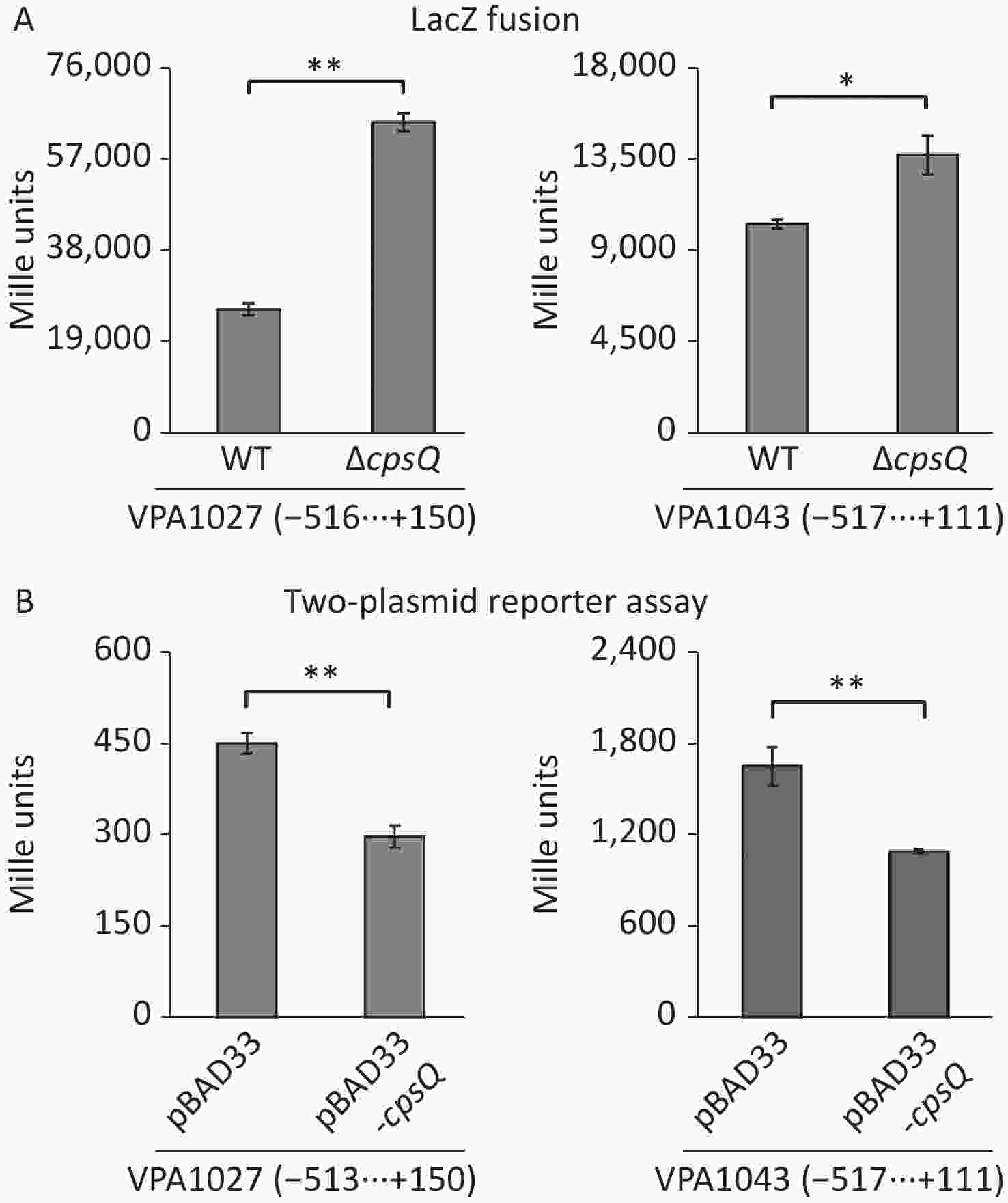
Figure 3. The negative and positive numbers indicate the nucleotide positions upstream and downstream of indicated genes, respectively. Paired Student’s t-test was used to calculate statistical significance, with a P-value less than 0.05 indicating significant differences. **P < 0.01; *P < 0.05. (A) LacZ fusion. The regulatory DNA region of each target gene was cloned into the pHRP309 plasmid and then transferred into ΔcpsQ and WT to determine the promoter activities in cellular extracts. (B) Two-plasmid reporter assay. E. coli 100 λpir (Epicentre) bearing pBAD33 or pBAD33-cpsR and a recombinant lacZ plasmid were grown in LB broth containing 0.1% L-arabinose to mid-log phase, and then aliquots were collected and assayed for lacZ expression using the β-galactosidase assay.
In conclusion, our findings showed that CpsQ regulated the transcription of 567 genes, including polar flagella, T6SS2, c-di-GMP metabolism-associated, putative regulatory, antioxidative, and outer membrane protein-encoding genes. CpsQ might directly repress the T6SS2-associated operons VPA1027-1024 and VPA1043-1028. Collectively, this study confirmed the new regulatory roles of CpsQ in V. parahaemolyticus.
-
Table S1. Selected genes from the CpsQ regulon
Locus Name FoldChange Product Lateral flagella VPA0261 2.077 flagellar export chaperone FlgN Polar flagellum VP0774 0.358 protein-glutamate O-methyltransferase VP0776 flgC 0.300 flagellar basal body rod protein FlgC VP0777 flgD 0.212 flagellar hook assembly protein FlgD VP0778 flgE 0.463 flagellar hook protein FlgE VP0780 0.180 flagellar basal body rod protein FlgF VP0781 flgG 0.243 flagellar basal-body rod protein FlgG VP0782 flgH 0.274 flagellar basal body L-ring protein FlgH VP0783 0.218 flagellar basal body P-ring protein FlgI VP0784 flgJ 0.253 flagellar assembly peptidoglycan hydrolase FlgJ VP0785 flgK 0.281 flagellar hook-associated protein FlgK VP0786 flgL 0.280 flagellar hook-associated protein FlgL VP0788 flaC 0.254 flagellin VP2111 0.231 OmpA family protein VP2225 0.458 chemotaxis protein CheW VP2227 0.441 ParA family protein VP2229 0.391 chemotaxis protein CheA VP2231 cheY 0.486 chemotaxis response regulator CheY VP2232 0.478 RNA polymerase sigma factor FliA VP2233 0.415 MinD/ParA family protein VP2237 fliR 0.483 flagellar type III secretion system protein FliR VP2241 fliN 0.265 flagellar motor switch protein FliN VP2242 fliM 0.460 flagellar motor switch protein FliM VP2244 0.465 flagellar hook-length control protein FliK VP2246 fliI 0.380 flagellar protein export ATPase FliI VP2249 fliF 0.432 flagellar M-ring protein FliF VP2251 0.414 sigma-54 dependent transcriptional regulator VP2252 0.444 PAS domain-containing protein VP2256 fliD 0.483 flagellar filament capping protein FliD Scv exopolysaccharide VP1464 0.300 O-antigen ligase family protein VP1469 scvE 3.975 sigma-54 dependent transcriptional regulator Type IV pilin VP2524 pilB 3.295 type IV-A pilus assembly ATPase PilB c-di-GMP VP1255 0.375 GGDEF-only VP1483 0.481 GGDEF-only VP1881 tpdA 0.421 EAL-only VP1979 3.129 EAL-only VPA0360 0.320 GGDEF-only VPA0518 2.100 hybrid T3SS1 VP1657 vopB 2.380 type III secretion system translocon subunit VopB T3SS2 VPA1326 2.725 hypothetical protein VPA1327 vopT 2.644 T3SS effector ADP-ribosyltransferase toxin VopT VPA1329 3.741 conjugal transfer protein TraA VPA1345 2.128 hypothetical protein VPA1346 vopA 4.834 type III secretion system YopJ family effector VopA T6SS1 VP1416 0.462 hypothetical protein T6SS2 VPA1024 5.741 hypothetical protein VPA1025 2.905 PAAR domain-containing protein VPA1026 vgrG 10.472 type VI secretion system tip protein VgrG VPA1027 10.854 type VI secretion system tube protein Hcp VPA1028 tssH 8.675 type VI secretion system ATPase TssH VPA1029 tssG 14.030 type VI secretion system baseplate subunit TssG VPA1030 tssF 13.979 type VI secretion system baseplate subunit TssF VPA1031 tssE 5.256 type VI secretion system baseplate subunit TssE VPA1032 7.011 protein of avirulence locus VPA1033 tssC 10.549 type VI secretion system contractile sheath large subunit VPA1034 tssC 3.575 type VI secretion system contractile sheath large subunit VPA1035 tssB 7.754 type VI secretion system contractile sheath small subunit VPA1036 tssA 9.924 type VI secretion system protein TssA VPA1037 6.385 protein phosphatase 2C domain-containing protein VPA1038 tagF 8.848 type VI secretion system-associated protein TagF VPA1039 tssM 5.244 type VI secretion system membrane subunit TssM VPA1040 tssL 4.422 type VI secretion system protein TssL%2C long form VPA1041 tssK 6.433 type VI secretion system baseplate subunit TssK VPA1042 tssJ 3.863 type VI secretion system lipoprotein TssJ VPA1043 tagH 4.076 type VI secretion system-associated FHA domain protein TagH VPA1044 4.890 serine/threonine protein kinase VPA1045 3.486 response regulator VPA1046 4.304 hypothetical protein Extracellular proteases VPA1071 2.654 S8 family serine peptidase Putative regulatory genes VP0080 2.651 sigma-54 dependent transcriptional regulator VP0358 0.399 DeoR family transcriptional regulator VP0838 seqA 0.318 replication initiation negative regulator SeqA VP1032 torR 0.439 two-component system response regulator TorR VP1382 2.345 LysR family transcriptional regulator VP1469 scvE 3.975 sigma-54 dependent transcriptional regulator VP1649 2.449 GntR family transcriptional regulator VP1906 0.415 MarR family transcriptional regulator VP1939 2.411 transcriptional regulator VP2037 0.421 chemotaxis protein CheV, response regulator VP2229 0.391 chemotaxis protein CheA, histidine kinase VP2231 0.486 chemotaxis response regulator CheY, response regulator VP2251 0.414 sigma-54 dependent transcriptional regulator VP2252 0.444 PAS domain-containing protein VP2299 glnB 0.430 nitrogen regulatory protein P-II VP2603 3.631 LysR family transcriptional regulator VP2710 scrP 2.352 helix-turn-helix transcriptional regulator VPA0358 0.229 helix-turn-helix transcriptional regulator VPA0420 0.486 TetR/AcrR family transcriptional regulator VPA0431 0.455 chemotaxis protein, response regulator VPA0619 3.870 MerR family DNA-binding transcriptional regulator VPA0663 2.174 AraC family transcriptional regulator VPA0717 0.253 LysR family transcriptional regulator VPA0740 3.494 LysR family transcriptional regulator VPA0746 0.414 chemotaxis protein CheV, response regulator VPA0826 pgtB 0.388 phosphoglycerate transport regulatory protein, histidine kinase VPA1045 3.486 response regulator VPA1162 0.405 response regulator VPA1423 2.961 AraC family transcriptional regulator VPA1562 2.684 TetR/AcrR family transcriptional regulator VPA1729 0.407 helix-turn-helix transcriptional regulator Antioxidative genes VPA1418 katE1 2.095 catalase VPA1683 ahpC1 2.349 alkyl hydroperoxide reductase subunit C Outer membrane proteins VP1008 0.059 porin VP1454 0.424 porin family protein VP1634 2.687 TolC family outer membrane protein VP2176 aqpZ 2.701 aquaporin Z VP2362 0.344 outer membrane protein OmpK VP2385 2.115 aquaporin VP2467 3.833 porin VPA0222 2.670 porin Table S2. Primers used in this study
Target Primers (forward/reverse, 5'-3') Construction of mutant cpsQ GTGACTGCAGGGTTCTCCAAGGCGATATG/CTTGTGGCTTGCGTCCTATGCTTTTCCGTGTACTGTTC GAACAGTACACGGAAAAGCATAGGACGCAAGCCACAAG/GTGAGCATGCCACCAGTTAGACGATCATTG GTGACTGCAGGGTTCTCCAAGGCGATATG/GTGAGCATGCCACCAGTTAGACGATCATTG qPCR VP0785 GCCGTCAGTCAGTGATTC/GTAGAGGACAGGTTGAGTTC VP0788 AATAAAGCGACCAACGAGCTG/TGCCACATCAAGACCACGAGA VP1469 GACAGGTCGTGATGCCATTC/GGCGATGATGACCGAAGTG VPA1027 TAAAGGTGAAGCGACAGCG/AATCATATAGGCGTGTTGC VPA1043 TGACCATAACGAGTTTCCAC/TTTAATCAATTCGCCGTGAG VPA1044 ATAGCAGCGATAGCGGAG/TTTGAGACAGTTTTGTATCC VPA1327 ATAGCAGCGATAGCGGAG/TTTGAGACAGTTTTGTATCC VP1255 GTAAGCTCGTCATCACACCTG/GCCGTACTTAGCCCACCTTCG VP1483 TCAAAGTGATCGACGGACCA/AATTACCTCGTCACCCGTTG VP1979 TTAAAGCCAGCGATGTAAACCC/GGCGAATCTGTTCTAACGCAAA VPA0360 TCGTTCTTTACCTACGCCTTA/TGCCAATAACACTCGATAGAGC VPA0518 GAAACATTAACGCAGCAAGCC/TCACCAATGTAGTTCCCGTTG VP1906 AAAGCTGAGCAAAGAGTCGGG/GTTGCTTGACTCATATTGGTG VPA0663 AGGCGATTCAGTTATGCGAAA/GTGGCTTCCAGTTTTTGGGTC VPA0717 CTATATTTAACCCAACCAGCC/GATCGAAGACTTCAGCCCCTA VPA1418 GATTTAGTCGGCAACAACAC/ATCCCAGTTGTTTGTCGAGC VPA1683 AGACCACTACGAAGAGCTAC/GTACTGGATCTTGCCGATTG VP1071 TCAGAACAATGGCATCTCGAC/ATGGCTTGCGTAAGTTTGGTG VP1008 AGGCAACATCTACGACAACGG/AGCCCAAAGGTCAACGTCTAC VP2362 ACCTAGCGTCAGACAAAGGC/TGAACTGGACCGAAAGACAGG LacZ fusion VPA1027 GCGCGTCGACTATTACCTTACTTGCCTCTCGG/GCGCGAATTCTGCTTCACGGTCCATTGC VPA1043 GCGCGTCGACTTTGTTGATAGGTGGTATTGTG/ATATGAATTCTGAGCGTCCGAAGGTTAC
doi: 10.3967/bes2024.049
Characterization of the CpsQ Regulon Reveals Its Role in the Transcription of Type VI Secretion System 2 Genes in Vibrio parahaemolyticus
-
-
S2. CpsQ did not affect the swimming motility of V. parahaemolyticus. (A) Swimming motility of WT and ΔcpsQ was evaluated by measuring the diameter of swimming areas in a semi-solid agar. (B) qPCR. Relative mRNA levels of each target gene were compared between WT and ΔcpsQ. Results were analyzed using paired Student’s t-test. ns, P > 0.05.
Figure 3. The negative and positive numbers indicate the nucleotide positions upstream and downstream of indicated genes, respectively. Paired Student’s t-test was used to calculate statistical significance, with a P-value less than 0.05 indicating significant differences. **P < 0.01; *P < 0.05. (A) LacZ fusion. The regulatory DNA region of each target gene was cloned into the pHRP309 plasmid and then transferred into ΔcpsQ and WT to determine the promoter activities in cellular extracts. (B) Two-plasmid reporter assay. E. coli 100 λpir (Epicentre) bearing pBAD33 or pBAD33-cpsR and a recombinant lacZ plasmid were grown in LB broth containing 0.1% L-arabinose to mid-log phase, and then aliquots were collected and assayed for lacZ expression using the β-galactosidase assay.
S1. Selected genes from the CpsQ regulon
Locus Name FoldChange Product Lateral flagella VPA0261 2.077 flagellar export chaperone FlgN Polar flagellum VP0774 0.358 protein-glutamate O-methyltransferase VP0776 flgC 0.300 flagellar basal body rod protein FlgC VP0777 flgD 0.212 flagellar hook assembly protein FlgD VP0778 flgE 0.463 flagellar hook protein FlgE VP0780 0.180 flagellar basal body rod protein FlgF VP0781 flgG 0.243 flagellar basal-body rod protein FlgG VP0782 flgH 0.274 flagellar basal body L-ring protein FlgH VP0783 0.218 flagellar basal body P-ring protein FlgI VP0784 flgJ 0.253 flagellar assembly peptidoglycan hydrolase FlgJ VP0785 flgK 0.281 flagellar hook-associated protein FlgK VP0786 flgL 0.280 flagellar hook-associated protein FlgL VP0788 flaC 0.254 flagellin VP2111 0.231 OmpA family protein VP2225 0.458 chemotaxis protein CheW VP2227 0.441 ParA family protein VP2229 0.391 chemotaxis protein CheA VP2231 cheY 0.486 chemotaxis response regulator CheY VP2232 0.478 RNA polymerase sigma factor FliA VP2233 0.415 MinD/ParA family protein VP2237 fliR 0.483 flagellar type III secretion system protein FliR VP2241 fliN 0.265 flagellar motor switch protein FliN VP2242 fliM 0.460 flagellar motor switch protein FliM VP2244 0.465 flagellar hook-length control protein FliK VP2246 fliI 0.380 flagellar protein export ATPase FliI VP2249 fliF 0.432 flagellar M-ring protein FliF VP2251 0.414 sigma-54 dependent transcriptional regulator VP2252 0.444 PAS domain-containing protein VP2256 fliD 0.483 flagellar filament capping protein FliD Scv exopolysaccharide VP1464 0.300 O-antigen ligase family protein VP1469 scvE 3.975 sigma-54 dependent transcriptional regulator Type IV pilin VP2524 pilB 3.295 type IV-A pilus assembly ATPase PilB c-di-GMP VP1255 0.375 GGDEF-only VP1483 0.481 GGDEF-only VP1881 tpdA 0.421 EAL-only VP1979 3.129 EAL-only VPA0360 0.320 GGDEF-only VPA0518 2.100 hybrid T3SS1 VP1657 vopB 2.380 type III secretion system translocon subunit VopB T3SS2 VPA1326 2.725 hypothetical protein VPA1327 vopT 2.644 T3SS effector ADP-ribosyltransferase toxin VopT VPA1329 3.741 conjugal transfer protein TraA VPA1345 2.128 hypothetical protein VPA1346 vopA 4.834 type III secretion system YopJ family effector VopA T6SS1 VP1416 0.462 hypothetical protein T6SS2 VPA1024 5.741 hypothetical protein VPA1025 2.905 PAAR domain-containing protein VPA1026 vgrG 10.472 type VI secretion system tip protein VgrG VPA1027 10.854 type VI secretion system tube protein Hcp VPA1028 tssH 8.675 type VI secretion system ATPase TssH VPA1029 tssG 14.030 type VI secretion system baseplate subunit TssG VPA1030 tssF 13.979 type VI secretion system baseplate subunit TssF VPA1031 tssE 5.256 type VI secretion system baseplate subunit TssE VPA1032 7.011 protein of avirulence locus VPA1033 tssC 10.549 type VI secretion system contractile sheath large subunit VPA1034 tssC 3.575 type VI secretion system contractile sheath large subunit VPA1035 tssB 7.754 type VI secretion system contractile sheath small subunit VPA1036 tssA 9.924 type VI secretion system protein TssA VPA1037 6.385 protein phosphatase 2C domain-containing protein VPA1038 tagF 8.848 type VI secretion system-associated protein TagF VPA1039 tssM 5.244 type VI secretion system membrane subunit TssM VPA1040 tssL 4.422 type VI secretion system protein TssL%2C long form VPA1041 tssK 6.433 type VI secretion system baseplate subunit TssK VPA1042 tssJ 3.863 type VI secretion system lipoprotein TssJ VPA1043 tagH 4.076 type VI secretion system-associated FHA domain protein TagH VPA1044 4.890 serine/threonine protein kinase VPA1045 3.486 response regulator VPA1046 4.304 hypothetical protein Extracellular proteases VPA1071 2.654 S8 family serine peptidase Putative regulatory genes VP0080 2.651 sigma-54 dependent transcriptional regulator VP0358 0.399 DeoR family transcriptional regulator VP0838 seqA 0.318 replication initiation negative regulator SeqA VP1032 torR 0.439 two-component system response regulator TorR VP1382 2.345 LysR family transcriptional regulator VP1469 scvE 3.975 sigma-54 dependent transcriptional regulator VP1649 2.449 GntR family transcriptional regulator VP1906 0.415 MarR family transcriptional regulator VP1939 2.411 transcriptional regulator VP2037 0.421 chemotaxis protein CheV, response regulator VP2229 0.391 chemotaxis protein CheA, histidine kinase VP2231 0.486 chemotaxis response regulator CheY, response regulator VP2251 0.414 sigma-54 dependent transcriptional regulator VP2252 0.444 PAS domain-containing protein VP2299 glnB 0.430 nitrogen regulatory protein P-II VP2603 3.631 LysR family transcriptional regulator VP2710 scrP 2.352 helix-turn-helix transcriptional regulator VPA0358 0.229 helix-turn-helix transcriptional regulator VPA0420 0.486 TetR/AcrR family transcriptional regulator VPA0431 0.455 chemotaxis protein, response regulator VPA0619 3.870 MerR family DNA-binding transcriptional regulator VPA0663 2.174 AraC family transcriptional regulator VPA0717 0.253 LysR family transcriptional regulator VPA0740 3.494 LysR family transcriptional regulator VPA0746 0.414 chemotaxis protein CheV, response regulator VPA0826 pgtB 0.388 phosphoglycerate transport regulatory protein, histidine kinase VPA1045 3.486 response regulator VPA1162 0.405 response regulator VPA1423 2.961 AraC family transcriptional regulator VPA1562 2.684 TetR/AcrR family transcriptional regulator VPA1729 0.407 helix-turn-helix transcriptional regulator Antioxidative genes VPA1418 katE1 2.095 catalase VPA1683 ahpC1 2.349 alkyl hydroperoxide reductase subunit C Outer membrane proteins VP1008 0.059 porin VP1454 0.424 porin family protein VP1634 2.687 TolC family outer membrane protein VP2176 aqpZ 2.701 aquaporin Z VP2362 0.344 outer membrane protein OmpK VP2385 2.115 aquaporin VP2467 3.833 porin VPA0222 2.670 porin S2. Primers used in this study
Target Primers (forward/reverse, 5'-3') Construction of mutant cpsQ GTGACTGCAGGGTTCTCCAAGGCGATATG/CTTGTGGCTTGCGTCCTATGCTTTTCCGTGTACTGTTC GAACAGTACACGGAAAAGCATAGGACGCAAGCCACAAG/GTGAGCATGCCACCAGTTAGACGATCATTG GTGACTGCAGGGTTCTCCAAGGCGATATG/GTGAGCATGCCACCAGTTAGACGATCATTG qPCR VP0785 GCCGTCAGTCAGTGATTC/GTAGAGGACAGGTTGAGTTC VP0788 AATAAAGCGACCAACGAGCTG/TGCCACATCAAGACCACGAGA VP1469 GACAGGTCGTGATGCCATTC/GGCGATGATGACCGAAGTG VPA1027 TAAAGGTGAAGCGACAGCG/AATCATATAGGCGTGTTGC VPA1043 TGACCATAACGAGTTTCCAC/TTTAATCAATTCGCCGTGAG VPA1044 ATAGCAGCGATAGCGGAG/TTTGAGACAGTTTTGTATCC VPA1327 ATAGCAGCGATAGCGGAG/TTTGAGACAGTTTTGTATCC VP1255 GTAAGCTCGTCATCACACCTG/GCCGTACTTAGCCCACCTTCG VP1483 TCAAAGTGATCGACGGACCA/AATTACCTCGTCACCCGTTG VP1979 TTAAAGCCAGCGATGTAAACCC/GGCGAATCTGTTCTAACGCAAA VPA0360 TCGTTCTTTACCTACGCCTTA/TGCCAATAACACTCGATAGAGC VPA0518 GAAACATTAACGCAGCAAGCC/TCACCAATGTAGTTCCCGTTG VP1906 AAAGCTGAGCAAAGAGTCGGG/GTTGCTTGACTCATATTGGTG VPA0663 AGGCGATTCAGTTATGCGAAA/GTGGCTTCCAGTTTTTGGGTC VPA0717 CTATATTTAACCCAACCAGCC/GATCGAAGACTTCAGCCCCTA VPA1418 GATTTAGTCGGCAACAACAC/ATCCCAGTTGTTTGTCGAGC VPA1683 AGACCACTACGAAGAGCTAC/GTACTGGATCTTGCCGATTG VP1071 TCAGAACAATGGCATCTCGAC/ATGGCTTGCGTAAGTTTGGTG VP1008 AGGCAACATCTACGACAACGG/AGCCCAAAGGTCAACGTCTAC VP2362 ACCTAGCGTCAGACAAAGGC/TGAACTGGACCGAAAGACAGG LacZ fusion VPA1027 GCGCGTCGACTATTACCTTACTTGCCTCTCGG/GCGCGAATTCTGCTTCACGGTCCATTGC VPA1043 GCGCGTCGACTTTGTTGATAGGTGGTATTGTG/ATATGAATTCTGAGCGTCCGAAGGTTAC -
[1] Li LZ, Meng HM, Gu D, et al. Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol Res, 2019; 222, 43−51. doi: 10.1016/j.micres.2019.03.003 [2] Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol, 2009; 17, 109−18. doi: 10.1016/j.tim.2008.12.004 [3] Zhou DS, Yan XJ, Qu F, et al. Quorum sensing modulates transcription of cpsQ-mfpABC and mfpABC in Vibrio parahaemolyticus. Int J Food Microbiol, 2013; 166, 458−63. doi: 10.1016/j.ijfoodmicro.2013.07.008 [4] Kimbrough JH, Cribbs JT, McCarter LL. Homologous c-di-GMP-binding Scr transcription factors orchestrate biofilm development in Vibrio parahaemolyticus. J Bacteriol, 2020; 202, e00723-19. [5] Sun FJ, Zhang YQ, Qiu YF, et al. H-NS is a repressor of major virulence gene loci in Vibrio parahaemolyticus. Front Microbiol, 2014; 5, 675. [6] Gao H, Zhang YQ, Yang L, et al. Regulatory effects of cAMP receptor protein (CRP) on porin genes and its own gene in Yersinia pestis. BMC Microbiol, 2011; 11, 40. doi: 10.1186/1471-2180-11-40 [7] Martínez-Méndez R, Camacho-Hernández DA, Sulvarán-Guel E, et al. A trigger Phosphodiesterase modulates the global c-di-GMP pool, motility, and biofilm formation in Vibrio parahaemolyticus. J Bacteriol, 2021; 203, e0004621. [8] Makino K, Oshima K, Kurokawa K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet, 2003; 361, 743−9. doi: 10.1016/S0140-6736(03)12659-1 [9] Zhang YQ, Qiu Y, Xue XF, et al. Transcriptional regulation of the virulence genes and the biofilm formation associated operons in Vibrio parahaemolyticus. Gut Pathog, 2021; 13, 15. doi: 10.1186/s13099-021-00410-y [10] Qiu Y, Hu LF, Yang WH, et al. The type VI secretion system 2 of Vibrio parahaemolyticus is regulated by QsvR. Microb Pathog, 2020; 149, 104579. doi: 10.1016/j.micpath.2020.104579 -
 23444+Supplementary Materials.pdf
23444+Supplementary Materials.pdf

-




 下载:
下载:





 Quick Links
Quick Links